Abstract
Circadian rhythms of many body functions in mammals are controlled by a master pacemaker residing in the hypothalamic suprachiasmatic nucleus (SCN) that synchronises peripheral oscillators. The SCN and peripheral oscillators share several components of the molecular clockwork and comprise transcriptional activators (BMAL1 and CLOCK/NPAS2) and inhibitors (mPER1/2 and mCRY1/2). Here we compared the ontogenetic maturation of the clockwork in the SCN and pars tuberalis (PT). The PT is a peripheral oscillator that strongly depends on rhythmic melatonin signals. Immunoreactions for clock gene proteins were determined in the SCN and PT at four different timepoints during four differential stages of mouse ontogeny: foetal (embryonic day 18), newborn (2-day-old), infantile (10-day-old), and adult. In the foetal SCN levels of immunoreactions of all clock proteins were significantly lower as compared to adult levels except for BMAL1. In the newborn SCN the clock protein immunoreactions had not yet reached adult levels, but the infantile SCN showed similar levels of immunreactions as the adult. In contrast, immunoreactions for all clock gene proteins in the foetal PT were as intense as in newborn, infantile, and adult and showed the same phase. As the foetal pineal gland is not yet capable of rhythmic melatonin production, the rhythms in clock gene proteins in the foetal PT are presumably dependent on the maternal melatonin signal. Thus, our data provide the first evidence that maternal melatonin is important for establishing and maintaining circadian rhythms in a foetal peripheral oscillator.
Keywords: circadian rhythms, clock genes, development, vasopressin, clockwork, PT, peripheral oscillator
Introduction
In mammals circadian rhythms in overt body functions which enable the organism to anticipate and to adapt to the solar cycle are controlled by a master circadian rhythm generator located in the hypothalamic suprachiasmatic nucleus (SCN) that synchronises peripheral oscillators (Reppert & Weaver, 2002). The molecular clockworks in the master and in peripheral oscillators are under the control of feedback loops of clock genes (reviewed by Reppert & Weaver, 2002). The positive regulators BMAL1 and CLOCK/NPAS2 (DeBruyne et al., 2007) activate transcription of negative regulators, such as mPER1/2 and mCRY1/2, as well as clock-controlled output genes like Avp (Jin et al., 1999). The mPER and mCRY proteins inhibit CLOCK:BMAL1-mediated transcription in a circadian manner (Lee et al., 2001; Etchegaray et al., 2003). Circadian rhythms in clock gene expression in the SCN and in some peripheral oscillators like the liver are self-sustained in the absence of rhythmic input (Yoo et al., 2004). In contrast, other peripheral oscillators like the kidney require rhythmic input to sustain rhythmicity and their rhythms damp out rapidly in vitro (Yoo et al., 2004). These differences suggest that peripheral oscillators have different capacities for autonomous oscillation and some are more dependent on rhythmic input signals than others. An excellent model for a strongly input-dependent oscillator is the pars tuberalis (PT) of the anterior pituitary. The rhythmic input needed to maintain rhythmic clock gene expression in the PT is the hormone melatonin (Messager et al., 2001; von Gall et al., 2002, 2005; Jilg et al., 2005).
The aim of this study was to test the hypothesis that the development of clockwork properties differs among the self-sustained circadian rhythm generator in the SCN and input-dependent oscillators. Therefore, we compared the composition of the clockwork in the SCN and the melatonin-dependent PT oscillator by determining the levels of immunoreactions for BMAL1, CLOCK, mPER1/2, and CRY1/2 at four different ontogenetic stages and four circadian timepoints.
In foetus we found a few SCN cells displaying circadian rhythms in mPER1- and mPER2-immunoreactions. Cells displaying a circadian rhythm in the mCRY2 immunoreaction were first observed in the newborn SCN. Ten days after birth the immunoreactions for these clock gene proteins in the SCN showed adult patterns. In contrast, high-amplitude rhythms in immunoreactions for mPER1, mPER2, mCRY1 and mCRY2 were already detected in the foetal PT. These data show that the clockwork in the melatonin-dependent peripheral PT oscillator matures during foetal life, whereas the clockwork in the circadian rhythm generator of the SCN matures gradually during postnatal development. Thus, our study provides the first evidence of a different ontogenetic maturation in the clockwork of a self-sustained circadian rhythm generator and an input-dependent circadian oscillator.
Materials and Methods
Chemicals were obtained from Sigma-Aldrich (St. Louis, MO) if not indicated otherwise.
Animals
All animal experimentation reported in this manuscript was conducted in accordance with the European Communities Council Directive (89/609/EEC). Timed pregnant female C3H/HeN mice (6–10 weeks old) were obtained 8 days after mating (E8) with C3H/HeN males from Charles River Wiga, Sulzfeld, Germany and kept in our animal facility. The mice were housed in individual cages at constant room temperature on a 12:12 hours light (250 μW/cm2)/dark (<15 μW/cm2) cycle and had access to food and water ad libitum. For studies in E18 mice, the timed pregnant mice (E17) were transferred to constant darkness one day before experimentation. For studies in P2 and P10 mice, the dams and the litter were transferred to constant darkness one day or 9 days after delivery, respectively. Predicted lights-off was defined as circadian time (CT) 12. Five animals of each developmental stage and time point were analysed.
Morphological analysis of the developing SCN
Timed pregnant (adult), newborn (P2), infantile (P10) and foetal (E18) C3H mice were decapitated at CT 06 (n=3/ontogenetic stage). Whole heads of foetal and newborn mice and brains of infantile and adult mice were fixed with 4% paraformaldehyde for 24 hours. Thereafter, the specimens were cyroprotected with 20% sucrose overnight and sectioned in the coronal plane at 40 μm on a freezing microtome. Fifteen free-floating consecutive sections of the anterior hypothalamic region were prepared from three mice of each developmental stage and stained with Mayer′s haematoxylin solution (AppliChem, Darmstadt, Germany). All sections were stained together to minimise variability. Sections were observed with a Zeiss microscope (Axioplan; 1003) equipped with a video camera connected to a computerized image analysis system (VIDAS, Kontron, Eching, Germany). NIH Image J software was used for image analyses. The mean nuclear area of SCN cells was determined by measuring 50 cell nuclei in the plane of the nucleolus in three animals of each developmental stage. The relative optical density (O.D.) of neuropil areas devoid of cell nuclei was determined as background staining and used to define a threshold that was kept constant for all further analyses. To determine the mean total number of cells within the intermediate portion of one of the paired SCN, the total area of stained cell nuclei with a relative O.D. above threshold was divided by the mean nuclear area (E18: 31×31 pixels; P2, P10 and adult: 40×40 pixels). Differences in the number of cell nuclei for the four different developmental stages were determined by one-way ANOVA followed by Bonferroni′s posthoc test.
