Abstract
The molecular mechanisms of pulmonary fibrosis are poorly understood. We have used oligonucleotide arrays to analyze the gene expression programs that underlie pulmonary fibrosis in response to bleomycin, a drug that causes lung inflammation and fibrosis, in two strains of susceptible mice (129 and C57BL/6). We then compared the gene expression patterns in these mice with 129 mice carrying a null mutation in the epithelial-restricted integrin β6 subunit (β6−/−), which develop inflammation but are protected from pulmonary fibrosis. Cluster analysis identified two distinct groups of genes involved in the inflammatory and fibrotic responses. Analysis of gene expression at multiple time points after bleomycin administration revealed sequential induction of subsets of genes that characterize each response. The availability of this comprehensive data set should accelerate the development of more effective strategies for intervention at the various stages in the development of fibrotic diseases of the lungs and other organs.
Keywords: cluster analysis, oligonucleotide arrays, αvβ6, bleomycin, pulmonary fibrosis
Pulmonary fibrosis is a progressive and largely untreatable group of disorders that affects up to 100,000 people in the United States (1). Current therapies, aimed principally at inhibiting lung inflammation that often precedes fibrosis, are effective only in a minority of affected individuals, and there are currently no proven therapies targeting the fibrotic process itself (2, 3). Much of the information regarding the development of pulmonary fibrosis has been acquired with a well characterized animal model in which fibrosis is induced by a single intratracheal administration of the cytotoxic drug, bleomycin (4). Bleomycin, like in most of the conditions associated with pulmonary fibrosis, initially induces lung inflammation that is followed by a progressive destruction of the normal lung architecture (5). Previous studies of gene expression in bleomycin-induced fibrosis have provided some clues to pathophysiology (6–10) but have generally been limited to analysis of one or a few genes, and therefore have provided little information about the coordinated pattern of gene expression involved in this response.
To understand the molecular basis for pulmonary fibrosis, in more detail, we have analyzed changes in gene expression in response to bleomycin by using oligonucleotide microarrays that permit a simultaneous and quantitative measurement of the expression of thousands of genes (11, 12). A comprehensive profile of the pulmonary response to bleomycin was generated with arrays containing probe sets for approximately 6,000 murine genes and expressed sequence tags (ESTs) (12). Changes in gene expression were monitored after bleomycin treatment of wild-type C57BL/6 mice, 129 mice, and 129 mice homozygous for a null mutation of the integrin β6 subunit gene (β6−/−). We have previously shown that β6−/− mice develop inflammation, but do not develop fibrosis in response to bleomycin (13). In this report, data obtained with the three strains of mice have allowed us to distinguish genes involved in the inflammatory response to bleomycin (induced in all three lines) from genes that specifically contribute to the fibrotic response (induced to a lesser degree in β6−/− mice). Analysis of the time course of induction of these genes has identified sequential genetic programs that characterize bleomycin-induced inflammation and fibrosis.
Materials and Methods
Bleomycin Treatment.
129 mice.
Bleomycin (Mead Johnson) dissolved in saline (0.05 unit in 60 μl of saline) or saline only was injected through a 27-gauge needle directly into trachea of mice under methoxyflourane anesthesia (Mallinckrodt). The protocols were approved by the Committee on Experimental Animal Research. Mice were sacrificed 7 or 14 days after bleomycin or 7 days after saline injection. A separate group of control animals was sacrificed without any injection, and at least three animals were included in each experimental group. Lungs from each animal were dissected and frozen immediately in liquid nitrogen. The right lung was used for total RNA isolation, while the left was kept frozen for future analysis. Frozen lung tissue was homogenized in ice-cold Trizol (Life Technologies, Grand Island, NY) and total RNA extracted for double-stranded cDNA synthesis.
C57BL/6 mice.
Female C57BL/6 mice (approximately 10 wk old) were anesthetized with methoxyfluorane by using a 24-gauge intubation needle inserted into the trachea through the oral cavity. A total of 50 μl of bleomycin (0.08 unit in 0.9% saline; Sigma) or saline only was slowly injected. Mice were sacrificed 2, 5, 7, or 14 days after bleomycin or 2 days after saline injection, and their lungs were frozen in liquid nitrogen. Total RNA was isolated from pooled lungs of six mice per time point and used to prepare twice purified poly(A) mRNA (Oligotex; Qiagen, Chatsworth, CA) for use as template for double-stranded cDNA synthesis.
