Abstract
The aryl hydrocarbon receptor (AHR) plays a central role in 2,3,7,8-tetrachlorodibenzo-p-dioxin (dioxin) hepatotoxicity, regulation of xenobiotic metabolism, and hepatovascular development. Each of these processes appears to be dependent on binding of the AHR to dioxin- responsive elements (DREs) within the genome. The Cyp1a1 and Cyp1a2 loci represent linked genes thought to play important roles in AHR biology. In the mouse, 8 DREs are located in the 14-kb intergenic region between the Cyp1a1 and Cyp1a2 genes. Seven of these DREs, collectively known as the DRE cluster (DREC), are located 1.4 kb upstream of the Cyp1a1 transcriptional start site and 12.6 kb upstream of the Cyp1a2 start site. To investigate the role of the DREC in each aspect of AHR biology, we generated a DREC-deficient mouse model through homologous recombination. Using this mouse model, we demonstrate that the DREC controls the adaptive up-regulation of both Cyp1a1 and Cyp1a2 genes in vivo. Using selected aspects of acute hepatic injury as endpoints, we also demonstrate that DREC null mice are more sensitive to dioxin-induced hepatotoxicity than WT mice. The results of parallel toxicologic studies using individual Cyp1a1 and Cyp1a2 null mice support the observation that up-regulation of these P450s is not the cause of many aspects of dioxin hepatotoxicity. Finally, we observed normal closure of the ductus venosus (DV) in DREC null mice. Given the 100% penetrance of patent DV in Ahr null mice, these results indicate that Cyp1a1 and Cyp1a2 do not play a dominant role in AHR-mediated vascular development.
Keywords: AHR, Cyp1a1, Cyp1a2, cytochrome P450, DRE
The aryl hydrocarbon receptor (AHR) is a basic helix–loop–helix (BHLH)/Per-Arnt-Sim (PAS) protein that is activated by various xenobiotic ligands (1–4). Ligand activation results in the shedding of chaperone proteins, increased affinity for the nuclear compartment, and dimerization of the AHR to another BHLH-PAS protein, the AHR nuclear translocator (ARNT) (5). The resultant heterodimeric transcription factor then recognizes cognate genomic enhancers within the genome and up-regulates a battery of phase I and phase II xenobiotic metabolism enzymes. This pathway explains the adaptive metabolism of certain xenobiotics, because this set of regulated genes includes those for specific cytochrome P450-dependent monooxygenases (CYPs) that decrease the biological residency of many important toxicants [e.g., polycyclic aromatic hydrocarbons (PAHs)] (2–4). The AHR also is the primary mediator of the toxic response to recalcitrant environmental pollutants, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (dioxin). Receptor-mediated toxic endpoints to these ligands include tumor promotion, chloracne, thymic involution, teratogenesis, and hepatotoxicity (6). More recently, the AHR has been shown to be an essential “developmental pathway” in the vascular remodeling of the liver and eye; for example, experiments with mutant mouse models indicate that the AHR is essential for the postnatal closure of a hepatovascular portocaval shunt known as the ductus venosus (DV) (2, 7–10). For simplicity, we define the 3 aspects of AHR biology as the adaptive, toxic, and developmental arms.
Evidence from recombinant mouse models supports the idea that the upstream signaling events are similar in the adaptive, toxic, and developmental pathways; that is, each pathway is dependent on chaperone binding by the AHR, activation of the AHR, dimerization of the AHR with ARNT, and the binding of this complex to genomic enhancer elements known as “dioxin-responsive enhancers” (DREs; 5′-TNGCGTG-3′) (1, 2, 11). Using the mouse liver as a model system for each aspect of AHR signaling, we turned our attention to identifying those DRE-regulated transcripts that may explain each aspect of this biology.
The AHR-responsive genes Cyp1a1 and Cyp1a2 encode major hepatic CYPs that have long been studied for their role in the adaptive metabolism of xenobiotics (12, 13). These prototypes of the adaptive metabolic response also are logical candidates for roles in the toxic and developmental aspects of AHR biology. Our initial interest in studying these 2 DRE-regulated loci was triggered by 3 observations: (i) Cyp1a1 and Cyp1a2 are among most inducible AHR target genes in the mammalian liver, a tissue in which all aspects of AHR signaling are represented (1, 14, 15); (ii) the Cyp1a1 and Cyp1a2 loci are common biomarkers of dioxin exposure and have been used in risk assessment by various scientific teams and regulatory agencies (16–18); and (iii) various potential mechanisms have been proposed for dioxin toxicity, including the concept of the up-regulation of one or both of these P450s as a causal step (19–21).
Because Cyp1a1 and Cyp1a2 are colocalized within the mammalian genome, creating a mouse harboring null alleles at both the Cyp1a1 and Cyp1a2 loci is technically challenging. In the mouse, these loci are separated by only 14 kb of noncoding genomic DNA, with opposing transcriptional directions (Fig. 1A) (22, 23). A comparative genomic analysis of this intergenic region reveals that the 1.4-kb region upstream from the transcriptional start site of mouse Cyp1a1 is highly conserved with that of human CYP1A1 (22–24). Because of its conservation across various species, the conserved DRE cluster (DREC) is widely considered to be the primary enhancer underlying Cyp1a1 induction via the AHR (22–28). In contrast, the proximal upstream region of Cyp1a2 is not well conserved across species, and the AHR-mediated regulation of Cyp1a2 is not clearly understood (22, 29).
