Abstract
The widely accepted oxidative stress theory of aging postulates that aging results from accumulation of oxidative damage. Surprisingly, data from the longest-living rodent known, naked mole-rats [MRs; mass 35 g; maximum lifespan (MLSP) > 28.3 years], when compared with mice (MLSP 3.5 years) exhibit higher levels of lipid peroxidation, protein carbonylation, and DNA oxidative damage even at a young age. We hypothesize that age-related changes in protein structural stability, oxidation, and degradation are abrogated over the lifespan of the MR. We performed a comprehensive study of oxidation states of protein cysteines [both reversible (sulfenic, disulfide) and indirectly irreversible (sulfinic/sulfonic acids)] in liver from young and old C57BL/6 mice (6 and 28 months) and MRs (2 and >24 years). Furthermore, we compared interspecific differences in urea-induced protein unfolding and ubiquitination and proteasomal activity. Compared with data from young mice, young MRs have 1.6 times as much free protein thiol groups and similar amounts of reversible oxidative damage to cysteine. In addition, they show less urea-induced protein unfolding, less protein ubiquitination, and higher proteasome activity. Mice show a significant age-related increase in cysteine oxidation and higher levels of ubiquitination. In contrast, none of these parameters were significantly altered over 2 decades in MRs. Clearly MRs have markedly attenuated age-related accrual of oxidation damage to thiol groups and age-associated up-regulation of homeostatic proteolytic activity. These pivotal mechanistic interspecies differences may contribute to the divergent aging profiles and strongly implicate maintenance of protein stability and integrity in successful aging.
Keywords: cysteine oxidation, Heterocephalus glaber, mechanisms of aging, proteasome activity, protein homeostasis
The naked mole-rat (MR; Bathyergidae; Heterocephalus glaber), is the longest-living rodent known with a maximum lifespan (MLS) of >28.3 years (1). Evolving in a protected underground milieu, extrinsic mortality is low and this mouse-sized (35 g) rodent lives 8 times longer than laboratory mice. MRs have a similar longevity quotient [LQ = 5; i.e., ratio of observed MLS to that predicted by body mass (2)] to that of humans, another very long-living mammal. MRs show many signs of attenuated aging (3); they exhibit no age-related changes in body composition, physiology, and molecular function from 2 to >20 years (4–6). Old animals (>24 years; >85% MLS), nevertheless, can be differentiated on sight in that they are less active and have a thin translucent parchment-like skin (1). These data support the premise that the MR is an excellent animal model for studying molecular and biochemical mechanisms involved in successful slow aging and sustained health span (7).
The oxidative stress theory of aging predicts that differential rates of aging among species may be caused by inherent differences in oxidative damage accrual (8). Although widely accepted (9–12), there are a growing number of exceptions to this theory, often contingent on the specific species, strain, and/or tissue under investigation (7, 13–15). Data from long-lived species (such as birds, bats, and MRs), and transgenic mice with altered expression of antioxidants, commonly suggest that oxidative damage is not directly correlated with MLS (15, 16). The MR is one of the notable exceptions to the oxidative stress theory of aging (7, 14, 17, 18). It has similar levels of reactive oxygen species (17, 18) and antioxidant defenses (19) when compared with shorter-living rodents; however, even at a young age it exhibits higher levels of lipid peroxidation, protein carbonylation, and DNA oxidative damage than do mice (14). These measures of oxidative stress do not change with age (20). In addition, MR cells appear to be inherently protected from most types of cytotoxic insults, maintaining greater viability than other rodents (17, 21). These data suggest that the oxidative damage, per se, may not effect longevity, but rather resilience to damage and the mechanisms facilitating this may be a far more important determinant of aging.
Proteins are one of the prime targets for oxidative damage (22), and cysteine residues are particularly sensitive to oxidation because the thiol group in cysteine can be oxidized to both reversible [sulfenic acid (SOH), disulfide bond formation (S—S)] and irreversible oxidative states [sulfinic (SO2H) and sulfonic acids (SO3H)] (23, 24). Cysteine residues are important physiologically because they are often found at catalytic and regulatory sites in proteins and enzymes (24). The thiol groups in cysteines are strongly buffered against oxidation by keeping the internal cellular environment in a relatively reduced state (25, 26). However, these intracellular conditions may change with age. Redox ratios in plasma reportedly decrease with age in humans (27, 28), increasing vulnerability of thiol groups to oxidative damage.