Preparation of Labeled cRNA and Hybridization to Microarrays.
Double-stranded cDNA was synthesized with a cDNA synthesis kit (Life Technologies Superscript cDNA Synthesis System) by using an oligo(dT)24 primer with a T7 RNA polymerase promoter site added to its 3′ end (Genset Corp., La Jolla, CA). The isolated cDNA was used for in vitro transcription (Ambion T7 Megascript system) in the presence of biotin-11-CTP and biotin-16-UTP (Enzo Diagnostics). A total of 25–50 μg of the cRNA product in buffer [40 mM Tris⋅acetate (pH 8.1)/100 mM potassium acetate/30 mM magnesium acetate] was fragmented at 94°C for 35 min. It was then used as a hybridization mix with Herring sperm DNA (0.1 mg/ml; Sigma), plus four control bacterial and phage cRNA (1.5 pM BioB, 5 pM BioC, 25 pM BioD, and 100 pM Cre) samples to serve as internal controls for hybridization efficiency as directed by the manufacturer (Affymetrix, Santa Clara, CA).
Aliquots of the hybridization murine cRNA mixtures (10 μg cRNA in 200 μl hybridization mix) were hybridized to a Mu6500 and each genechip array was washed and scanned (Hewlett Packard, GeneArray scanner G2500A) according to procedures developed by manufacturer (Affymetrix).
Analysis of genechip Data.
Scanned output files were visually inspected for hybridization artifacts and then analyzed with genechip 3.1 software (Affymetrix). Arrays were scaled to an average intensity of 100 and analyzed independently. The method of determination of whether each RNA species represented on the array was detectable has been previously described (11, 14). The expression value (average difference) for each gene was determined by calculating the average of differences of intensity (perfect match intensity minus mismatch intensity) between its probe pairs.
The expression analysis files created by genechip 3.1 software were transferred to a database (Microsoft Access) and linked to Internet genome databases (e.g., NHLBI, Swiss Prot, and GeneCards). Mean intensity for each experimental condition was defined as the mean of average differences of individual mice in the group. Fold changes were determined by dividing the mean intensity of each experimental condition by the mean intensity of the comparison group. Since the pattern of gene expression 7 days after saline injection was not substantially different from the pattern in uninjected animals, we pooled the values obtained from the uninjected animals with those from the saline-injected animals.
A value of 20 was assigned to all intensity measurements below 20. For further data mining and presentation we used spotfire pro 3.0 (Spotfire, Göteborg, Sweden). For cluster analysis we used gene cluster and treeview programs (15). Genes with at least one mean intensity value above 100 and a 2-fold difference in one pair-wise comparison were included in the cluster analysis. Since we calculated the fold ratios by using the mean values of the average differences of several mice, a change was not considered substantial if it was caused only by a single outstanding value.
Isolation of Alveolar Macrophages.
Ten-week-old 129 strain β6−/− or β6+/+ mice were anesthetized with methoxyfluorane, and lungs were lavaged with 4 ml (5 × 0.8 ml) of pyrogen-free PBS. Lavage fluid from six animals in each experimental group was pooled and centrifuged at 800 rpm at 4°C. The cell pellet was resuspended in erythrocyte (RBC) lysis buffer (Sigma) for 10 min, followed by recentrifugation and resuspension in RPMI 1640 medium. Cells were plated on plastic dishes (Falcon 3047) and after 30 min washed to remove nonadherent cells (16) and then lysed with 1 ml of Trizol reagent (GIBCO/BRL).
Reverse Transcription (RT)–PCR.