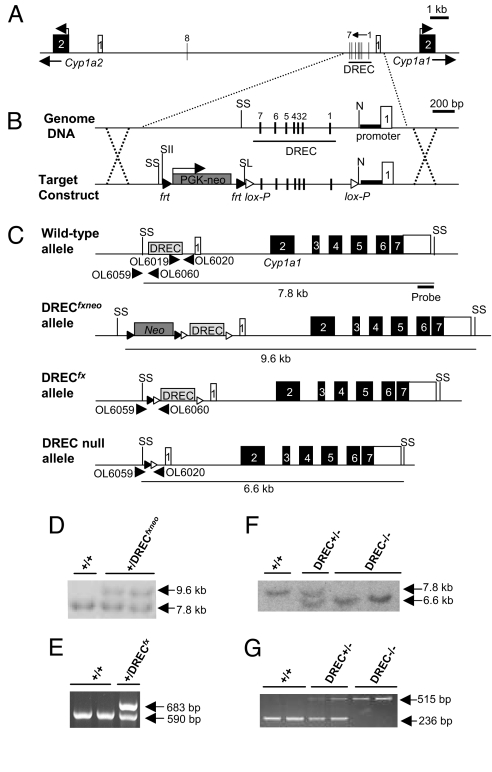
Fig. 1.
Generation of DREC conditional null mice. (A) Genomic structure of mouse Cyp1a1 and Cyp1a2 genes on chromosome 9. The Cyp1a1–Cyp1a2 junction region contains 8 consensus DRE sequences (vertical bars, DREs 1–8). The DREC containing 7 of these DREs (DREs 1–7) is located in the enhancer upstream region of the Cyp1a1 gene within −1.4 kb of the transcriptional start site. Open box, exon 1 of Cyp1a1 or Cyp1a2; closed box, exon 2 of Cyp1a1 and Cyp1a2. (B) Targeting strategy for excision of DREC. White arrowhead, lox-P sites; black arrowhead, frt sites; bold line, Cyp1a1 proximal promoter; Neo, neomycin-resistant gene cassette; N, NcoI; SL, SalI; SII, SacII; SS, SspI site. (C) Genotyping strategies for WT, DRECfxneo, DRECfx, and DREC null alleles. (D) Southern blot analysis of tail DNA from WT and DRECfxneo heterozygous alleles. The 7.6-kb and 9.4-kb SspI-digested bands correspond to WT and DRECfxneo alleles, respectively. (E) PCR genotyping for WT and DRECfx heterozygous alleles. The 590-bp and 683-bp amplified bands were generated from WT and DRECfx alleles, respectively. (F) Southern blot analysis of tail DNA from WT, DREC+/−, and DREC−/− mice. The 7.8-kb and 6.6-kb bands digested by SspI correspond to WT and DREC−/− alleles, respectively. (G) PCR genotyping for WT, DREC+/−, and DREC−/− mice. The 236-bp and 515-bp amplified bands were generated from WT and DREC−/− alleles, respectively.
Based on the conservation of the DREC across species, we hypothesized that the DREC proximal to the Cyp1a1 locus also controls AHR-mediated regulation of the Cyp1a2 locus 12.6 kb away. Recently published in vitro data supports this hypothesis (30). If this model is correct, then deletion of the DREC would generate a mouse model allowing examination of the collective action of Cyp1a1 and Cyp1a2 in both the toxicologic and developmental functions of the AHR. To test this idea, we used homologous recombination to generate mice lacking the DREC upstream of Cyp1a1. Using this model, we demonstrate that the DREC does in fact regulate both Cyp1a1 and Cyp1a2 expression in vivo and that these 2 gene products do not cause many common dioxin-induced toxic endpoints and do not play major roles in the AHR developmental pathway, as defined by closure of the DV.
Results
Generation of Conditional DREC Null Mice.
Sequence analysis of the mouse Cyp1a1 and Cyp1a2 loci revealed that the Cyp1a1 and Cyp1a2 genes are separated by 13,927 bp of noncoding genome DNA (i.e., ≈14 kb), and that the transcriptional directions oppose each other (Fig. 1A) (22). The junction region between Cyp1a1 and Cyp1a2 was found to harbor 8 consensus DRE sequences (i.e., TNGCGTG, DREs 1–8), 7 of which (DREs 1–7) are located in the −488 to −1,379 upstream region of Cyp1a1 (Fig. 1A). Collectively, these 7 proximal DREs are referred to as the DRE cluster (DREC) (22–28).
To examine the role of the DREC in AHR biology, lox-P sites were inserted at its boundaries, and the neomycin resistance gene cassette (Neo) was included with flanking frt sites for later excision, with the appropriate recombinases expressed as transgenes (Fig. 1B) (31). After homologous recombination, embryonic stem (ES) cells harboring the targeted allele were selected for the presence of Neo by treatment with G418, and homologous recombination was confirmed by Southern blot analysis (Fig. 1C). Screening of 288 ES cells yielded 5 homologous recombination events (corresponding to clones P1–5); ES clone P2 was used to generate chimeric mice. Germ-line transmission of the DRECfxneo allele was confirmed by Southern blot analysis (Fig. 1D). The chimeric mice were then bred with mice harboring a ROSA26-Flp transgene for excision of the Neo cassette (32, 33). Excision of Neo was confirmed by PCR genotyping. Mice harboring the conditional DRECfx allele were subsequently bred with mice harboring a CMV-Cre transgene for excision of the DREC (Fig. 1E) (34–36). The resulting mice carrying the DREC null allele were detected using Southern blot analysis and PCR genotyping, and these mice were backcrossed at least 4 times to the C57BL/6J background before any experiments were conducted (Fig. 1 F and G).
For all experiments, mice heterozygous for the DREC null allele were intercrossed to generate the WT (+/+), heterozygous DREC null (DREC+/−), and homozygous DREC null (DREC−/−) mice. The birth ratios of the WT, DREC+/−, and DREC−/− mice were consistent with simple Mendelian segregation of a viable null allele [i.e., WT, 29% (32/109); DREC+/−, 49% (53/109); DREC−/−, 22% (24/109)]. Outward phenotypes, including male/female ratio, fertility, body weight increase, and tissue weights (liver, lung, kidney, heart, thymus, and spleen) did not differ significantly across the 3 genotypes (data not shown).