Immunohistochemistry
Timed pregnant (adult), foetal (E18), newborn (P2), and infantile (P10) C3H mice were killed at CT00, CT06, CT12 and CT18 by decapitation (n=5 per timepoint and ontogenetic stage). Whole heads of foetal and newborn mice and brains of adult mice were fixed with 4% paraformaldehyde for 12 h, cryoprotected with 20% sucrose overnight and sectioned in the coronal plane at 40 μm on a freezing microtome. Free-floating sections were processed for immunohistochemistry as previously described (von Gall et al., 1998) using an avidin–biotin labelling method (Vector Laboratories, Burlingame, CA, USA) and diaminobenzidine as the chromogen. For each experiment all sections were processed together to minimize variability. Polyclonal antibodies raised in guinea pig against BMAL1, CLOCK, mPER1, mPER2, mCRY1 and mCRY2 as characterized by Lee et al., (2001, 2004) were used at a dilution of 1:500. To assess immunoreactions in the SCN and the PT, the number of immunoreactive cells and the relative optical density (O.D.) were used respectively as described by Nuesslein-Hildesheim et al. (2000), due to differences in the cellular distribution of clock gene proteins among these tissues. Images were captured through a VIDAS system (Kontron, Munich, Germany). Quantitative analysis of immunoreaction (IR) was accomplished using NIH ImageJ software in a blind manner. The number of immunoreactive cell nuclei in the intermediate aspect of the SCN was analysed as described (von Gall et al., 1998, 2003). The relative optical density (O.D.) of non-specific background staining in neuropil areas devoid of cell nuclei was used to define a threshold that was kept constant for all analyses. From each mouse two sections through the intermediate portion of the SCN were analysed. To obtain the number of immunoreactive cell nuclei, the total area of the IR with relative O.D. above threshold was divided by the mean nuclear area (E18: 31×31 pixels; P2, P10, and adult: 40×40 pixels). The nuclear localization of the IR was confirmed by immunocytochemical detection of clock gene proteins using anti-guinea pig secondary antibodies conjugated to Cy3 (Molecular probes, Eugene, OR) and Syto24 (Molecular probes, Eugene, OR) for nuclear staining (not shown).
The relative optical density of the nuclear IR in PT cell nuclei was determined as previously described (von Gall et al., 2002, 2005, Jilg et al., 2005). Gray scale values between 0 (black) and 255 (white) of the specific nuclear IR of at least randomly chosen 20 cell nuclei within the PT region were averaged and corrected against the non-specific background staining in brain neuropil areas devoid of cell nuclei. Four PT sections per mouse were analysed for each protein and averaged to give a single value for each animal. Data are expressed as the relative O.D. of the nuclear IR relative to the highest nuclear staining intensity.
In situ hybridisation
The Avp cDNA fragment-containing plasmid (generously provided by Tom Sherman, Dept. Physiology and Biophysics, Georgetown University 37th and O Street, Washington, DC 20057) was linearised with the respective restriction enzymes and used as template for sense or anti-sense complementary RNA probes. 33P-labeled RNA probes were synthesised using an in vitro transcription kit (MAXIscript Kit; Ambion, Austin, TX). Probe quality and size were confirmed by 33P incorporation into TCA-precipitable material and by gel electrophoresis. In situ hybridisation was performed as previously described (Jin et al., 1999). Timed pregnant (adult), foetal (E18), and newborn (P2) C3H mice were killed at CT00, CT06, CT12 and CT18 by decapitation (n=5/timepoint and ontogenetic stage). Whole heads of foetal and newborn mice and brains of adult mice were removed immediately and frozen by immersion in cooled (−20°C) 2-methylbutane. Coronal sections through the SCN region (14 μm) were cut in a cryostat, mounted on glass slides (SuperFrost® Plus, Braunschweig, Germany), fixed in 4% paraformaldehyde, deproteinated with 0.2 N HCl, acetylated using 0.25% acetyl anhydride in 0.1 M triethanolamine, and finally air dried. Probes (50 μl at 107 cpm/ml) were applied to each slide. Slides were coverslipped and incubated in humidified chambers overnight at 55°C. Slides were then washed in 2x SSC, then treated with 10 μg/ml RNAse, and washed again with 2x SSC and twice more with 0.1x SSC at 53°C, and finally twice more with 0.1x SSC at room temperature. After washing, slides were air dried and exposed to Kodak BioMax MR film for 14 days.
For densitometric analyses of the hybridisation signals the films were digitised by an Epson GT-15000 scanner and image analysis was performed with NIH Image J software. Gray scale units (0=black, 255=white) of the specific autoradiographic signals within the SCN region were corrected against the non-specific background signals in neuropil areas surrounding the SCN. Four SCN sections per mouse were analysed and averaged to give a single value for each animal. The location of the SCN was confirmed by staining the sections with haematoxylin. Data are expressed as percent of relative O.D. (relative to the highest autoradiographic signal intensity).
Statistical analyses
Data are presented as the mean +/− SEM of five animals per time point and ontogenetic stage. Statistical analysis was performed using Graph Pad Prism (Graph Pad, San Diego, CA, USA) and Cosinor 6.3 software (Richelieu, France). The presence of circadian rhythms was determined by a Single Cosinor test with a period of 24 h. Significant variations between time points within one developmental stage were determined using one-way analysis of variance (ANOVA), followed by Bonferroni’s post hoc test. Amplitudes of rhythms were analysed by a Single Cosinor test and significant differences between the amplitudes of developmental stages were analysed using one-way ANOVA followed by a Bonferroni’s post hoc test. Values were considered significantly different with p < 0.05.
Results
Morphological changes of the developing SCN
All data presented in this paragraph refer to the intermediate region of one of the paired SCN. The mean size (area) of SCN cell nuclei was 3.4+/−0.1 μm (31×31 pixels) in the foetus (E18) and 5+/−1 μm (40×40 pixels) in the newborn (P2), infantile (P10) and adult mice. The mean number of SCN cell nuclei within the intermediate portion of a single foetal SCN was significantly lower (p<0.001) in foetus (E18: 1075+/−22) than in newborn (P2: 1507+/−28), infantile (P10: 1488+/−52) and adult (1423+/−19) (Table 2). The mean numbers of SCN cell nuclei did not significantly differ among newborn, infantile and adult.
Table 2.
Comparison between the total number of SCN cells and the maximum number of SCN cells immunoreactive for clock gene proteins during ontogenetic development. Mean cell numbers are as determined in the intermediate portion of the SCN.
| A | |||||||
|---|---|---|---|---|---|---|---|
| SCN | E18 | P2 | P10 | Adult | |||
| BMAL1 | % Rhythmicity | 2% | 15% | 22% | 30% | ||
| Rhythm | No | No | No | No | |||
|
| |||||||
| CLOCK | % Rhythmicity | 34% | 14% | 5% | 37% | ||
| Rhythm | No | No | No | No | |||
|
| |||||||
| mPER1 | % Rhythmicity | 67% | 52% | 63% | 83% | ||
| Rhythm | Yes | Yes | Yes | Yes | |||
|
| |||||||
| mPER2 | % Rhythmicity | 51% | 72% | 62% | 72% | ||
| Rhythm | Yes | Yes | Yes | Yes | |||
|
| |||||||
| mCRY1 | % Rhythmicity | 31% | 8% | 55% | 51% | ||
| Rhythm | No | No | Yes | Yes | |||
|
| |||||||
| mCRY2 | % Rhythmicity | 3% | 51% | 62% | 59% | ||
| Rhythm | No | Yes | Yes | Yes | |||
|
| |||||||
| Avp | % Rhythmicity | 63% | 96% | 85% | |||
| Rhythm | Yes | Yes | Yes | ||||
|
| |||||||
|
B | |||||||
| PT | E18 | P2 | P10 | Adult | |||
|
| |||||||
| BMAL1 | % Rhythmicity | 45% | 20% | 9% | 29% | ||
| Rhythm | No | No | No | No | |||
|
| |||||||
| CLOCK | % Rhythmicity | 35% | 23% | 30% | 31% | ||
| Rhythm | No | No | No | No | |||
|
| |||||||
| mPER1 | % Rhythmicity | 90% | 76% | 72% | 88% | ||
| Rhythm | Yes | Yes | Yes | Yes | |||
|
| |||||||
| mPER2 | % Rhythmicity | 95% | 63% | 79% | 95% | ||
| Rhythm | Yes | Yes | Yes | Yes | |||
|
| |||||||
| mCRY1 | % Rhythmicity | 60% | 55% | 81% | 82% | ||
| Rhythm | Yes | Yes | Yes | Yes | |||
|
| |||||||
| mCRY2 | % Rhythmicity | 81% | 68% | 51% | 1% | ||
| Rhythm | Yes | Yes | Yes | No | |||
Clock gene proteins in the SCN during ontogenetic development
To determine the clockwork in the circadian rhythm generator of the SCN during ontogenetic development, we analysed levels of BMAL1-, CLOCK-, mPER1-, mPER2-, mCRY1- and mCRY2-IR in the intermediate portion of the SCN in foetal (E18), newborn (P2), infantile (P10), and adult (6–10-weeks old) C3H mice at four timepoints during the circadian cycle (CT00, 06, 12, 18).