RT-PCR was performed as described previously (17). Briefly, total RNA was isolated and reverse-transcribed (Superscript II; GIBCO/BRL). Aliquots of each sample were diluted serially, and the abundance of hypoxanthine phosphoribosyltransferase (HPRT) among the samples of interest was normalized to identical concentrations of a 450-bp modified HPRT insert in a polycompetitor plasmid (pLOC; gift of Richard Locksley) added to each reaction (5 ng/μl). Primers for the amplification of a 650-base fragment of macrophage metalloelastase (MME) were (5′ to 3′) AGCATCTTAGAGCAGTGCCC (forward) and ATGTTGGTGGCTGGACTCCC (reverse) (18). Samples were subjected to 30 rounds of PCR at an annealing temperature of 60°C. PCR products were separated in a 2.5% agarose gel by electrophoresis and visualized with ethidium bromide staining.
Results
To identify patterns of gene expression in response to bleomycin, we performed cluster analysis as previously described (15) by using mean values of the average difference intensities for each gene. We chose 470 genes that had a mean intensity of >100 arbitrary units in at least one condition and differed in intensity by at least 2-fold in at least one pairwise comparison. We then performed cluster analysis, treating each possible pairwise comparison of mean values as a single data point. With this approach, we hoped to identify groups of genes that were regulated in a coordinate fashion, both at baseline and after treatment with bleomycin. Cluster analysis of these genes is shown in Fig. 1. The full clustering and data for all 470 genes can be seen on our web site (http://medicine.ucsf.edu/divisions/lbc/).
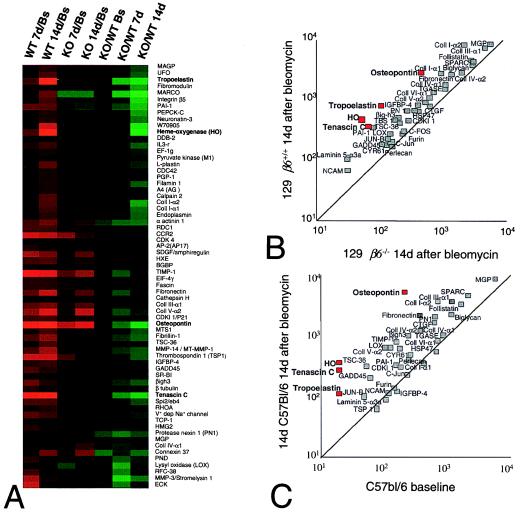
Figure 1.

Cluster analysis of 470 genes that changed >2-fold and had at least one mean intensity value of 100 in at least one of the experimental conditions. The data are presented as a ratio of the mean of the average intensities from all animals in each experimental group. Genes that were present at higher levels in the examined group (increased) are shown in progressively brighter shades of red, depending on the fold difference, and genes that were expressed at lower levels are shown in progressively brighter shades of green. Genes shown in black were not different between the two groups being compared. Columns identify the pair-wise comparison being made between wild-type (WT) or β6−/− (KO) mice examined at baseline (Bs) or at 7 days (7d) or 14 days (14d) after treatment with intratracheal bleomycin. Baseline values included both untreated mice (n = 4) and mice sacrificed 7 days after treatment with intratracheal saline (n = 3). All other values were the mean of three average intensities. Cluster analysis was performed on log transformed values of the fold ratios with gene cluster and presented with treeview. The letters on the right define distinct clusters that are discussed in the text or presented in the following figures or on our web site.
One large cluster of genes (cluster B; Figs. 1 and 2A) was expressed at similar levels at baseline in wild-type and β6−/− mice (Figs. 1 and 2A, KO/WTBs). It was dramatically induced by bleomycin in wild-type mice (Figs. 1 and 2A, WT 7d/Bs and WT 14d/Bs), but was induced to a lesser degree in β6−/− mice (Figs. 1 and 2A, KO 7d/Bs and KO 14d/Bs). These genes could thus provide clues to critical steps in the development of fibrosis. The cluster was composed of 66 genes that included 16 genes involved in the formation of the extracellular matrix, 10 genes involved in regulation of cellular responses to the extracellular matrix, and 3 genes known to be induced by DNA damage (GADD45, CDKI 1, and DDB2). Quantitative comparison among these genes identified a small subgroup, including osteopontin, tenascin-C, tropoelastin, and the stress-induced protein, heme-oxygenase (HO), which were most dramatically induced by bleomycin. These same genes were the most different between wild-type and β6−/− mice after bleomycin treatment (Fig. 2B). Expression of 78% of these genes was also increased in C57BL/6 mice, including each of the four genes described above (Fig. 2C).