Deletion of the DREC Region Leads to Decreased Induction of Both Cyp1a1 and Cyp1a2 by Dioxin.
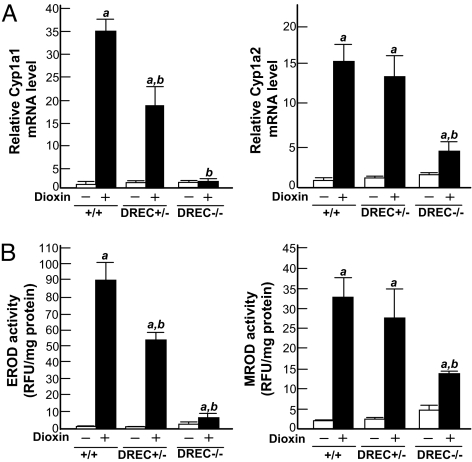
To examine the effect of DREC deletion on Cyp1a1 and Cyp1a2 induction in vivo, we measured hepatic mRNA levels by Northern blot analysis after treating the mice with DMSO or dioxin [Fig. 2A; supporting information (SI) Fig. S1]. The hepatic Cyp1a1 mRNA level was increased ≈35-fold in the WT mice given dioxin, whereas induction of Cyp1a1 was increased ≈18-fold in the DREC+/− mice and essentially eliminated in the DREC−/− mice (Fig. 2A). Although Cyp1a2 induction did not differ significantly in the WT and DREC+/− mice given dioxin, it was reduced by ≈78% in the DREC−/− mice compared with the WT mice (Fig. 2A). The disruption of Cyp1a1 and Cyp1a2 induction by DREC deletion was confirmed by measurement of corresponding protein levels (Western blot) and enzyme activities (Figs. 2B and Fig. S1). In the DREC−/− mice given dioxin, the protein levels of Cyp1a1 and Cyp1a2 in liver microsomes coincided with their mRNA levels (Fig. S1). Correspondingly, liver microsomal ethoxyresorufin-O-deethylase (EROD) (reflecting Cyp1a1 enzyme activity) and methoxyresorufin-O-deethylase (MROD) (reflecting Cyp1a2 enzyme activity) activities were significantly decreased in these mice compared with WT mice (91% vs 61%) (Fig. 2B). In the dioxin-treated DREC+/− mice, EROD activity was 34% lower than that in WT mice, whereas MROD activity did not differ significantly (Fig. 2B).
Fig. 2.
Loss of Cyp1a1 and Cyp1a2 induction in dioxin-treated DREC−/− mice. Total RNAs were isolated from the livers of WT, DREC+/−, and DREC−/− mice. The mice were given a single injection of DMSO vehicle alone or 64 μg/kg of dioxin in DMSO and were killed 5 days later. (A) Relative fold induction of Cyp1a1 and Cyp1a2 mRNA by dioxin. The mRNA levels were quantified by the Molecular Dynamics Storm system, and the measured Cyp1a1 and Cyp1a2 mRNA levels were normalized to the GAPDH mRNA level. Results are expressed as relative mRNA level compared with DMSO-treated WT mice. (B) EROD activity (indicating Cyp1a1 enzyme activity) and MROD activity (indicating Cyp1a2 activity), expressed as relative fluorescence units per mg of microsome protein. Each group contained more than 4 mice. Open bars, DMSO treatment; closed bars, dioxin treatment. Error bars represent SE. a, Significantly different relative to the DMSO-treated WT mice (P < .05). b, Significantly different relative to the dioxin-treated WT mice (P < .05).
Normal Hepatic Vascular Development in DREC−/− Mice.
To explore whether the DREC plays a role in DV closure, we perfused the portal vein with Trypan Blue dye and monitored flow through the parenchyma (closed DV) or directly through to the vena cava (patent DV) (10). As shown in Table 1, the frequency of patent DV in DREC null allele mice was indistinguishable from that in WT littermates. Ahr null mice were used as positive controls (2). All of the Ahr null mice exhibited a patent DV, whereas no WT or DREC−/− mice did so.
Table 1.
Rate of DV patency in WT, DREC−/−, and AHR−/− mice
| Genotype | Patent DV (%) | n |
|---|---|---|
| WT | 0 | 0/10 |
| DREC−/− | 0 | 0/9 |
| AHR−/− | 100 | 5/5 |
Impact of DREC Deletion on Dioxin-Induced Toxicity.
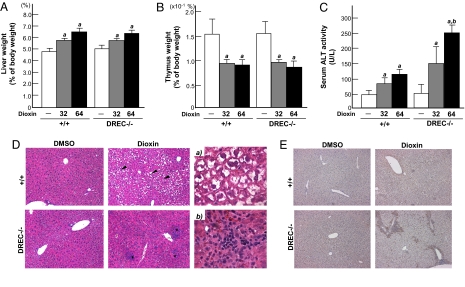
To investigate whether deletion of the DREC affects dioxin-induced toxicity, we compared several classic endpoints (7, 8, 20, 21, 37, 38). The dioxin-treated WT and DREC−/− mice displayed similar increases in liver weight (dioxin dose of 32 μg/kg, 113% ± 9% and 110% ± 4%; dose of 64 μg/kg, 132% ± 11% and 128% ± 7%) and decreases in thymus weight (dioxin dose of 32 μg/kg, 64% ± 13% and 65% ± 7%; dose of 64 μg/kg, 60% ± 11% and 52% ± 10%) (Fig. 3 A and B). The 2 groups exhibited no significant differences in lung, heart, kidney, spleen, or total body weights (data not shown).
Fig. 3.