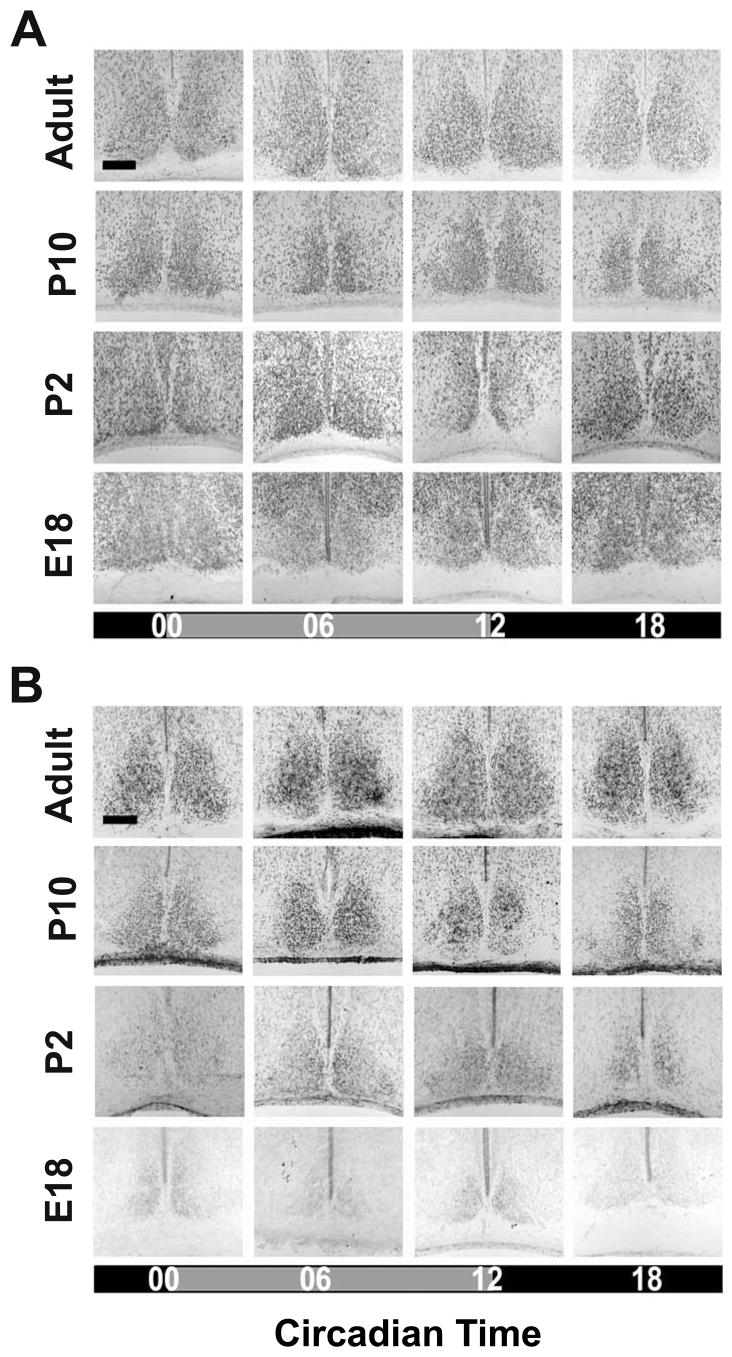
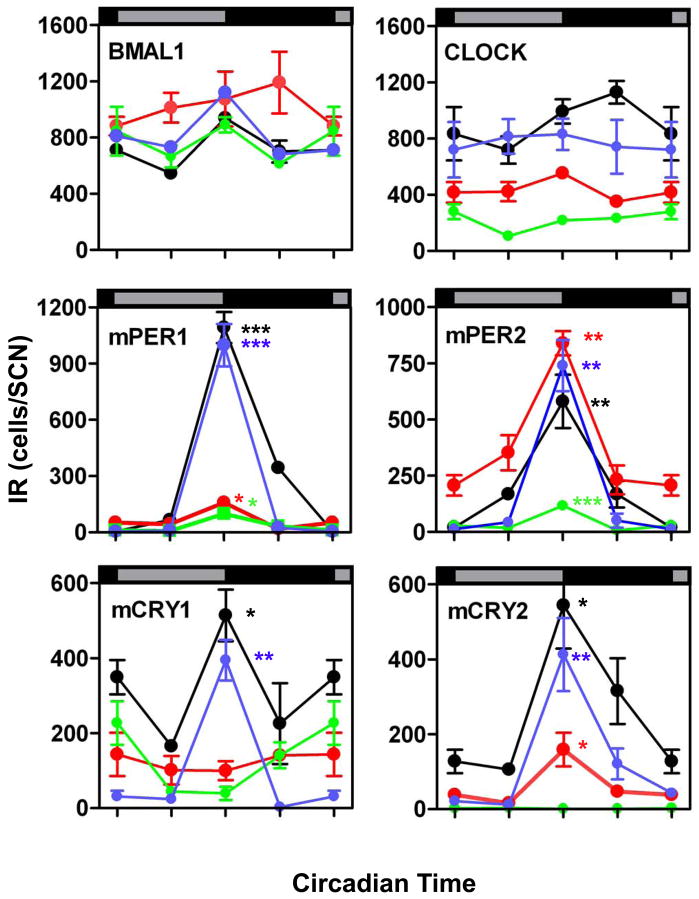
At all developmental stages investigated the number of SCN cells with nuclear BMAL1-and CLOCK-IR did not show a circadian rhythm (Tab. 1A). Furthermore, a high number of cells with nuclear BMAL1-IR was already observed in the foetal SCN and this number remained constant during further development (Figs. 1 and 4). In contrast, the number of SCN cells with nuclear CLOCK-IR was significantly lower in E18 compared to P2 (p<0.001) and significantly lower in P2 compared to adult (p<0.001) (Figs. 1 and 4). The number of SCN cells with CLOCK-IR was not significantly different between infantile and adult (Figs. 1 and 4).
Table 1.
Analyses of circadian rhythms in the SCN (A) and the PT (B). Chronobiometric parameters of the Single Cosinor test with a period of 24 h were used to determine the percentage and the existence of a circadian rhythm.
| Maximum number of SCN cells immunoreactive for | |||||||
|---|---|---|---|---|---|---|---|
| Total number of SCN cells | BMAL1 | CLOCK | PER1 | PER2 | CRY1 | CRY2 | |
| Fetal | 1075+/−22 | 889+/−54 | 278+/−53 | 98+/−21 | 116+/−5 | 227+/−58 | 4+/−1 |
| Newborn | 1507+/−28 | 1072+/−196 | 551+/−26 | 158+/−33 | 839+/−53 | 143+/−57 | 158+/−44 |
| Infantile | 1488+/−52 | 1120+/−25 | 829+/−111 | 1098+/−112 | 340+/−113 | 395+/−54 | 413+/−98 |
| Adult | 1423+/−19 | 936+/−40 | 991+/−87 | 1092+/−82 | 580+/−117 | 514+/−68 | 544+/−115 |
Figure 1. BMAL1- and CLOCK-immunoreactivity in the SCN during ontogenetic development.