Figure 2.
(A) Enlargement of Cluster B from Fig. 1 of genes that were not different at baseline and were induced preferentially in wild-type mice. The name of each gene is shown to the right of each row. Genes shown in bold text were induced more than 8-fold in response to bleomycin. (B) Because many of the genes in cluster B had been previously shown to be induced by TGF-β, expression of 53 known TGF-β-inducible genes was compared 14 days after bleomycin treatment in β6−/− mice (x axis) and wild-type (β6+/+) mice (y axis). Genes induced at least 8-fold by bleomycin are shown bold in these panels, and the mean intensity values are shown by red boxes. The middle diagonal in B and C is the line of identity. (C) Comparison of expression of the same 53 TGF-β-inducible genes at baseline (x axis) and 14 days after bleomycin (y axis) in wild-type C57BL/6 mice.
Because of the presumed significant role of transforming growth factor (TGF)-β in fibrosis and the presence of multiple TGF-β-inducible genes in cluster B, we compared the expression of a group of 53 genes known to be induced by TGF-β 14 days after bleomycin treatment in wild-type and β6−/− mice (Fig. 2B). The overwhelming majority of TGF-β-inducible genes were expressed at higher levels after bleomycin in wild-type mice than in β6−/− mice. Up-regulation of these TGF-β-inducible genes also occurred in C57BL/6 mice (Fig. 2C). Despite this dramatic evidence of TGF-β effect, increases in TGF-β1, -β2, and -β3 mRNA by bleomycin were not detectable in any of the mouse strains (data not shown).
Another cluster (Cluster A, Figs. 1 and 3A) included 63 genes that were expressed at higher levels at baseline in β6−/− mice (Figs. 1 and 3A, KO/WT Bs) and were also induced by bleomycin. At both 7 and 14 days after bleomycin, the absolute level of expression of most of the genes in this cluster was the same or higher in β6−/− mice as in wild-type mice (Figs. 1 and 3A, KO/WT 7d, KO/WT 14d). This cluster is dominated by genes known to be involved in inflammatory responses and includes complement components (5), serine proteases (4), serum amyloid proteins (3), chemokines and chemokine receptors (4), genes restricted to leukocytes (9), and others associated with inflammation (6). In this cluster of genes, 83% were also induced in C57BL/6 mice.
Figure 3.
(A) Enlargement of Cluster A from Fig. 1. The name of each gene is shown to the right of each row. Genes shown in bold text were, at baseline, more than 8-fold higher in β6−/− mice than in β6+/+ mice. (B) Scatter plot comparing the values of all the genes in β6+/+ (x axis) and β6−/− mice (y axis). Genes shown in bold were expressed at least 8-fold higher in β6−/− mice compared with β6+/+ mice, and their mean intensity values are shown by red circles. The middle diagonal in B and C is the line of identity. The upper and lower diagonals represent 2-fold increases and decreases, respectively. (C) Scatter plot comparing the expression of 47 genes restricted to leukocytes in β6+/+ mice and β6−/− mice. (D) Ethidium bromide stained 2.5% agarose gel demonstrating the results of noncompetitive RT-PCR with primers specific for MME from macrophages purified by adherence from bronchoalveolar lavage of β6+/+ mice or β6−/− mice. (Bottom) Parallel competitive RT-PCR with primers and a plasmid competitor specific for HPRT.
To determine whether the increase in leukocyte-related genes could be explained entirely by a difference in baseline cellular composition of the lungs of wild-type and β6−/− mice, we conservatively identified genes whose expression is largely restricted to leukocytes, and compared their baseline expression in wild-type and β6−/− mice (Fig. 3C). A vast majority of these genes were increased 4-fold or less in β6−/− mice, consistent with the 2- to 4-fold increase in numbers of macrophages, lymphocytes, eosinophils, and neutrophils that we have previously described for these mice (13, 19, 20). However, one gene, the macrophage-restricted metalloproteinase, MME was increased more than 20-fold. RT-PCR of RNA from whole lung and alveolar macrophages confirmed that this dramatic induction reflected a true increase in mRNA expression rather than simply a difference in cell type (Fig. 3D). Four other members of this cluster, the acute phase reactant serum amyloid 3 (SAA3), lipocalin 2, a neutrophil gelatinase-associated protein that is expressed also in carcinomas and normal epithelial tissues (21–23), BRP 39, a cartilage glycoprotein, and a C-C chemokine, C10, were also among the genes induced most dramatically in β6−/− mice (Fig. 3B).