Chronic oxidative stress may lead to protein misfolding. The intricate tertiary-folded structure of proteins is maintained with the assistance of various heat shock proteins and molecular chaperones (29). Impairment of functional structure can accelerate formation of toxic protein oligomers or aggregates that contribute to pathologies commonly observed in neurodegenerative diseases such as Alzheimer's, Parkinson's, and Huntington diseases (30). Thus, we hypothesize that attenuated accumulation of oxidized proteins and maintenance of protein structural stability may be important determinants for slow aging.
In this article, we test this hypothesis by determining whether MRs, when compared with shorter-lived mice, (i) exhibit attenuated age-related changes in protein oxidation (e.g., cysteine oxidation), (ii) have proteins that are more resistant to urea-induced unfolding, and (iii) maintain protein homeostasis as measured by ubiquitination and proteasome activity with age. We report that when compared with mice MRs show (i) no age-related changes in cysteine modification (from reversible to irreversible oxidation state), (ii) resistance to protein unfolding, and (iii) attenuated accumulation of ubiquitinated proteins and sustained proteasomal function during aging. Our comparative findings elucidate a key mechanistic difference that may contribute to disparate longevity among species and strongly implicate maintenance of protein stability in successful aging.
Results
Attenuated Age-Related Changes in Cysteine Oxidation in MRs.
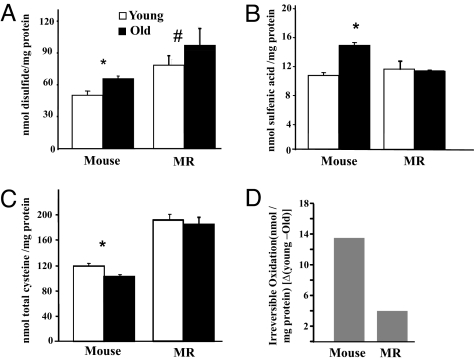
The thiol group in cysteine is sensitive to oxidation and exhibits various oxidation states, including reversible and irreversible states. We have developed 3 distinct fluorescence-based assays to quantify the various states of cysteine oxidation; 2 measure reversible states (disulfide and sulfenic acid) (31) and the third measures total cysteine and indirectly indicates the levels of irreversible oxidation [sulfinic/sulfonic/others (23, 32)] that we have used in liver tissues of young and old MRs and mice. The disulfide content detected in young MRs tends (P > 0.07) to be higher than that found in young mice (Fig. 1A). Disulfide content in mouse liver proteins is elevated (≈28%; P < 0.05) with age, whereas age-associated accumulation is not evident in MR samples.
Fig. 1.
Age-related changes in cysteine oxidation in cytosolic liver homogenates from young (open bars) and old (solid bars) mice and MRs. Data are the means of 8 (mice) and 10 (MRs) ± SEM. The * denotes values that are significantly (P ≤ 0.05) different from young mice as analyzed by nonparametric ANOVA. (A) Disulfide bond content increases 1.3-fold in mice with age, but is unchanged (#, P = 0.07) with age in MRs. (B) Sulfenic acid levels increase 1.5-fold with age in mice (P = 0.04), but are unchanged with age in MRs. (C) Total cysteine content in MRs is 1.6-fold higher than in mice (P = 0.05). Whereas total cysteine declines significantly with age in mice, no age-related changes are evident in MRs. (D) Irreversible oxidation increases 3.4-fold more with age in mice than in MRs. Levels of irreversible cysteine oxidation are obtained from the differences (Δ) between the values of total cysteine in young and old mice and MRs.
Similarly, sulfenic acid in MR is unchanged with age, whereas in mice it increases with age (Fig. 1B). These results show that reversibly oxidized cysteine groups accumulate over a 1.7-year period in mice, whereas age-related accrual is absent over a 25-year interval in MRs.
We also measured total cysteine and indirectly the levels of the irreversible oxidized products of cysteine (−SO2H/−SO3H) (32, 33). Because the fluorescent thiol alkylating agent [6-iodoacetmidofluorescein (6-IAF)] we use binds selectively to −SH groups and not to oxidized thiols, a lower fluorescence signal (thus lower levels of incorporation of 6-IAF to the thiol group) indicates increased levels of irreversible oxidation. Young MRs have a 1.6-fold more total cysteine than do young mice (Fig. 1C) that is sustained over the 24-year age interval in our MR samples (Fig. 1C). This titer decreases significantly (≈12%) with age in mice, indicative of an overall increase in irreversibly oxidized cysteine residues (Fig. 1D). Lack of age-related changes over a 24-year period in either reversible or irreversible cysteine oxidation suggests that MRs have evolved efficient means of maintaining protein thiols.