Impact of DREC deletion on dioxin-induced toxicity. (A–C) Mice were given a single injection of DMSO or 32 or 64 μg/kg of dioxin and were killed 5 days later. Liver weight (A) and thymus weight (B) expressed as percentage of total body weight. (C) Serum ALT activity. Each group contains more than 4 mice. Open bars, DMSO treatment; gray closed bars, 32 μg/kg dioxin treatment; black closed bars, 64 μg/kg dioxin treatment. Error bars represent SE. a, Significantly different relative to the DMSO-treated WT mice (P < .05). b, Significantly different relative to the dioxin-treated WT mice (P < .05). (D and E) Mice were given a single injection of DMSO or 64 μg/kg of dioxin and were killed 5 days later. Liver sections were stained with H&E and immunostained with F4/80 antibody. (D) H&E. The black arrowhead indicates the region of hydropic degeneration; * indicates focal inflammation. (Magnification: ×100.) (a) WT given dioxin. (Magnification: ×400.) (b) DREC−/− given dioxin. (Magnification: ×400.) (E) F4/80. The brown spots indicate F4/80-positive cells. (Magnification: ×100.)
Hepatic injury is a characteristic dioxin-induced toxic endpoint (20, 21, 37, 38). Serum alanine-aminotransferase (ALT) activity, a sensitive marker of hepatic injury, was increased by 2.0-fold in the WT mice given a dioxin dose of 32 μg/kg and by 2.8-fold in those given a dioxin dose of 64 μg/kg (Fig. 3C). The DREC−/− mice given 64 μg/kg of dioxin exhibited significantly increased (by 2.5-fold) ALT activity compared with the similarly treated WT mice (Fig. 3C). As an independent measure of hepatotoxicity, we analyzed liver sections using H&E staining for general pathology, Oil Red O staining for lipid accumulation, and F4/80 immunostaining to confirm macrophage infiltration (39) (Fig. 3 D and E and Figs. S2–S4). The H&E staining revealed that dioxin-treated DREC−/− mice displayed areas of focal inflammation consisting of macrophages, lymphocytes, and necrotic cells, whereas the dioxin-treated WT mice had fewer of these infiltrates and the vehicle controls had no infiltrates (Fig. 3D and Fig. S2). In liver sections of dioxin-treated DREC null mice, F4/80-positive cells were prominent, whereas relatively few F4/80-positive cells were observed in liver sections of the dioxin-treated WT mice (Fig. 3E and Fig. S3). In these latter mice, H&E-stained liver sections exhibited severe hydropic degeneration in zone 2 (Fig. 3D and Fig. S2). In contrast to effects on hepatic inflammation, the dioxin-treated DREC−/− mice exhibited little hydropic degeneration (Fig. 3D and Fig. S2). Finally, in the dioxin-treated WT and DREC−/− mice, liver sections stained with Oil Red O displayed similar levels of lipid droplets in zones 1 and 2 (Fig. S4).
Individual Cyp1a1 and Cyp1a2 Null Alleles also Protect Against Dioxin-Induced Acute Hepatic Injury.
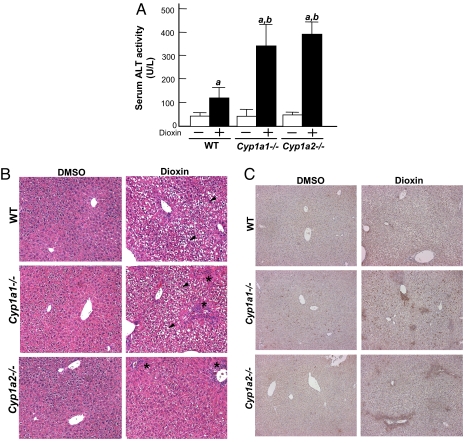
To support the concept that enhanced dioxin-induced hepatic injury in DREC−/− mice is due to decreased Cyp1a expression, we repeated the experiments with both Cyp1a1−/− and Cyp1a2−/− mice generated by another laboratory (40, 41). We found dioxin-responsive increases in serum ALT activity by 3.0-fold in the Cyp1a1−/− mice and by 3.3-fold in the Cyp1a2−/− mice compared with the WT (C57BL/6J) mice given dioxin (Fig. 4A). In addition, histological analysis of Cyp1a1−/− and Cyp1a2−/− mouse livers revealed increased numbers of focal inflammatory cells compared with WT mice in response to dioxin (Fig. 4B and Fig. S5). The numbers of F4/80-positive cells also were increased in the livers of both dioxin-treated Cyp1a1−/− and Cyp1a2−/− mice compared with WT mice (Fig. 4C and Fig. S6). Finally, the livers of the Cyp1a2−/− mice exhibited little hydropic degeneration, whereas the livers of the Cyp1a1−/− mice showed similar levels of hydropic degeneration in zone 2 as those of the WT mice (Fig. 4B and Fig. S5).
Fig. 4.
Dioxin-induced hepatic injury in Cyp1a1−/− and Cyp1a2−/− mice. The mice were given a single injection of DMSO or 64 μg/kg of dioxin and were killed 5 days later. (A) Serum ALT activity. Each group contains more than 5 mice. (B and C) Liver sections stained with H&E and immunostained with F4/80 antibody. (B) H&E. The black arrowhead indicates the region of hydropic degeneration; * indicates focal inflammation. (Magnification: ×100.) (C) F4/80. The brown spots indicate F4/80-positive cells. (Magnification: ×100.)
Discussion
The AHR is a central mediator of adaptive xenobiotic metabolism, dioxin-induced toxicity, and vascular development (1–4). Our previous experiments with recombinant mouse models have led us to 3 conclusions: (i) Each of these pathways requires similar upstream signaling steps (i.e., nuclear translocation of the AHR, dimerization of the AHR with ARNT, and binding of the dimeric complex to DREs) (2, 5); (ii) we have little understanding of the identity of the DRE-driven genes important for dioxin toxicity and vascular development; and (iii) the mouse liver provides a tissue in which each of the 3 biological processes can be investigated.