Cluster analysis also demonstrated that protection of β6−/− mice from bleomycin-induced pulmonary fibrosis was not simply due to blunting of all cellular responses to the drug. One cluster of genes (Cluster C, Fig. 1) was expressed at similar levels in wild-type and β6−/− mice at baseline and induced to a similar degree by bleomycin. Another cluster (Cluster D, Fig. 1) was preferentially induced by bleomycin in β6−/− mice.
To describe the time course of gene induction by bleomycin, data from the detailed time course experiments performed in C57BL/6 mice were analyzed. Overall, 63% of the genes whose expression level was increased after bleomycin in wild-type 129 mice were also seen at an increased level in at least one time point after bleomycin in C57BL/6 mice. For the genes in clusters A and B, this number was substantially higher amounting to 78% of the genes in cluster A and 83% of the genes in cluster B. The temporal patterns of expression observed for subsets of genes in each of these clusters are shown in Fig. 4. In each of these clusters, we were able to identify groups of genes with similar temporal patterns of expression. In some cases, the similar temporal pattern was closely linked by a known function or transcriptional regulation. For example, five of the eight genes from cluster A that were maximally increased at the earliest time point studied (Fig. 4A) are known components of interferon-activated signaling including STAT-1 and four interferon-inducible genes that are all downstream of STAT1 activation (IFI 1–8d, IFI 6–16, ISG15, and IRF-7) (24, 25). Together, the observed temporal pattern of expression of the genes in cluster A indicates that in response to bleomycin, the earliest action includes activation of an interferon response pathway and the serum amyloid proteins (Fig. 4A). This is followed by an induction of the C-C chemokines, MCP-1 and MCP-3 (Fig. 4B), a delayed activation of complement factor genes and cathepsins (Fig. 4C), and an even later induction of the chemokines C-10 and MIP-1γ (Fig. 4D).
Figure 4.
Time course of expression after saline and at days 2, 5, 7, and 14 after bleomycin, in C57BL/6 mice of selected genes from Clusters A (A–D) and B (E and F) that were also induced by bleomycin in this strain. Each panel shows data for genes that followed a similar temporal pattern of induction. Values for each gene are plotted as a percentage of maximal induction for that gene, which is considered 100%.
Cluster B also included coordinately expressed subsets of genes with distinct temporal patterns. All of the extracellular matrix components in this cluster were induced progressively throughout the 14-day period examined (Fig. 4H). The matrix-degrading proteinases and their inhibitors were also expressed coordinately, with an early peak at 5 days and a later peak at 14 days (Fig. 4G). The genes in cluster B that peaked at the earliest time point (2 days, Fig. 4E) contained two genes, GADD45 and p21, whose expression has previously been shown to be coordinately regulated, as for example in response to p53 and/or DNA damage (26, 27). Interestingly, TGF-β-inducible genes were spread among all of the temporal clusters, suggesting that factors other than the activation of TGF-β participate in controlling the patterns of observed expression.
An additional insight from this study was the large number of genes whose expression was decreased after treatment with bleomycin (Fig. 1). Genes that are expressed at decreased levels during induction of a disease model could be as important in understanding pathogenesis as genes that are induced, and these genes have been previously largely ignored. Genes with diverse functions are represented in this group, and it is not immediately apparent how these functions are biologically linked. However, the pattern of decreased gene expression was also conserved between strains, with 71% of the genes inhibited in 129 mice also inhibited in C57BL/6 mice. The complete listing of these genes is also available at our web site (http://medicine.ucsf.edu/divisions/lbc/).