Proteins from MRs Are Resistant in Vitro Unfolding.
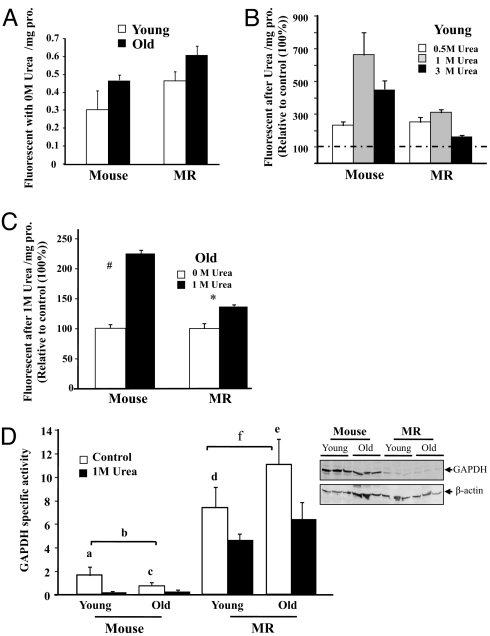
Cysteine thiol groups are critical components for structural maintenance of proteins (24). Because we observed marked interspecies differences in levels of thiol modification, we questioned whether they may influence the stability and structural state of proteins by measuring the relative resistance of proteins to urea-induced unfolding. Urea-induced denaturation exposes the hydrophobic pockets on the surface of proteins. Hydrophobic sites can be photolabeled by the apolar fluorescent probe 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid (BisANS). The intensity of BisANS fluorescence is indicative of the number of hydrophobic pockets exposed and thereby changes in protein conformation can be monitored (34). No significant differences in the fluorescence intensity of untreated (baseline) liver protein samples are evident both among species and with age, although levels of fluorescence tend to be higher in MRs samples than in mice (Fig. 2A). We set these species baseline levels arbitrarily to 100% when testing changes in fluorescence intensity in response to urea-induced unfolding. Increasing concentrations of urea (from 0 to 3 M) result in greater exposure of hydrophobic sites. BisANS incorporation peaks at 1 M and thereafter declines, indicating that surface hydrophobic pockets have collapsed because of denaturation induced by excessively high levels of urea (Fig. 2B). We therefore assessed both interspecies differences and age effects at a urea concentration of 1 M. At this concentration, the young mouse proteome shows more than twice as much BisANS incorporation as that of young MRs, indicating that MRs are more resistant to urea-induced unfolding (Fig. 2B). Age-related interspecific differences are also evident (Fig. 2C). Old MR samples show only a 50% increase in fluorescent intensity after 1 M urea treatment, whereas the old mouse proteome exhibited a 220% increase from basal levels. Clearly the MR proteome is significantly more resistant to unfolding and even 26-year-old MRs are better able to maintain protein homeostasis than can young mice.
Fig. 2.
Proteins from both young and old MRs are extremely resistant to protein unfolding, whereas those from mice are markedly more susceptible. Protein unfolding is measured by incorporation of the BisANS fluorescence probe when hydrophobic pockets are exposed and is expressed as fluorescent units/mg protein as a percentage of control (100%). Data are mean ± SEM (n = 6). The # (P = 0.0038) and * (P = 0.048) denote those values that are significantly different from untreated samples when analyzed by nonparametric ANOVA. (A) Basal protein unfolding is unchanged with age (P = 0.094) in mice and MRs. (B) Young mice have markedly higher levels of protein unfolding after urea treatment than do young MRs with maximal BisANS incorporation at 1 M urea. (C) BisANS incorporation in response to1 M urea is abrogated with age, although old mice show less resistance to unfolding than do old MRs. (D) GAPDH activity declines less with urea treatment in MRs than it does in mice regardless of age, indicating that MRs are better able to maintain protein structure and function than mice. A representative Western blot of GAPDH expression is shown. Letters a–e represent significant differences among comparative datasets (a = 0.03; b = 0.05; c = 0.05; d = 0.05; e = 0.05), while f (representing age-related differences in untreated GAPDH activity in MRs) is not significant (P = 0.24).