To identify essential genes in dioxin toxicity and vascular development, we initially focused on the roles of the Cyp1a1 and Cyp1a2 loci. Our experimental design, using deletion of the DREC cluster proximal to the Cyp1a1 promoter, arose from the initial hypothesis that this region also regulates Cyp1a2. We postulated that generation of a DREC null mouse would provide in vivo validation of this regulatory model and also provide a Cyp1a1/Cyp1a2 double-null mouse model for toxicology and developmental studies. Toward this end, we generated mice lacking the DREC using homologous recombination and the Cre-lox/Flp-frt system (32–36).
Using the resulting DREC null model, we generated in vivo evidence that the DREC regulates both Cyp1a1 and Cyp1a2 promoters (22, 29, 42). These data are consistent with the concept that in the liver, the DREC is the only AHR regulatory element for Cyp1a1 induction by dioxin and the primary regulatory element for the Cyp1a2 locus that is 12.4 kb away. Identifying the AHR regulatory element for Cyp1a2 has proven difficult, because the proximal enhancer region of Cyp1a2 does not contain obvious DRE sequences or other putative AHR-responsive elements (22, 29). While our work was in progress, in vitro evidence was published supporting a role of the DREC in human CYP1A1 and CYP1A2 regulation (30). The remaining dioxin-induced regulation of Cyp1a2 in DREC null mice suggests that an additional cryptic DRE or an as-yet unidentified AHR-dependent regulatory element plays a secondary role (43). We propose that this AHR regulatory element might be the eighth DRE (DRE 8) identified in the Cyp1a1–Cyp1a2 intergenic region (4.2 kb upstream from Cyp1a2) (Fig. 1A).
We have reported previously that disruption of the Ahr, Arnt, or Ara9 loci leads to abnormal hepatic vascular development, manifested as a patent DV, a fetal hepatic vascular structure that shunts blood flow from the umbilical vein to the inferior vena cava (7–10, 44). Conditional alleles of the Ahr have provided evidence suggesting that the AHR signaling in endothelial cells is essential for DV closure (37). Using this DREC null model, we explored whether the Cyp1a1 and/or Cyp1a2 loci might play important roles in AHR-mediated vascular development (Table 1). Our finding of normal DV closure in the DREC null mice, as well as in individual Cyp1a1 and Cyp1a2 null mice (data not shown), is consistent with the idea that vascular development must be regulated by an AHR-regulated pathway independent of Cyp1a1 and Cyp1a2.
Our most unexpected finding was that Cyp1a1 and Cyp1a2 protect against many aspects of acute dioxin hepatotoxicity. Initially, we predicted that if either Cyp1a1 or Cyp1a2 induction were an underlying cause of dioxin-induced hepatotoxicity, then deletion of DREC should lead to decreases in those endpoints. This hypothesis is supported by previously reported findings indicating that disruption of Cyp1a1 or Cyp1a2 led to decreased dioxin-induced hepatic injury in subchronic toxicity studies (20, 21). To our surprise, our data are consistent with the concept that the Cyp1a loci are not causal, but actually are protective for the basic aspects of dioxin-induced acute hepatocellular damage and hepatic inflammation. This conclusion is based on the observation that the loss of Cyp1a1 and/or Cyp1a2 induction through the DREC corresponds to exaggerated dioxin-induced hepatic injury, as indicated by increased serum ALT activity and histological evidence of focal inflammation. Dioxin-induced hepatic inflammation is of particular interest, because this endpoint is characteristic of hepatic injury that may be related to dioxin-induced chronic toxicity, such as hepatic cancer (6). In this regard, we have recently reported that dioxin-induced hepatic inflammation is mediated by inflammatory cytokine pathways, such as TNF, IL-1, and IL-1–like cytokines (39).
In contrast to dioxin-induced hepatic inflammation, induction of Cyp1a2, but not of Cyp1a1, through the DREC appears to play a causal role in dioxin-induced hepatocellular hydropic degeneration. Interestingly, this dioxin-induced hepatocellular hydropic degeneration is not linked to elevated serum ALT activity or hepatic inflammation. We predict that it is caused by an overload of intracellular water, but is not related to an accumulation of fatty acids (45). Exposure to dioxin has been reported to change the proliferation of smooth endoplasmic reticulum (46, 47), which could lead to influx of H2O, Na+, and Ca2+ into the hepatocytes. Consistent with results from other laboratories is the observation that the Cyp1a1 and Cyp1a2 loci play little, if any, role in dioxin-induced thymic involution and hepatomegaly (20, 21).
In conclusion, using the DREC deletion mouse model, we have demonstrated that the DREC plays an important role in the AHR-mediated adaptive regulation of both Cyp1a1 and Cyp1a2. In terms of the adaptive regulation of Cyp1a1 and Cyp1a2, the DREC acts as a common, long-range enhancer of both genes in vivo. This regulatory mechanism can help explain the broad spectrum of gene expression induced by dioxin exposure. With respect to dioxin-induced hepatotoxicity, we have demonstrated that Cyp1a1 and Cyp1a2 induction through the DREC is a protective response against acute dioxin-induced hepatocellular necrosis and hepatic inflammation. In contrast, Cyp1a2 induction through the DREC appears to play a causal role in dioxin-induced hepatocellular hydropic degeneration. But hydropic degeneration apparently is not linked to the more important inflammatory and necrotic changes induced by dioxin. Together, our findings suggest that additional DRE-regulated genes are important in dioxin toxicology and AHR-mediated development and that these promoters may lie great distances from DRE clusters.
Materials and Methods
Generation of Conditional DREC Null Mice.