Discussion
The results of this study identify a large number of changes in pulmonary gene expression induced by bleomycin. From cluster analysis of expression patterns in wild-type and β6−/− 129 mice, we have identified distinct groups of genes that are induced in association with the inflammatory and fibrotic responses to this drug. The majority of genes in both clusters were also induced by bleomycin in C57BL/6 mice. In this strain, a detailed time course analysis revealed that the large number of genes in each of these clusters could be grouped into a series of smaller clusters that were each expressed with a distinct temporal pattern. These findings provide a framework for designing interventions that could prevent the development or progression of fibrosis at various stages of disease development.
One of the most informative clusters of genes identified was cluster B. Based on its pattern of expression, i.e., little difference between wild-type and β6−/− mice at baseline (Figs. 1 and 2A, KO/WT Bs), but preferential induction by bleomycin in wild-type mice (Figs. 1 and 2A, WT 7d/Bs and WT 14d/Bs vs. KO 7d/Bs and KO 14d/Bs), this cluster is likely to contain genes that play a critical role in the fibrotic response to bleomycin. The functional properties of many genes in this cluster are known, including most of the extracellular matrix proteins within the 470 genes subjected to cluster analysis, and genes involved in modification of the matrix and in cellular responses to the matrix. This provides further evidence that these genes are critical to the development of bleomycin-induced pulmonary fibrosis. Interestingly, of the five genes induced early after bleomycin in this cluster (Fig. 4E), two are well-characterized genes known to be induced by DNA damage, i.e., GADD 45 and p21 (27, 28). These genes demonstrated a unique pattern of early induction and then a persistent expression (Fig. 4E) in C57BL/6 mice, and maximal at the earliest time point examined in wild-type 129 mice. In contrast, these genes were minimally induced in β6−/− 129 mice, suggesting that the expression of these genes may be important in initiating the cascade of events that leads to pulmonary fibrosis. Our observation that most of the known TGF-β-inducible genes present on the array set are preferentially induced in wild-type 129 and C57BL/6 mice compared with β6−/− mice (Fig. 2 B and C) supports our recent finding that the integrin αvβ6 itself plays a role in the activation of latent TGF-β1 complexes (13). However, the up-regulation of TGF-β-inducible genes occurred with several distinct temporal patterns, an indication that factors other than activation of TGF-β must be involved. These observations also indicate a limitation of the global analysis of gene expression as a method for identifying proteins that play critical roles in biological responses. This method cannot directly identify proteins that participate in biological responses solely or principally as a result of posttranslational modification. Nonetheless, by simultaneously analyzing the expression patterns of large numbers of genes, it is possible to detect the downstream effects of such events and possibly identify the proteins and pathways involved.
A major advantage of the availability of our experimental results is the opportunity for other investigators to extract useful additional information from the data that are currently available on our web site. With regard to the genes that we have identified as associated specifically with the development of fibrosis (Cluster B), it should now be possible to design experiments utilizing inhibitors of the proteins encoded by these genes and/or mice expressing null mutations to directly examine their role in matrix modification and allow identification of potential new targets for the treatment of fibrotic diseases of the lungs and other tissues. Mice expressing null mutations of some of the genes in clusters A and B have already been generated [osteopontin (29, 30), heme-oxygenase (29–31), PAI-1 (32)]. In fact, mice expressing a null mutation for the plasminogen activator inhibitor 1 (PAI-1, cluster B, Figs. 1 and 2A) have already demonstrated protection from bleomycin-induced pulmonary fibrosis.
Global analysis of gene expression applied to complex multicellular organs or organisms provides a general expression pattern of the tissue but is limited in distinguishing changes in transcriptional regulation from changes in cellular composition of the organ being studied. A related limitation is the inability to ascribe changes in gene expression to events in any particular cell type. These limitations are relevant to the findings in this report. For example, we know that at baseline the lungs of β6−/− mice contain increased numbers of macrophages, lymphocytes, neutrophils, and eosinophils (13, 19, 20) and that bleomycin induces significant recruitment of each of these cell types to the lung (33, 34). Thus, it is likely that some of the genes that are differentially expressed between wild-type and β6−/− mice or between baseline and after treatment with bleomycin represent differences in cellular composition rather than differences in transcriptional regulation. However, one advantage of simultaneously analyzing the expression patterns of large numbers of genes is that the genes analyzed will include several that are restricted in their expression to specific cells. The expression of a group of cellularly restricted genes can thus be used to estimate changes in cellular content. Thus, for example, by comparing values obtained for a large number of leukocyte-restricted genes in wild-type and β6−/− mice (Fig. 3 B and C), we have identified at least one gene, MME, that was clearly induced out of proportion to any difference that could be attributed simply to a difference in macrophage number.