Representative coronal sections showing BMAL1- (A) and CLOCK- (B) immunoreaction (IR) in the SCN of fetal (E18), newborn (P2), infantile (P10), and adult (6–10 week-old) C3H mice. Mice were sacrificed during early or mid-subjective day (CT00, CT06) and early or mid-subjective night (CT12, CT18). Scale bars: 150 μm.
Figure 4. Number of cells with BMAL1-, CLOCK-, mPER1-, mPER2-, mCRY1-, and mCRY2-immunoreactive cells in the intermediate portion of the SCN in fetal (green), newborn (red), infantile (blue), and adult (black) mice.
Data are expressed as the mean +/− SEM of five animals per timepoint and ontogenetic stage. Data points at CT 00/24 are double-plotted. Asterisks indicate significant differences between peak values at CT12 and trough values at CT00 (*=p<0.05, **=p<0.01, ***=p<0.001; one-way ANOVA).
mPER1- and mPER2-IRs were both detected in SCN cell nuclei of foetal, newborn, infantile and adult mice (Fig. 2). In the foetal and the newborn SCN mPER1- and mPER2-IR were predominantly found in the dorsomedial region (Fig. 2). In all developmental stages the number of SCN cells with nuclear mPER1- and mPER2-IR showed significant circadian rhythms with a peak at CT12 (Fig. 4 and Tab. 1A). However, the amplitude of the mPER1 rhythm was significantly lower in E18 compared to P2 (p<0.01) and in P2 compared to adult (p<0.001) (Figs. 2 and 4). The amplitude of the mPER1 rhythm was not significantly different between infantile and adult. The amplitude of the mPER2 rhythm was also significantly lower in E18 compared to P2 and adult (p<0.05). However, the amplitude of the mPER2 rhythm in newborn and infantile was as high as in adult.
Figure 2. mPER1- and mPER2-immunoreactivity in the SCN during ontogenetic development.
Representative coronal sections showing cells with mPER1- (A) and mPER2- (B) immunoreaction (IR) in the SCN of fetal (E18), newborn (P2), infantile (P10), and adult (6–10 week-old) C3H mice. Mice were sacrificed during early or mid-subjective day (CT00, CT06) and early or mid-subjective night (CT12, CT18). Scale bars: 150 μm.
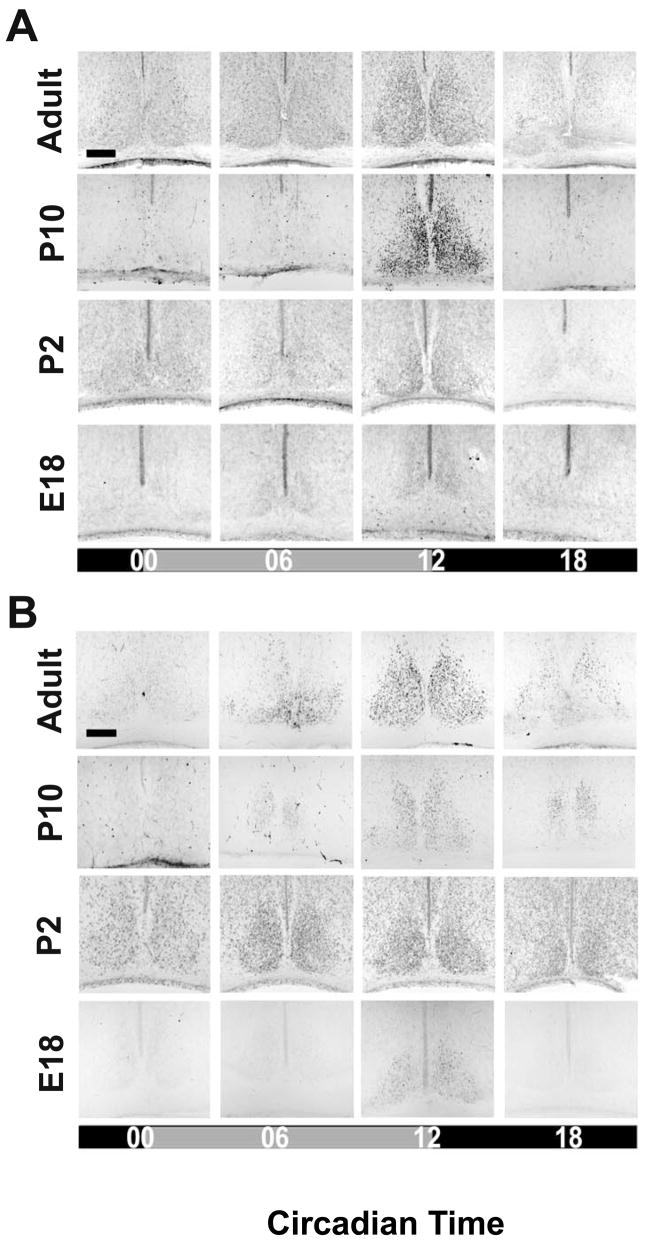
The number of SCN cells with nuclear mCRY1- and mCRY2-IRs showed a significant circadian rhythm in infantile and adult mice peaking at CT12 (p<0.05) (Figs. 3 and 4). The amplitudes of the mCRY1 and mCRY2 rhythms were not significantly different between the infantile and adult SCN. SCN cells with nuclear mCRY1-IR were also found in foetus and newborn, but their number remained constant throughout the circadian cycle. The number of SCN cells with mCRY1-IR was significantly lower in foetal and newborn mice compared to adult (p<0.001) (Figs. 3 and 4). SCN cells with nuclear mCRY2-IR were not detected in the foetus, but were found in the newborn (Fig. 3). Here they were predominantly present in the dorsomedial region and they showed a circadian rhythm peaking at CT12 (Fig. 4, Tab. 1A). The amplitude of the circadian mCRY2 rhythm was significantly lower in the newborn SCN than in the infantile and the adult SCN (p<0.05).
Figure 3. mCRY1- and mCRY2-immunoreactivity in the SCN during ontogenetic development.
Representative coronal sections showing cells with mCRY1- (A) and mCRY2- (B) immunoreaction (IR) in the SCN of fetal (E18), newborn (P2), infantile (P10), and adult (6–10 week-old) C3H mice. Mice were sacrificed during early or mid-subjective day (CT00, CT06) and early or mid-subjective night (CT12, CT18). Scale bars: 150 μm.
Rhythmic Avp mRNA expression in the SCN during ontogenetic development
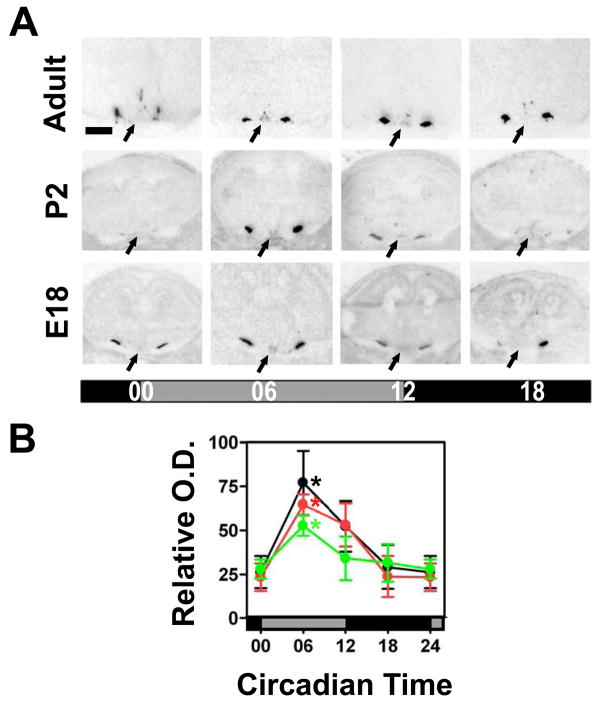
To evaluate whether the immature circadian pacemaker in the foetal SCN was sufficient to drive rhythmic expression of output genes, we investigated Avp expression by in situ hybridisation. In adult, foetus, and newborn Avp expression was strong and constitutive in the supraoptic nucleus (SON) and much weaker, but rhythmic in the SCN (Fig. 5). In the adult SCN Avp mRNA levels peaked at CT06 which is in line with a previous study (Jin et al., 1999). Both the foetal and the newborn SCN showed circadian rhythms in Avp mRNA levels with the same phase as the adult SCN (Fig. 5). The amplitude of the circadian Avp mRNA rhythm was significantly lower in the foetal compared to the adult SCN (p<0.01); however, this amplitude was not significantly different between the newborn and the adult SCN.