The Cyp1a1 gene, including its upstream region, was isolated from the bacterial artificial chromosome clone 17278 (BAC17278) derived from a BAC library constructed from mouse ES-129/Sv DNA (Genome Systems) (22). The 1,149-bp DNA fragment (−254 to –1,403 upstream of Cyp1a1) containing DREs 1–7 (the DREC) was inserted into the plasmid PL1169, which contains an adjacent lox-P site and Neo flanked by frt sites, as described previously (Fig. 1B) (31). The first lox-P site was inserted into the 5′-end of the Cyp1a1 promoter using the NcoI site located 80 bp upstream of the promoter, and the second lox-P site was inserted 24 bp upstream of DRE 7 from the transcriptional start site of Cyp1a1. The frt sites–Neo construct was inserted upstream of the second lox-P site using the SacII and SalI sites in PL1169 (Fig. 1B). The targeting construct was then electroporated into GS1 ES cells (Genome Systems) as described previously (8). The neomycin-resistant ES clones were selected by G418 treatment, and Southern blot analysis was used to detect homologous recombinations using SspI-digested genomic DNA and a 645-bp DNA probe corresponding to bases +4451 to +5096 of the Cyp1a1 gene (Fig. 1C). Chimeric mice were generated by microinjection of the targeted ES clones into C57BL/6J blastocysts (9). The resultant chimeric mice were backcrossed to C57BL/6J mice for determination of germ-line transmission, using coat color as a phenotype. Germ-line transmission was confirmed by SspI-digested tail DNA using the same probe described earlier (Fig. 1D).
To excise the Neo cassette, the founder mice (DRECfxneo) were crossed to mice harboring the ROSA26-FLP allele [strain name, Gt(ROSA)26Sortm1(FLP1)Dym; Jackson Laboratory], which express FLP recombinase constitutively (32, 33). The excision of the Neo cassette was confirmed by PCR genotyping using forward primer OL6059 (5′-ATCACCTGTTTCCCCCATAGC-3′) and reverse primer OL6060 (5′-CGTGGGTGAGGCTCTTAAAGG-3′). PCR was carried out for 28 cycles (95 °C for 30 s, 60 °C for 45 s, and 72 °C for 45 s) in a reaction mixture containing 2.5 U of Taq polymerase (Promega), 50 mM KCl, 10 mM Tris-HCl (pH 9.0 at 25 °C), 1.5 mM MgCl2, 1% Triton X-100, 0.2 mM dNTPs, and 0.2 μM of each primer. A 590-bp PCR product was amplified from the WT allele, and a 683-bp PCR product was amplified from the conditional DREC allele (DRECfx) lacking the Neo cassette (Fig. 1E).
The resultant heterozygous DRECfx mice were crossed with cytomegalovirus (CMV)-Cre [strain name, B6.C-Tg(CMV-cre)1Cgn/J], in which the P1 bacteriophage cyclization recombination recombinase (Cre) was expressed ubiquitously. This crossing produced mice with the DREC null allele (34–36). Excision of the DREC was determined by Southern blot analysis of SspI-digested tail DNA using the 645-bp Cyp1a1 flanking region probe described earlier (Fig. 1F) and by PCR using the forward primer for the WT allele (OL6019; 5′-AATGGAGGCCCCAGTACTTAC-3′), the forward primer for DREC null allele (OL6059; 5′-ATCACCTGTTTCCCCCATAGC-3′), and the common reverse primer (OL6020; 5′-ACAGCCTTAGGGAGTGCTCTA-3′). These primers amplified a 236-bp band from the WT allele and a 515-bp band from the DREC null allele (Fig. 1G). PCR was performed under the same conditions described above. Mice carrying the DREC null allele were backcrossed to C57BL6/J mice for 4 generations (N4) before any experiments were conducted. These N4 backcrossed heterozygous DREC null (+/−) mice were interbred to generate offspring that were used in evaluation of these phenotypes and toxicity studies by dioxin.
Animals.
The mice were housed in a selective pathogen-free facility on corncob bedding with food and water ad libitum following the protocol established by University of Wisconsin Medical School's Animal Care and Use Committee. The Cyp1a1 and Cyp1a2 null mice were generous gifts from Dr. D.W. Nebert (40, 41). Before the experiments, these Cyp1a1 and Cyp1a2 null mice, as well as the CMV-Cre and ROSA26-FLP mice, were backcrossed to C57BL/6J mice for > 10 generations in our facility. All strains of mice were selected for homozygosity for the AHRb1 allele (48). In experiments using Cyp1a1 and Cyp1a2 null mice, C57BL/6J mice were used as WT controls.
Treatment and Toxicity Studies.
Eight-week-old male mice were injected with a single i.p. dose of dioxin (either 32 or 64 μg/kg total body weight) dissolved in DMSO or of DMSO alone as vehicle control. Five days after the injection, the mice were killed by CO2 euthanasia. Serum was prepared, and the ALT activity was measured at the University of Wisconsin's Veterinary Medical Teaching Hospital. Liver, heart, lung, spleen, thymus, and kidney were removed and weighed. The liver was used for preparation of total RNA and microsomes. At the same time, slices of the left liver lobe were fixed in 10% formalin in PBS or embedded in frozen section compound (Surgipath). Paraffin-embedded sections were stained with H&E and immunostained with F4/80 antibody, and frozen sections were stained with Oil Red O as described previously (39, 49).
Assessment of DV Status.
DV patency was confirmed by perfusion of the liver with 0.4% Trypan Blue Dye as described previously (10). The AHR null mice served as positive controls for DV patency (2, 10).
Northern Blot Analysis.
Total RNA was isolated from the liver of each mouse using a Qiagen RNeasy kit. Ten μg of total RNA was subjected to electrophoresis in a 0.8% agarose gel containing 18% formaldehyde and then transferred to Hybond-N + membrane (GE Healthcare Bioscience). The membrane was hybridized with 32P-labeled cDNA probes for detection of Cyp1a1, Cyp1a2, and GAPDH mRNA as described previously (37). Quantitative determination of gene expression was done using the Molecular Dynamics Storm system (GE Healthcare Bioscience).