We assessed whether resistance to urea-induced protein unfolding is accompanied by sustained protein function by measuring the activity of a key glycolytic enzyme, GAPDH when liver samples are treated with 1 M urea. We specifically chose this enzyme because its catalytic site has critical thiol groups that are vulnerable to unfolding (35). Activity of GAPDH decreased by 40% with age (4 months–2 years) in mice, whereas it was sustained (P > 0.24) over a 24-year period in MRs (Fig. 2D). After urea treatment specific activity of GAPDH declined to similar very low levels in both young and old mice. Both young and old MRs showed only a 40% decline and maintained a significantly higher level of activity than observed in mice. These data show that proteins of MRs are extremely resilient and effectively maintain both structure and function when faced with unfolding stressors.
Age-Related Increase in Protein Ubiquitination Is Diminished in MRs.
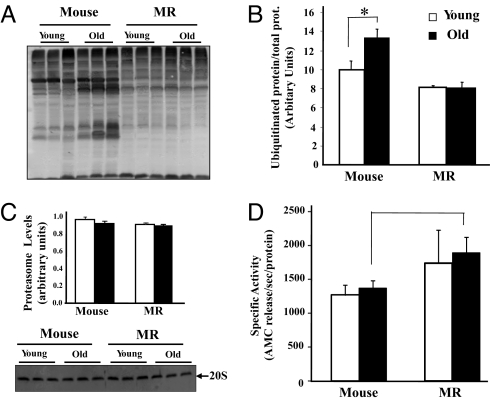
Polyubiquitination is the rate-limiting step for degradation of the oxidized/misfolded proteins by proteasome (36). We measured relative amounts of protein ubiquitination by Western blot analyses. Ubiquitin levels in young mice and MR were similar (P = 0.09) (Fig. 3A). Levels increased with age in mice, whereas no age-related changes from 2 to 26 years were evident in MR samples (Fig. 3B and Fig. S1). Absence of age-related accumulation of oxidized thiol groups and ubiquitinated proteins in MR most likely reflects rapid removal of oxidized and/or misfolded protein in this species.
Fig. 3.
Ubiquitinated proteins increase with age in mice, but are similar in samples from 2-year-old and 26-year-old MRs. (A) A representative Western blot of ubiquitinated proteins is shown. (B) Levels of ubiquitinated protein as measured by Western blot analysis in cytosolic protein homogenates from the livers of young (open bars) and old (solid bars) mice and MRs are shown. Data (mean ± SEM; n = 3) were analyzed by nonparametric ANOVA. The * denotes values that are significantly (P ≤ 0.05) different from young mice. (C) The levels of proteasome (20S) are similar in both mouse and MRs. The total amount of 20S proteasome subunit is measured by Western blot analysis. (D) Proteasome activity is greater in old MRs than in old mice (*, P = 0.052). Chymotrypsin-like proteasome activity is measured in cytosolic protein homogenates from livers of young (open bars) and old (solid bars) mice and MRs by using the 20S proteasome fluormetric (AMC) assay kit. Data (mean ± SEM; n = 3) were analyzed by nonparametric ANOVA.
We measured age-related changes in 20S proteasomal chymotrypsin-like activity (CLPA) in liver proteins from young and old by fluorescent-labeled substrate cleavage. Rates of activity were normalized to protein levels of 20S (Fig. 3C and refs. 37 and 38). 20S protein content did not change with age in either species (Fig. 3C). CLPA tended to be greater in young MR than in young mice (P = 0.12), but was significantly greater (P = 0.05) in old MRs relative to old mice (Fig. 3D).
Discussion
Our data show that MR protein structure and integrity are better maintained during aging than that of mice. There are 4 salient findings in this study. When compared with mice MRs show (i) higher levels of total cysteine, (ii) no age-related change in cysteine oxidation over >2 decades, an exceptionally long period for a mouse-sized rodent, (iii) remarkable resistance to protein unfolding, and (iv) lower levels of protein ubiquitination and higher levels of CLPA.