Figure 5. Avp mRNA expression in the SCN during ontogenetic development.
(A) Representative coronal sections showing the Avp hybridisation signal in the SCN (arrows) of fetal (E18), newborn (P2) and adult (6–10 week-old) C3H mice. Scale bar: 750 μm. (B) Relative optical density (O.D.) within the SCN region of fetal (green), newborn (red), and adult (black) C3H mice. Data are expressed as the mean +/− SEM of five animals per timepoint and ontogenetic stage. Data points at CT00/24 are double-plotted. Asterisks indicate significant differences between peak levels at CT06 and trough levels at CT00 (*p<0.05, one-way ANOVA).
Circadian rhythms in clock gene proteins in the foetal PT
To assess the ontogenetic development of the clockwork in the melatonin-dependent peripheral oscillator in the PT, we analysed BMAL1-, CLOCK-, mPER1-, mPER2-, mCRY1- and mCRY2-IR in the PT of foetal (E18), newborn (P2), infantile (P10) and adult C3H mice at four circadian timepoints during the circadian cycle (CT00, 06, 12, 18).
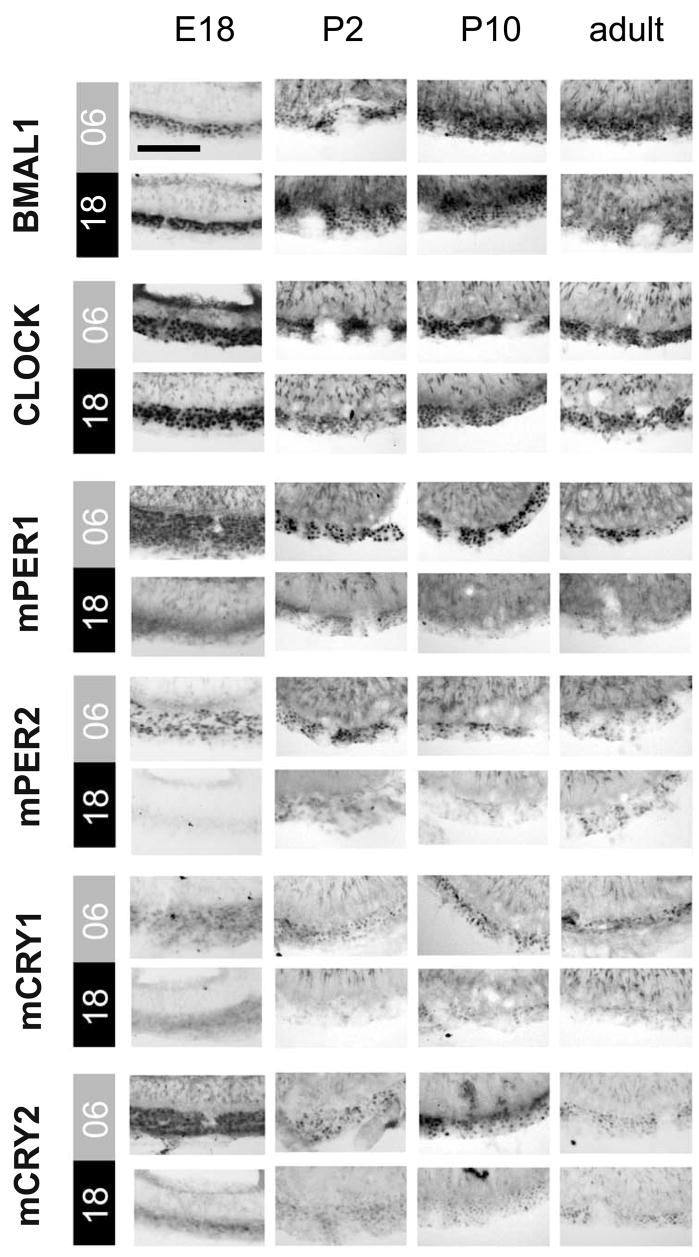
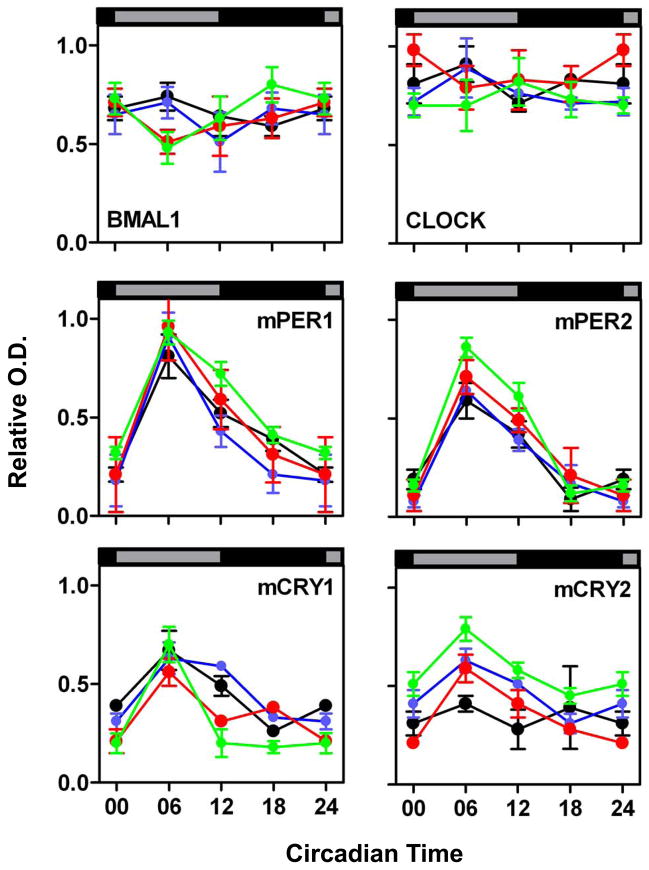
In the PT cells of all four developmental stages the six clock proteins showed a nuclear localisation throughout the circadian cycle (Fig. 6) and the intensities of the immunoreactions for the positive regulators BMAL1- and CLOCK were constantly high (Figs. 6 and 7). Immunoreactions for the negative regulators mPER1, mPER2, and mCRY1 showed circadian rhythms in the PT at all four developmental stages and peaked during subjective mid-day (CT06) (Figs. 6 and 7, Tab. 1B). The levels of BMAL1 and CLOCK and the amplitudes of mPER1, mPER2, and mCRY1 were not significantly different for the four developmental stages. Finally, mCRY2-IR was rhythmic in the foetal, newborn and infantile PT but not in the adult PT.
Figure 6. BMAL1-, CLOCK-, mPER1-, mPER2-, mCRY1-, and mCRY2-immunoreactivity in the PT.