Protein Assays.
Liver microsomes were prepared as described previously (8). For the Western blot analysis, 50 μg of microsomal protein was loaded onto 7.5% SDS/PAGE gels, electrophoresed, and transferred to Immobilon-P transfer membrane (Millipore). The membrane was incubated with anti-rat Cyp1a1 and Cyp1a2 antibodies (Human Biologics) as described previously (2). The EROD and MROD activities were measured by spectrofluorimetric analysis (excitation, 510 nm; emission, 590 nm) (8, 50). All data were normalized to protein concentrations determined using the BCA protein assay kit (Thermo).
Statistical Analysis.
All statistical data are presented as mean ± SE. Intergroup comparisons were performed using one-way ANOVA (37). Differences among groups were deemed statistically significant when the P value was < .05.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants R01-ES-013566–01, P01-CA-22484–27, and P30-CA-014520–29. We thank Joe Warren for the microinjections; Ruth Sullivan for her pathological expertise; Edward Glover, Jacqueline A. Walisser, Brian E. McIntosh, and Linh Nguyen for their supporting experiments; and Anna L. Shen and Emily A. Stevens for their manuscript review.
Footnotes
Conflict of interest statement: C.A.B. has served as a scientific consultant to Dow Chemical Co. on issues related to dioxin toxicity.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809613106/DCSupplemental.
References
- 1.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci U S A. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitlock JP, Jr, et al. Induction of drug-metabolizing enzymes by dioxin. Drug Metab Rev. 1997;29:1107–1127. doi: 10.3109/03602539709002245. [DOI] [PubMed] [Google Scholar]
- 4.Nebert DW, Gonzalez FJ. P450 genes: Structure, evolution, and regulation. Annu Rev Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [DOI] [PubMed] [Google Scholar]
- 5.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: Sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 6.Pohjanvirta R, Tuomisto J. Short-term toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in laboratory animals: Effects, mechanisms, and animal models. Pharmacol Rev. 1994;46:483–549. [PubMed] [Google Scholar]
- 7.Lahvis GP, et al. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor–deficient mice. Proc Natl Acad Sci U S A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunger MK, et al. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 9.Walisser JA, Bunger MK, Glover E, Bradfield CA. Gestational exposure of Ahr and Arnt hypomorphs to dioxin rescues vascular development. Proc Natl Acad Sci U S A. 2004;101:16677–16682. doi: 10.1073/pnas.0404379101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harstad EB, Guite CA, Thomae TL, Bradfield CA. Liver deformation in Ahr-null mice: Evidence for aberrant hepatic perfusion in early development. Mol Pharmacol. 2006;69:1534–1541. doi: 10.1124/mol.105.020107. [DOI] [PubMed] [Google Scholar]
- 11.Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson DR, et al. P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–43. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Yang SK. Stereoselectivity of cytochrome P-450 isozymes and epoxide hydrolase in the metabolism of polycyclic aromatic hydrocarbons. Biochem Pharmacol. 1988;37:61–70. doi: 10.1016/0006-2952(88)90755-1. [DOI] [PubMed] [Google Scholar]
- 14.Hayes KR, et al. EDGE: A centralized resource for the comparison, analysis, and distribution of toxicogenomic information. Mol Pharmacol. 2005;67:1360–1368. doi: 10.1124/mol.104.009175. [DOI] [PubMed] [Google Scholar]
- 15.Hayes KR, et al. Hepatic transcriptional networks induced by exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chem Res Toxicol. 2007;20:1573–1581. doi: 10.1021/tx7003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whysner J, Williams GM. 2,3,7,8-Tetrachlorodibenzo-p-dioxin mechanistic data and risk assessment: Gene regulation, cytotoxicity, enhanced cell proliferation, and tumor promotion. Pharmacol Ther. 1996;71:193–223. doi: 10.1016/0163-7258(96)00068-x. [DOI] [PubMed] [Google Scholar]
- 17.Andersen ME, Barton HA. The use of biochemical and molecular parameters to estimate dose-response relationships at low levels of exposure. Environ Health Perspect. 1998;106:349–355. doi: 10.1289/ehp.98106s1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz M, Appel KE. Carcinogenic risks of dioxin: Mechanistic considerations. Regul Toxicol Pharmacol. 2005;43:19–34. doi: 10.1016/j.yrtph.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Ma Q, Lu AY. CYP1A induction and human risk assessment: An evolving tale of in vitro and in vivo studies. Drug Metab Dispos. 2007;35:1009–1016. doi: 10.1124/dmd.107.015826. [DOI] [PubMed] [Google Scholar]
- 20.Uno S, et al. Cyp1a1(-/-) male mice: Protection against high-dose TCDD-induced lethality and wasting syndrome, and resistance to intrahepatocyte lipid accumulation and uroporphyria. Toxicol Appl Pharmacol. 2004;196:410–421. doi: 10.1016/j.taap.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Smith AG, et al. Protection of the Cyp1a2(-/-) null mouse against uroporphyria and hepatic injury following exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 2001;173:89–98. doi: 10.1006/taap.2001.9167. [DOI] [PubMed] [Google Scholar]