It is an intriguing observation that the proteome of MRs exhibits such high levels of cysteine (Fig. 1C). We do not know with certainty the functional significance of this finding; for cysteine-rich proteins tend to be more vulnerable to oxidation. However, it is unlikely that the MR proteome differs this much from that of other rodents, especially because we routinely and mostly successfully use commercially available antibodies for protein analyses. More likely certain cysteine-rich proteins are in great abundance in MRs; indeed MRs have more cysteine-rich GST (3-fold) and GSH (1.4-fold) than mice (Fig. S2A), suggesting that detoxification processes are more enhanced in these animals. An alternate plausible explanation may be that the larger cysteine content shields the critical domains of proteins (such as the catalytic or ligand binding sites) from oxidation and thereby contributes to better maintenance of protein structure and function. If this were indeed true, it is possible that inherent cysteine content may serve as a determinant of longevity. A comprehensive comparative proteomic study of cysteine content of soluble proteins from animals with disparate longevity could test this hypothesis.
Mice show a 12% decline in total cysteine with age, pinpointing accrued irreversible cysteine oxidation. We and others have shown that oxidation is not random and rather attacks specific proteins. GAPDH is particularly susceptible to oxidation (32, 35); for example a 16% change in disulfide content of the entire mouse proteome is associated with a 36% decline in age-related GAPDH activity. Thus, even small changes in oxidative damage in the whole proteome may lead to biologically significant decreases in the myriad of activities regulated by cysteine-rich proteins. Knowing that cysteines are found at the catalytic and regulatory domains of many proteins, a 12% increase in irreversible oxidation in cysteine may lead to inactivation of critical enzymes that participate in redox regulatory pathways, such as inflammation, metabolism, or apoptosis, all of which change during aging.
Although oxidative damage of MRs at a young age is higher than that of mice, high steady-state levels of both irreversible and reversible oxidative damage are maintained throughout life. Mice, however, show pronounced age-related increases in protein damage and concomitant decrements in function. Lack of age-related changes in oxidative damage in MRs implies that thiols in MR proteins are better protected from oxidation than those of mice. Lack of age-related increases in cysteine oxidation provide some support for the oxidative stress theory of aging, although extremely high steady-state levels exceeding those of very old mice lead us to question the role of oxidative damage in lifespan determination. High steady-state levels of protein oxidation imply that MRs have a high tolerance threshold of oxidation before induced damage affects functionality. Only when this threshold is exceeded do MRs expend energy in removing oxidative moieties. One possible reason MRs tolerate such high levels of damage even at a young age may be caused by the inherent structural stability of MR proteins. The greater cysteine content in the MR proteome may shield critical peptides and contributes to their resistance to unfolding and better maintenance of protein structure and function.
Our data examining the relationship between structure (BisANS) and activity for GAPDH clearly demonstrate that MRs are better able to maintain their protein structural and functional integrity than can mice (Fig. 2D). Interestingly, Western blot analysis of GAPDH showed that MRs have 10 times less GAPDH than mice in liver protein homogenates. It is unlikely that this order of magnitude difference reflects interspecies differences in GAPDH antibody affinity as we used a highly conserved peptide sequence (residues 317–334) from the C-terminal region of human GAPDH to raise the antibody used in this study. Despite the lower level of GAPDH expression in MR liver, activity is high and the catalytic domain of the MR enzyme is structurally better protected than that of mice, as indicated by the 2.5-fold difference in functional inactivation after 1 M urea treatment (young mouse 90% and young MR 40% decline). Old mice show very low levels of GADPH activity even in the absence of urea treatment, and these decline even further (40%) after urea treatment. This decreased sensitivity to urea compared with young mice may reflect that proportionately more of the GAPDH is lesioned in the old animal. The similar protein content of GAPDH in young and old mice coupled with the marked decline in basal activity in the old cohort support the premise that only a fraction of this enzyme is biologically active. No age-related change in activity or protein content was evident in MRs with or without urea treatment.
Resistance to unfolding and concomitant maintenance of functionality of MR proteins may indicate superior molecular chaperone activity and thus better protein protection. However, Western blot analysis of Hsp70 and Hsc70 levels in liver homogenates showed no interspecies or age-related difference in MRs. It is possible that there may be alternate, as yet unidentified, specific sets of chaperones in the MR and other long-living species that provide superior protection by facilitating structural stability and protecting proteins from unfolding and aggregating. An alternative explanation is that MRs have a better repair system for proteins that could also explain the sustained steady state of protein oxidation with age. Surprisingly, our data based on levels of 2 key repair proteins (methionine sulfoxide reductase A and glutaredoxin) do not support this premise (Fig. S2 B and C). Rather, more efficient removal and replacement of damaged proteins may contribute to augmented protein stability.