A reassuring aspect of our results was the considerable overlap between the genes that were induced (and inhibited) by bleomycin in two genetically distinct strains of mice, 129 and C57BL/6. The experiments in 129 and C57BL/6 mice were performed by different investigators, at different times, with differences in the experimental protocol (see Materials and Methods). Despite these differences, similar genes were increased after bleomycin in both strains, suggesting that the method of analysis used is valid and that the patterns of gene expression we identified are relevant to fibrosis of the lung and other tissues.
Tissue fibrosis represents a final common consequence for a large variety of disease processes in many organs. It is a common cause of organ dysfunction and a major cause of morbidity and mortality. Despite the medical significance of fibrosis, there are currently no approved treatments to specifically target the fibrotic process itself. This may in part be the result of the inherent difficulty in dissecting out the fibrotic response from the accompanying inflammatory response. Our approach has compared the gene expression patterns in mice that develop pulmonary fibrosis in response to bleomycin (wild-type 129) with mice from the same genetic background that do not (β6−/− 129). This approach has allowed us to identify a group of genes that are likely to be directly relevant to the fibrotic process. The results presented in this paper and the availability of our large data set on the internet should provide additional insight into the disease process and accelerate the development of effective and specific interventions for the treatment of fibrosis of the lungs and other organs.
Acknowledgments
We thank Dr. Harold Van Wart and Dr. Elsie Eugui for their enthusiastic support and Yong Kim for help with experimental methods during the course of this work. Dr. Eric Brown provided helpful comments on the manuscript. This work was supported by Roche Bioscience and in part by National Institutes of Health Grants HL/A133259, HL47412, HL53949, HL 47660, and HL09793.
Abbreviations
- RT
reverse transcription
- TGF
transforming growth factor
- MME
macrophage metalloelastase
- HPRT
hypoxanthine phophoribosyltransferase
Footnotes
Current address: 76 Bolingbroke Grove, London, SW11 6HB, United Kingdom.
References
- 1.Coultas D B, Zumwalt R E, Black W C, Sobonya R E. Am J Respir Crit Care Med. 1994;150:967–972. doi: 10.1164/ajrccm.150.4.7921471. [DOI] [PubMed] [Google Scholar]
- 2.Ryu J H, Colby T V, Hartman T E. Mayo Clin Proc. 1998;73:1085–1101. doi: 10.4065/73.11.1085. [DOI] [PubMed] [Google Scholar]
- 3.King T E., Jr Ann Intern Med. 1998;129:806–812. doi: 10.7326/0003-4819-129-10-199811150-00012. [DOI] [PubMed] [Google Scholar]
- 4.Snider G L, Hayes J A, Korthy A L. Am Rev Respir Dis. 1978;117:1099–1108. doi: 10.1164/arrd.1978.117.6.1099. [DOI] [PubMed] [Google Scholar]
- 5.Shen A S, Haslett C, Feldsien D C, Henson P M, Cherniack R M. Am Rev Respir Dis. 1988;137:564–571. doi: 10.1164/ajrccm/137.3.564. [DOI] [PubMed] [Google Scholar]
- 6.Haase M, Koslowski R, Lengnick A, Hahn R, Wenzel K W, Schuh D, Kasper M, Muller M. Virchows Arch. 1997;431:441–448. doi: 10.1007/s004280050121. [DOI] [PubMed] [Google Scholar]
- 7.Lucey E C, Ngo H Q, Agarwal A, Smith B D, Snider G L, Goldstein R H. Lab Invest. 1996;74:12–20. [PubMed] [Google Scholar]