- 22.Nukaya M, Bradfield CA. Conserved genomic structure of the Cyp1a1 and Cyp1a2 loci and their dioxin-responsive elements cluster. Biochem Pharmacol. 2009;77:654–659. doi: 10.1016/j.bcp.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corchero J, Pimprale S, Kimura S, Gonzalez FJ. Organization of the CYP1A cluster on human chromosome 15: Implications for gene regulation. Pharmacogenetics. 2001;11:1–6. doi: 10.1097/00008571-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 25.Kress S, Reichert J, Schwarz M. Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. Eur J Biochem. 1998;258:803–812. doi: 10.1046/j.1432-1327.1998.2580803.x. [DOI] [PubMed] [Google Scholar]
- 26.Fujii-Kuriyama Y, Imataka H, Sogawa K, Yasumoto K, Kikuchi Y. Regulation of CYP1A1 expression. FASEB J. 1992;6:706–710. doi: 10.1096/fasebj.6.2.1537460. [DOI] [PubMed] [Google Scholar]
- 27.Strom DK, Postlind H, Tukey RH. Characterization of the rabbit CYP1A1 and CYP1A2 genes: Developmental and dioxin-inducible expression of rabbit liver P4501A1 and P4501A2. Arch Biochem Biophys. 1992;294:707–716. doi: 10.1016/0003-9861(92)90745-i. [DOI] [PubMed] [Google Scholar]
- 28.Zeruth G, Pollenz RS. Isolation and characterization of a dioxin-inducible CYP1A1 promoter/enhancer region from zebrafish (Danio rerio) Zebrafish. 2005;2:197–210. doi: 10.1089/zeb.2005.2.197. [DOI] [PubMed] [Google Scholar]
- 29.Owens RA, Nebert DW. Expression of the chloramphenicol acetyltransferase (CAT) reporter gene by the murine Cyp1a-2 (cytochrome P3(450)) promoter in hepatoma cell cultures. Biochem Biophys Res Commun. 1990;172:1109–1115. doi: 10.1016/0006-291x(90)91561-6. [DOI] [PubMed] [Google Scholar]
- 30.Ueda R, et al. A common regulatory region functions bidirectionally in transcriptional activation of the human CYP1A1 and CYP1A2 genes. Mol Pharmacol. 2006;69:1924–1930. doi: 10.1124/mol.105.021220. [DOI] [PubMed] [Google Scholar]
- 31.Lin BC, et al. Deletion of the aryl hydrocarbon receptor–associated protein 9 leads to cardiac malformation and embryonic lethality. J Biol Chem. 2007;282:35924–35932. doi: 10.1074/jbc.M705471200. [DOI] [PubMed] [Google Scholar]
- 32.Dymecki S. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- 34.Hoess R, Wierzbicki A, Abremski K. The role of the spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sternberg N, Hoess R. The molecular genetics of bacteriophage P1. Annu Rev Genet. 1983;17:123–154. doi: 10.1146/annurev.ge.17.120183.001011. [DOI] [PubMed] [Google Scholar]
- 36.Schwenk F, Baron U, Rajewsky KA. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor–dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci U S A. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walisser JA, Bunger MK, Glover E, Harstad EB, Bradfield CA. Patent ductus venosus and dioxin resistance in mice harboring a hypomorphic Arnt allele. J Biol Chem. 2004;279:16326–16331. doi: 10.1074/jbc.M400784200. [DOI] [PubMed] [Google Scholar]
- 39.Pande K, Moran SM, Bradfield CA. Aspects of dioxin toxicity are mediated by interleukin 1–like cytokines. Mol Pharmacol. 2005;67:1393–1398. doi: 10.1124/mol.105.010983. [DOI] [PubMed] [Google Scholar]
- 40.Liang HC, et al. Cyp1a2(−/−) null mutant mice develop normally but show deficient drug metabolism. Proc Natl Acad Sci U S A. 1996;93:1671–1676. doi: 10.1073/pnas.93.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalton TP, et al. Targeted knockout of Cyp1a1 gene does not alter hepatic constitutive expression of other genes in the mouse [Ah] battery. Biochem Biophys Res Commun. 2000;267:184–189. doi: 10.1006/bbrc.1999.1913. [DOI] [PubMed] [Google Scholar]
- 42.Jones KW, Whitlock JP., Jr Functional analysis of the transcriptional promoter for the CYP1A1 gene. Mol Cell Biol. 1990;10:5098–5105. doi: 10.1128/mcb.10.10.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quattrochi LC, Vu T, Tukey RH. The human CYP1A2 gene and induction by 3-methylcholanthrene: A region of DNA that supports AH receptor binding and promoter-specific induction. J Biol Chem. 1994;269:6949–6954. [PubMed] [Google Scholar]
- 44.Lin BC, Nguyen LP, Walisser JA, Bradfield CA. A hypomorphic allele of aryl hydrocarbon receptor-associated protein-9 produces a phenocopy of the AHR-null mouse. Mol Pharmacol. 2008;74:1367–1371. doi: 10.1124/mol.108.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crawford JM. In: Robbins and Cotran Pathologic Basis of Disease. 7th Ed. Kumar V, Abbas AK, Fausto N, editors. Philadelphia, PA: Saunders; 2005. pp. 880–882. [Google Scholar]
- 46.Lucier GW, et al. TCDD-induced changes in rat liver microsomal enzymes. Environ Health Perspect. 1973;5:199–209. doi: 10.1289/ehp.7305199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fowler BA, Lucier GW, Brown HW, McDaniel OS. Ultrastructural changes in rat liver cells following a single oral dose of TCDD. Environ Health Perspect. 1973;5:141–148. doi: 10.1289/ehp.7305141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas RS, Penn SG, Holden K, Bradfield CA, Rank DR. Sequence variation and phylogenetic history of the mouse Ahr gene. Pharmacogenetics. 2002;12:151–163. doi: 10.1097/00008571-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 49.Bunger MK, et al. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol Sci. 2008;106:83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamm JT, Ross DG, Richardson VM, Diliberto JJ, Birnbaum LS. Methoxyresorufin: An inappropriate substrate for CYP1A2 in the mouse. Biochem Pharmacol. 1998;56:1657–1660. doi: 10.1016/s0006-2952(98)00241-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.