Resistance to unfolding may also reflect interspecies differences in structural composition that render MR proteins more stable in a reducing environment. Because we used a total protein liver homogenate in tour assays, this seems most unlikely, for if true it would suggest that many, if not most, MR proteins must have a radically different protein structure. Alternately proteins could appear to be more stable if the cellular degradation machinery more rapidly and effectively removed those proteins that become misfolded or oxidized. In this case, proteasome activity and thus protein turnover would be high. Future studies need to examine age-related interspecific differences in rates of protein turnover, because this, too, may be critical to disparate species longevity.
MRs have substantially lower levels of ubiquitinated proteins than mice. It is unlikely that this reflects interspecies differences in ubiquitin antibody affinity because ubiquitin is one of the few proteins that is highly conserved across the animal kingdom with only 2–3 amino acid sequence differences across vertebrates and invertebrates (39). Further, because we observed intraspecific age-related changes in ubiquitination in both species, we can rule out species differences in protein recognition. Rather, it is more likely that low ubiquitination levels in both young and old MRs reflect less oxidized (40) or misfolded protein awaiting proteasomal degradation than in mice. Further support of this premise is the higher proteasome activity in MRs compared with mice and the 1.5-fold age-related increase in MR proteasome activity, facilitating increased damage removal, should age-related increases in translation errors prevail. These low levels of ubiquitination coupled with higher proteasome activity reveal that old MRs are able to better maintain their proteasome, and that old MRs must also exhibit a higher protein turnover than do old mice and can thereby better maintain protein homeostasis.
Collectively, our results strongly suggest that enhanced protein homeostasis coupled with tightly modulated protein-redox state may contribute significantly to the exceptional longevity of MRs and may be critical determinants for the observed abrogated aging in this species.
Materials and Methods
Animals.
Wild-type C57BL/6 mice were fed ad libitum a standard NIH-31 chow and maintained in microisolator cages on a 12-h dark/light cycle. MRs were from the well-characterized colonies maintained in R.B.'s laboratory at the University of Texas Health Science Center. The ages of the young and old animals used for this study were between 4 and 6 months and 26–28 months for mice and 2–4 years and 24–28 years for MRs. Animals were killed after anesthesia, and the liver tissue was immediately excised and placed on liquid nitrogen. All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center and the subcommittee for animal studies at Audie L. Murphy Memorial Veterans Hospital (San Antonio, TX).
Measurement of Cysteine Oxidation.
Cysteine oxidation was measured by detection of disulfides (31, 41). Briefly, liver tissues were homogenized in 50 mM potassium phosphate buffer (pH 8.0) containing 0.5 mM MgCl2, 1 mM EDTA, 150 mM iodoacetamide, and protease inhibitor mixture [500 μM 4-(2-aminoethyl)benzenesulfonyl fluoride, 150 nM aprotinin, 0.5 mM EDTA, and 1 μM leupeptin hemisulfate] and centrifuged for 30 min at 16,000 × g at 4 °C. For the disulfide assay, samples were incubated in phosphate buffer (pH 8.0) also containing 150 mM iodoacetamide, which reacts with free thiol groups; the free iodoacetamide was removed by protein precipitation with 10% trichloroacetic acid and washed 3 times with 100% ethanol/ethyl acetate (1:1). Samples were resuspended in 8 M urea and incubated with 1 mM DTT for 30 min at 37 °C to reduce disulfide bonds in the samples. Free thiol groups (−SH) arising from the reduced disulfides were labeled with 1 mM 6-IAF, a fluorescent-tagged iodoacetamide. For the sulfenic assay (reversible cysteine oxidation), cytosolic samples were incubated with 6 M Urea and 2 mM DTT for 1 h at 37 °C. Then free thiol groups arising from these reducing conditions were blocked with 200 mM iodoacetmide. AsO3 (5 mM) was added to reduce the reversible oxidation in cysteines, followed by 1 mM DTT. Free thiol groups arising from these reducing conditions were labeled with 1 mM 6-IAF. For determination of irreversible cysteine oxidation, we measured the total amount of cysteine indirectly (thiol and reversible oxidation) by incubating with 6 M Urea and 2 mM DTT for 1 h at 37 °C. Then 5 mM of AsO3 was added to reduce all reversible oxidation. Free thiol groups arising from these reducing conditions were labeled with 1 mM 6-IAF. Protein concentration was measured by using the Bradford method, and 10 μg of protein was subjected to gel electrophoresis. After electrophoresis, the image of the fluorescent protein and the total amount of protein (measured by Sypro Ruby staining) were captured by using the Typhoon 9400 scanner (emission filter 526 and 620 nm, respectively; Amersham Bioscience). The intensities of fluorescence and Sypro Ruby were calculated for each protein by using ImageQuant 5.0 software (Molecular Dynamics). Data were expressed as nmol of cystine oxidation/mg of protein.