Representative coronal sections showing immunoreactivity for all six clock gene proteins in the PT of fetal (E18), newborn (P2), infantile (P10), and adult C3H mice sacrificed at mid-subjective day (CT06) or mid-subjective night (CT18). Scale bar: 50 μm.
Figure 7. Relative optical density (O.D.) of BMAL1-, CLOCK-, mPER1-, mPER2-, mCRY1-, and mCRY2- immunoreactivity in the PT of fetal (green), newborn (red), infantile (blue), and adult (black).
Data are expressed as the mean +/− SEM of five animals per timepoint and ontogenetic stage. Data points at CT00/24 are double-plotted. Asterisks indicate significant differences between peak levels at CT06 and trough levels at CT18 (*, p<0.05, **: p<0.01, ***: p<0.001, one-way ANOVA).
Discussion
In this study we compared the ontogenetic maturation of the clockwork in the self-sustained circadian rhythm generator of the SCN and an input-dependent circadian oscillator, the melatonin-dependent peripheralPT. Our data show that the clockwork in the self-sustained circadian rhythm generator of the SCN matures gradually during postnatal life. In contrast, the melatonin-dependent peripheral PT oscillator matures during foetal life. These data suggest that the SCN that is capable of endogenous rhythm generation and peripheral circadian oscillators that are driven by rhythmic input mature differentially during mouse ontogeny.
Previous ontogenetic studies of the circadian clock have focused on the detection of clock gene mRNA levels by radioactive in situ hybridisation and have provided data at the level of the whole SCN nuclei (Shimomura et al., 2001; Ohta et al., 2002; 2003; Kovacikova et al., 2006; Sladek et al., 2007). In the present immunohistochemical study we investigated the ontogenetic maturation of the SCN clockwork by determining the number of cells that display immunoreactions for BMAL1, CLOCK, mPER1/2, and mCRY1/2 proteins at four different ontogenetic stages and four different circadian timepoints and compared these results with those obtained in the PT. This approach not only provides cellular and subcellular resolution with high sensitivity but also yields information on the relevant players in the regulation of circadian gene expression, the clock gene proteins.
In the foetal (E18) SCN, the number of cells with nuclear BMAL1- and CLOCK-IR remained constant at all four timepoints investigated. These data from the foetus are consistent with earlier studies in adult mice (Maywood et al., 2003; von Gall et al., 2003). The ratio between BMAL1-immunoreactive cells and the total number of SCN cells (both numbers determined in the intermediate portion of the SCN) was already high in the foetal SCN and remained at this constantly high level during later ontogenetic stages even though the total number of SCN cells was significantly lower in foetal than in newborn, infantile and adult mice. In contrast, the number of SCN cells displaying CLOCK-IR was much smaller in the foetal SCN and it increased in the postnatal stages. As shown for BMAL1, the number of cells with CLOCK-IR remained constant at all timepoints investigated. Only a few foetal SCN cells showed mPER1-and mPER2-IR and their number varied with the circadian timepoints. In the foetal SCN these PER rhythms peaked during day-night transition and were in the same phase as in the dam. A finding that has also been previously reported (Hastings et al., 1999; Bae et al., 2001). These data further confirm and go beyond other studies showing that the PER rhythms are synchronized between foetus and dam (Reppert & Schwartz, 1984; Reppert & Uhl, 1987;Shibata & Moore, 1987; Shimomura et al., 2001). Previous studies have shown that foetal SCN cells are not influenced by the foetal retina and retinohypothalamic tract (Reppert & Schwartz, 1986) and that synaptic connections between foetal SCN cells are very poor (Moore & Bernstein, 1989). Thus, maternal cues like the hormone melatonin are imperative to synchronize circadian rhythms in the foetal SCN via the fetomaternal unit (Reppert & Schwartz, 1986; Shibata & Moore, 1988; Jud & Albrecht, 2006) (Fig. 8). mCRY2 was absent from the foetal SCN and only a few SCN cells showed constitutive mCRY1-IR. However, these low mCRY1 levels appear to be sufficient to prevent ubiquitination and subsequent degradation of the mPER proteins; processes that would take place in the absence of mCRY (Yagita et al., 2000).
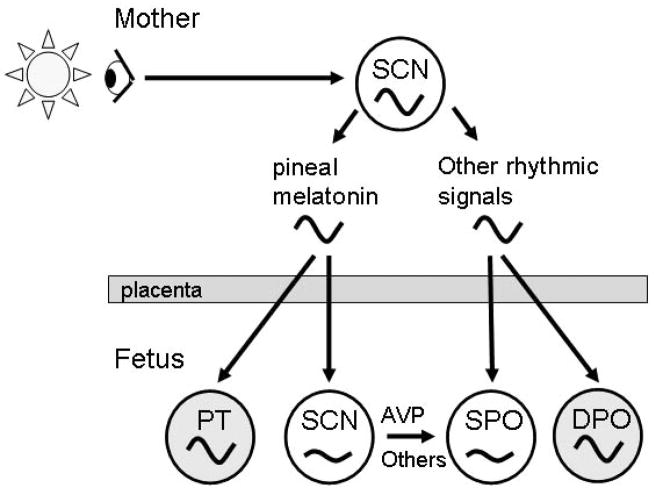
Figure 8. Model depicting differences in the ontogenetic development between the self-sustained circadian rhythm generator and input-dependent peripheral oscillators.
The mother generates circadian rhythms in melatonin and other hormones, body temperature and metabolites. These rhythmic maternal signals cross the placenta and synchronise the SCN and self-sustained peripheral oscillators (SPO) or input-dependent peripheral oscillators (DPO) like the PT in the fetus. AVP and other rhyhthmic signals generated by the primordial fetal SCN might also be important for the synchronisation of SPOs. The DPOs (high amplitude rhythms) mature earlier during ontogenetic development than the SPOs (low amplitude rhythms).
Our data suggest that the foetal SCN contains only a few pacemaker cells that comprise a primordial molecular oscillator based on rhythmic mPER and constitutive BMAL1, CLOCK, and mCRY1 levels. The CLOCK-, mPER- and CRY1-IRs were predominantly found in the dorsomedial part of the foetal SCN which is known to contain oscillator cells that are able to drive rhythmic output like AVP (Hamada et al., 2001; Antle et al., 2003). One can speculate that those SCN neurons that have a positive BMAL1-IR, but lack CLOCK-IR employ other dimerisation partners such as NPAS2 that stabilise BMAL1 in the absence of CLOCK (DeBruyne et al., 2006, 2007).
The clockwork in the foetal mouse SCN appears sufficient to drive a low amplitude rhythm in the output gene Avp. This observation is in accordance with earlier studies in rat (Reppert & Uhl, 1987). AVP secreted from the SCN in a rhythmic manner (Schwartz & Reppert, 1985) is an important diffusible factor in the adult circadian system and drives rhythmicity in the hypothalamic paraventricular nucleus (Tousson & Meissl, 2004), an essential centre for the control of both the neuroendocrine (Kalsbeek et al., 1992) and the autonomic nervous (Buijs & Kalsbeek 2001) systems. The early onset of rhythmic Avp expression during ontogeny suggests that AVP is important for synchronization of slave oscillators in the brain and periphery of the foetus (Fig. 8).
In the newborn (P2) SCN the number of cells with CLOCK- and mPER1-IR increased gradually. In contrast, the number of SCN cells with mPER2-IR was as high as in the adult. Interestingly, CLOCK-deficient mice show a strong reduction in the amplitude of mPER1 rhythms, but not of mPER2 rhythms (DeBruyne et al., 2006). These data suggest that mPer1 expression depends more strongly on the expression of CLOCK than mPer2 expression; a notion that is supported by our developmental data. The number of cells with mCRY1-IR was equally low in foetal and newborn SCN and did not vary with the circadian time. In contrast, the small number of SCN cells with mCRY2-IR varied in phase with the dam.