- 8.Shahzeidi S, Mulier B, de Crombrugghe B, Jeffery P K, McAnulty R J, Laurent G J. Thorax. 1993;48:622–628. doi: 10.1136/thx.48.6.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahzeidi S, Jeffery P K, Laurent G J, McAnulty R J. Eur Respir J. 1994;7:1938–1943. [PubMed] [Google Scholar]
- 10.Swiderski R E, Dencoff J E, Floerchinger C S, Shapiro S D, Hunninghake G W. Am J Pathol. 1998;152:821–828. [PMC free article] [PubMed] [Google Scholar]
- 11.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 12.Fambrough D, McClure K, Kazlauskas A, Lander E S. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 13.Munger J S, Huang X, Kawakatsu H, Griffiths M J, Dalton S L, Wu J, Pittet J F, Kaminski N, Garat C, Matthay M A, et al. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 14.Der S D, Zhou A, Williams B R G, Silverman R H. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams D O. Methods Enzymol. 1979;58:494–505. doi: 10.1016/s0076-6879(79)58164-6. [DOI] [PubMed] [Google Scholar]
- 17.Reiner S L, Zheng S, Corry D B, Locksley R M. J Immunol Methods. 1994;173:133. doi: 10.1016/0022-1759(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro S D, Griffin G L, Gilbert D J, Jenkins N A, Copeland N G, Welgus H G, Senior R M, Ley T J. J Biol Chem. 1992;267:4664–4671. [PubMed] [Google Scholar]
- 19.Huang X Z, Chen A, Agrez M, Sheppard D. Am J Respir Cell Mol Biol. 1995;13:245–251. doi: 10.1165/ajrcmb.13.2.7626292. [DOI] [PubMed] [Google Scholar]
- 20.Huang X Z, Wu J F, Cass D, Erle D J, Corry D, Young S G, Farese R V, Jr, Sheppard D. J Cell Biol. 1996;133:921–928. doi: 10.1083/jcb.133.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bundgaard J R, Sengelov H, Borregaard N, Kjeldsen L. Biochem Biophys Res Commun. 1994;202:1468–1475. doi: 10.1006/bbrc.1994.2096. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen B S, Borregaard N, Bundgaard J R, Timshel S, Sehested M, Kjeldsen L. Gut. 1996;38:414–420. doi: 10.1136/gut.38.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoesz S P, Friedl A, Haag J D, Lindstrom M J, Clark G M, Gould M N. Int J Cancer. 1998;79:565–572. doi: 10.1002/(sici)1097-0215(19981218)79:6<565::aid-ijc3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 24.Boehm U, Klamp T, Groot M, Howard J C. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 25.Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 26.Arriola E L, Lopez A R, Chresta C M. Oncogene. 1999;18:1081–1091. doi: 10.1038/sj.onc.1202391. [DOI] [PubMed] [Google Scholar]
- 27.El-Deiry W S. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 28.Landesman Y, Bringold F, Milne D D, Meek D W. Cell Signalling. 1997;9:291–298. doi: 10.1016/s0898-6568(97)89890-7. [DOI] [PubMed] [Google Scholar]
- 29.Liaw L, Birk D E, Ballas C B, Whitsitt J S, Davidson J M, Hogan B L. J Clin Invest. 1998;101:1468–1478. doi: 10.1172/JCI1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rittling S R, Denhardt D T. Exp Nephrol. 1999;7:103–113. doi: 10.1159/000020591. [DOI] [PubMed] [Google Scholar]
- 31.Yet S F, Perrella M A, Layne M D, Hsieh C M, Maemura K, Kobzik L, Wiesel P, Christou H, Kourembanas S, Lee M E. J Clin Invest. 1999;103:R23–R29. doi: 10.1172/JCI6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eitzman D T, McCoy R D, Zheng X, Fay W P, Shen T, Ginsburg D, Simon R H. J Clin Invest. 1996;97:232–237. doi: 10.1172/JCI118396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White D A, Kris M G, Stover D E. Thorax. 1987;42:551–552. doi: 10.1136/thx.42.7.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandler D B, Hyde D M, Giri S N. Am J Pathol. 1983;112:170–177. [PMC free article] [PubMed] [Google Scholar]