Our results with the newborn mouse SCN showed that it was capable of generating a circadian rhythm of the output gene Avp with an amplitude as high as in the adult. Similarly, the SCN of newborn hamster is capable of generating an adult pattern of expression for calbindin (Antle et al., 2005). However, as shown here for mice, the newborn SCN is still not fully mature since it has a significantly lower number of cells with CLOCK-, mPER1-, and mCRY2-IR compared to adult.
In the infantile (P10) SCN, the number of cells with BMAL1- and CLOCK-IR reaches adult levels (von Gall et al., 2003; Maywood et al., 2003; this study). In addition, the amplitude of the circadian rhythms in numbers of SCN cells with mPER1-, mPER2-, mCRY1-, and mCRY2- IR was as high as in the adult. This finding suggests that the mouse SCN is fully mature around postnatal day 10. During this postnatal period, the number of synapses between SCN neurons increases dramatically (Moore & Bernstein, 1989). Light induction of the Fos-like protein in the SCN occurs first at P4 (Munoz Llamosas et al., 2000) and the expression of clock genes starts to be affected by the photoperiod at P10 (Sumová et al. 2003). Thus, stabilisation of the SCN neuronal network and innervation by retinal afferents might facilitate maturation of the SCN circadian rhythm generator. However, it remains to be established whether the herein described differences in clock protein levels in the SCN during development reflect the maturation of the molecular clockwork in individual pacemaker cells or are due to the ontogenetic maturation of a neuronal network in the SCN pacemaker.
In the foetal PT immunoreactions for BMAL1 and CLOCK were similarly intense, suggesting that CLOCK is the prevailing interaction partner for BMAL1 in the foetal PT oscillator. mPER1, mPER2, and mCRY1-IRs showed a high-amplitude circadian rhythm in the foetal PT which was in the same phase as found in adult mice. In contrast to the adult PT (Jilg et al., 2005; this study), mCRY2-IR showed a circadian rhythm in the foetal, newborn and infantile PT which was in phase with the rhythms in mPER1-, mPER2-, and mCRY1-IR. This difference for the PT in adults as well as for all younger ontogenetic stages may suggest that mCRY2 loses its participation in the PT negative regulatory complex during adulthood. The levels of BMAL1-, CLOCK-, and mCRY2-IR and the amplitudes of the circadian rhythms in mPER1-, mPER2-, and mCRY1-IR reported here for the adult PT were slightly higher than in a previous study (Jilg et al., 2005). This difference is probably due to the differences in the genetic background of the animals (Jilg et al., 2005: genetically manipulated C57BL/6 chimeras bred into C3H/HeN; this study: pure C3H/HeN).
The foetal PT oscillator shows basically the same characteristics as the adult. This observation is remarkable because the PT oscillator is known to strongly depend on a rhythmic melatonin input signal (von Gall et al., 2002, 2005; Jilg et al., 2005). Rhythms in clock gene expression in the PT are known to be abolished a few days after removal of the pineal gland, the major source of melatonin (Messager et al., 2001; von Gall et al., 2002). Notably, the pineal gland of foetal rodents is not yet capable of rhythmic melatonin production (Deguchi 1975). Thus, the rhythms in clock gene proteins in the foetal PT are presumably driven by maternal melatonin that is known to cross the placenta (Klein 1972; Reppert et al., 1979; Yellon & Longo 1988).
Although melatonin receptors are present in both the foetal PT and the foetal SCN (hamster: Carlson et al., 1991; mice: unpublished observation), high-amplitude oscillations in clock gene proteins were found only in the foetal PT and not in the foetal SCN. In contrast to the homogenous population of PT cells, the SCN is highly specialised with pacemaker cells in the vasopressinergic dorsomedial (shell) part and photic input cells in the VIP-ergic ventrolateral (core) part (Moore et al., 2002). Our data therefore led us to speculate that only few pacemaker cells in the core part of the foetal SCN are synchronized by maternal melatonin. The input shell cells in the foetal SCN might not respond to melatonin and neuronal coupling between these two SCN subdivisions is weak in the foetus. In addition to the known role of maternal melatonin in synchronising the foetal SCN with the maternal phase (Reppert et al., 1988), maternal melatonin is important for the maturation and maintenance of circadian rhythms in the peripheral oscillator of the foetal PT (Fig. 8).
Currently, little is known about the maturation of the molecular clockwork in other peripheral oscillators. In the foetal monkey clock genes are differentially expressed different tissues. For example, in SCN, adrenal and thyroid Bmal1, Per2 and Cry1 are rhythmic, whereas in pineal and pituitary only Cry1/Bmal1 and Per2/Bmal1 are rhythmic (Torres-Farfan et al., 2006). Interestingly, circadian rhythms in clock genes in the foetal monkey SCN, but not the adrenal gland, are synchronised by maternal melatonin (Torres-Farfan et al., 2006). In the foetal liver only mCry1 and RevErba are rhythmic, whereas Per1, Per2, Cry1, and Bmal1 are not (Sladek et al., 2007). Similar to the SCN, the liver oscillator develops gradually during postnatal life (Sladek et al., 2004, 2007). Importantly, the adult SCN and liver show highly persistent oscillations of clock genes in the absence of rhythmic input (Yoo et al., 2004). Thus, we hypothesise that persistent/self-sustained pacemakers like the SCN and liver develop gradually during postnatal life, whereas input-dependent oscillators like the PT earlier during foetal life (Fig. 8).
In summary, this study has demonstrated that a few foetal mouse SCN cells show circadian rhythms in mPER-IR coincident with a low-amplitude circadian rhythm in Avp mRNA levels. The SCN matures gradually at the neonatal stage and shows an adult pattern of cells with clock gene protein immunoreactions at the infantile stage. In contrast, the melatonin-dependent PT oscillator shows high-amplitude circadian rhythms in clock gene proteins already in the foetus. Thus, the PT oscillator, which is directly dependent on rhythmic maternal signals, matures earlier than the SCN itself.
Acknowledgments
We thank Anja von Gall for expert technical assistance and Dr. Ruth Willmott for helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (GA 737/5-1) and the Dr. Paul und Cilli Weill-Stiftung (NA).
Abbreviations
- ANOVA
analysis of variance
- CT
circadian time
- DPO
input-dependent peripheral oscillators
- E
embryonic day
- IR
Immunoreaction, O.D., optical density
- P
postnatal day
- PT
Pars tuberalis
- SCN
suprachiasmatic nucleus
- SPO
self sustained peripheral oscillators
References
- Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: a network model of the brain clock. J Biol Rhythms. 2003;18:339– 350. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, LeSauter J, Silver R. Neurogenesis and ontogeny of specific cell phenotypes within the hamster suprachiasmatic nucleus. Brain Res Dev Brain Res. 2005;157:8–18. doi: 10.1016/j.devbrainres.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- Carlson LL, Weaver DR, Reppert SM. Melatonin receptors and signal transduction during development in Siberian hamsters (Phodopus sungorus) Brain Res Dev Brain Res. 1991;59:83–88. doi: 10.1016/0165-3806(91)90032-e. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi T. Ontogenesis of a biological clock for serotonin: acetyl coenzyme A N-acetyltransferase in pineal gland of rat. Proc Natl Acad Sci USA. 1975;72:2814–2818. doi: 10.1073/pnas.72.7.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg A, Moek J, Weaver DR, Korf HW, Stehle JH, von Gall C. Rhythms in clock proteins in the mouse pars tuberalis depend on MT1 melatonin receptor signalling. Eur J Neurosci. 2005;22:2845–2854. doi: 10.1111/j.1460-9568.2005.04485.x. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Jud C, Albrecht U. Circadian rhythms in murine pups develop in absence of a functional maternal circadian clock. J Biol Rhythms. 2006;21:149–154. doi: 10.1177/0748730406286264. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Buijs RM, van Heerikhuize JJ, Arts M, van der Woude TP. Vasopressin-containing neurons of the suprachiasmatic nuclei inhibit corticosterone release. Brain Res. 1992;580:62–67. doi: 10.1016/0006-8993(92)90927-2. [DOI] [PubMed] [Google Scholar]
- Klein DC. Evidence for the placental transfer of 3 H-acetyl-melatonin. Nat New Biol. 1972;237:117–118. doi: 10.1038/newbio237117a0. [DOI] [PubMed] [Google Scholar]
- Kovacikova Z, Sladek M, Bendova Z, Illnerova H, Sumova A. Expression of clock and clock-driven genes in the rat suprachiasmatic nucleus during late fetal and early postnatal development. J Biol Rhythms. 2006;21:140–148. doi: 10.1177/0748730405285876. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, O'Brien JA, Hastings MH. Expression of mCLOCK and other circadian clock-relevant proteins in the mouse suprachiasmatic nuclei. J Neuroendocrinol. 2003;15:329–334. doi: 10.1046/j.1365-2826.2003.00971.x. [DOI] [PubMed] [Google Scholar]
- Messager S, Garabette ML, Hastings MH, Hazlerigg DG. Tissue-specific abolition of Per1 expression in the pars tuberalis by pinealectomy in the Syrian hamster. Neuroreport. 2001;12:579–582. doi: 10.1097/00001756-200103050-00029. [DOI] [PubMed] [Google Scholar]
- Moore RY, Bernstein ME. Synaptogenesis in the rat suprachiasmatic nucleus demonstrated by electron microscopy and synapsin I immunoreactivity. J Neurosci. 1989;9:2151–2162. doi: 10.1523/JNEUROSCI.09-06-02151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY, Speh JC, Leak RH. Suprachiasmatic nucleus organization. Cell Tissue Res. 2002;309:89–98. doi: 10.1007/s00441-002-0575-2. [DOI] [PubMed] [Google Scholar]
- Nuesslein-Hildesheim B, O'Brien JA, Ebling FJ, Maywood ES, Hastings MH. The circadian cycle of mPER clock gene products in the suprachiasmatic nucleus of the siberian hamster encodes both daily and seasonal time. Eur J Neurosci. 2000;12:2856–2864. doi: 10.1046/j.1460-9568.2000.00173.x. [DOI] [PubMed] [Google Scholar]
- Ohta H, Honma S, Abe H, Honma K. Effects of nursing mothers on rPer1 and rPer2 circadian expressions in the neonatal rat suprachiasmatic nuclei vary with developmental stage. Eur J Neurosci. 2002;15:1953–1960. doi: 10.1046/j.1460-9568.2002.02016.x. [DOI] [PubMed] [Google Scholar]
- Ohta H, Honma S, Abe H, Honma K. Periodic absence of nursing mothers phase-shifts circadian rhythms of clock genes in the suprachiasmatic nucleus of rat pups. Eur J Neurosci. 2003;17:1628–1634. doi: 10.1046/j.1460-9568.2003.02584.x. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Chez RA, Anderson A, Klein DC. Maternal-fetal transfer of melatonin in the non-human primate. Pediatr Res. 1979;13:788–791. doi: 10.1203/00006450-197906000-00015. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Schwartz WJ. The suprachisamatic nuclei of the fetal rat: characterization of a functional circadian clock using C14-labeled desoxyglucose. J Neurosci. 1984;4:1677–1682. doi: 10.1523/JNEUROSCI.04-07-01677.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Uhl GR. Vasopressin messenger ribonucleic acid in supraoptic and suprachiasmatic nuclei: appearance and circadian regulation during development. Endocrinology. 1987;120:2483–2487. doi: 10.1210/endo-120-6-2483. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR, Rivkees SA. Maternal communication of circadian phase to the developing mammal. Psychoneuroendocrinology. 1988;13:63–78. doi: 10.1016/0306-4530(88)90007-8. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Reppert SM. Neural regulation of the circadian vasopressin rhythm in cerebrospinal fluid: a pre-eminent role for the suprachiasmatic nuclei. J Neurosci. 1985;5:2771–2778. doi: 10.1523/JNEUROSCI.05-10-02771.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Moore RY. Development of a fetal circadian rhythm after disruption of the maternal circadian system. Brain Res. 1988;469:313–317. doi: 10.1016/0165-3806(88)90194-0. [DOI] [PubMed] [Google Scholar]
- Shimomura H, Moriya T, Wakamatsu H, Akiyama M, Miyake Y, Shibata S. Differential daily expression of Per1 and Per2 mRNA in the suprachiasmatic nucleus of fetal and early postnatal mice. Eur J Neurosci. 2001;13:687–693. doi: 10.1046/j.0953-816x.2000.01438.x. [DOI] [PubMed] [Google Scholar]
- Sladek M, Sumova A, Kovacikova Z, Bendova Z, Laurinova K, Illnerova H. Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2004;101:6231–6236. doi: 10.1073/pnas.0401149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek M, Jindrakova Z, Bendova Z, Sumova A. Postnatal ontogenesis of the circadian clock within the rat liver. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1224–R1229. doi: 10.1152/ajpregu.00184.2006. [DOI] [PubMed] [Google Scholar]
- Torres-Farfan C, Rocco V, Monso C, Valenzuela FJ, Campino C, Germain A, Torrealba F, Valenzuela GJ, Seron-Ferre M. Maternal melatonin effects on clock gene expression in a nonhuman primate fetus. Endocrinology. 2006;147:4618–4626. doi: 10.1210/en.2006-0628. [DOI] [PubMed] [Google Scholar]
- Tousson E, Meissl H. Suprachiasmatic nuclei grafts restore the circadian rhythm in the paraventricular nucleus of the hypothalamus. J Neurosci. 2004;24:2983–2988. doi: 10.1523/JNEUROSCI.5044-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gall C, Duffield GE, Hastings MH, Kopp MD, Dehghani F, Korf HW, Stehle JH. CREB in the mouse SCN: a molecular interface coding the phase-adjusting stimuli light, glutamate, PACAP, and melatonin for clockwork access. J Neurosci. 1998;18:10389–10397. doi: 10.1523/JNEUROSCI.18-24-10389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gall C, Garabette ML, Kell CA, Frenzel S, Dehghani F, Schumm-Draeger PM, Weaver DR, Korf HW, Hastings MH, Stehle JH. Rhythmic gene expression in pituitary depends on heterologous sensitization by the neurohormone melatonin. Nat Neurosci. 2002;5:234–238. doi: 10.1038/nn806. [DOI] [PubMed] [Google Scholar]
- von Gall C, Weaver DR, Moek J, Jilg A, Stehle JH, Korf HW. Melatonin plays a crucial role in the regulation of rhythmic clock gene expression in the mouse pars tuberalis. Ann NY Acad Sci. 2005;1040:508–511. doi: 10.1196/annals.1327.105. [DOI] [PubMed] [Google Scholar]
- von Gall C, Noton E, Lee C, Weaver DR. Light does not degrade the constitutively expressed BMAL1 protein in the mouse suprachiasmatic nucleus. Eur J Neurosci. 2003;18:125–133. doi: 10.1046/j.1460-9568.2003.02735.x. [DOI] [PubMed] [Google Scholar]
- Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Yellon SM, Longo LD. Effect of maternal pinealectomy and reverse photoperiod on the circadian melatonin rhythm in the sheep, fetus during the last trimester of pregnancy. Biol Reprod. 1988;39:1093–1099. doi: 10.1095/biolreprod39.5.1093. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]