Measurements of Ubiquitinated Proteins.
Levels of ubiquitinated protein were measured in cytosolic protein homogenates from liver. Protein concentration was measured by BCA (bichronic acid) protein assay (Pierce), and equal amounts of protein (15 μg) were separated on a 12% SDS/PAGE and subjected to Western blot analysis. Mouse polyclonal antibody against ubiquitin (Santa Cruz Biotechnology) was used to identify the ubiquitinated proteins. Intensities of the bands in each sample (entire lane from the top to the bottom) were quantified by densitometry using ImageQuant version 5.0. Results were expressed as ubiquitinated protein normalized to the total amount of protein measured by Coomassie blue.
Determination of Resistance to Protein Unfolding.
Resistance to protein unfolding was measured by 3 M urea treatment followed by UV-induced photoincorporation of BisANS to proteins as described (34). Briefly, cytosolic protein extracts were diluted to 1 mg/mL in labeling buffer containing 50 mM Tris·HCl, 10 mM MgSO4 at pH 7.4, and protease inhibitors. Samples were treated with and without 3 M Urea for 2 h at room temperature, then 100 mM BisANS was added, and samples were immediately vortexed. Sample (200 μL) was added to a clear 96-well plate and incubated on ice (to minimize local heating) for 1 h under direct exposure of a 115-V, 0.16-Amp handheld longwave UV lamp (365 nm; UV Products model UVL-21). After photoincorporation of BisANS, proteins were then dissolved in Laemmli buffer and subjected to SDS/PAGE. After electrophoresis, gels were removed and illuminated with 365-nm UV light, and BisANS fluorescence was captured with an AlphaImage 3400 (Alpha Innotech) for quantification. Data were expressed as fluorescent units/mg protein.
20S Proteasome Activity Assay.
Liver samples were homogenized in homogenization buffer [50 mM Tris·CL (pH 8.0), 1 mM EDTA, 0.5 mM DTT], and protein concentrations were measured by BCA protein assay (Pierce). For each sample, 100 μg of total protein was assayed in triplicate in 96-well plates by using 20S proteasome fluormetric [7-amino-4-methylcoumarin (AMC)] assay kit per the vendor's instructions (Calbiochem). In brief, the release of free AMC from the fluorogenic peptide Suc-Leu-Leu-Val-Tyr-AMC was measured over time at 37 °C by using a microplate fluorescence spectrophotometer. 20S activity was calculated by the slope of free AMC release over time, after a ≈10-min period of normalization. Quantity of 20S proteasome was measured by Western blot analysis. Twenty micrograms of total cellular protein extract was subjected to SDS/PAGE followed by Western blot analysis using rabbit polyclonal antibody against 20S proteasome subunit (37, 38). Band intensities were measured by using Image QuaNT software package (Molecular Dynamics). 20S-specific activity was calculated by normalizing 20S activity to the quantity of 20S proteasome, measured by Western blot analysis. Data were expressed as AMC release/s per protein.
GAPDH Enzymatic Activity Assay.
GAPDH activity was measured as described by Rafter et al. (42) and Harting and Velick (43). Briefly, liver cytosolic protein extracts were diluted in 15 mM sodium pyrophosphate/30 mM sodium arsenate buffer at pH 8.5 to 0.05 mg of protein/mL, and 0.1 mL of tissue extract was added to a cuvette containing 2.6 mL of pyrophosphate/arsenate buffer containing 0.015 M sodium pyrophosphate, 0.03 M sodium arsenate at pH 8.5, 0.1 mL of 7.5 mM NAD, and 0.1 mL of 0.1 M DTT. Absorbance was monitored at 340 nm, and enzyme units were calculated by using the extinction coefficient of NADH, where 1 unit is equal to the amount of enzyme required to convert 1 μmoL of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate/min at 25 °C and pH 8.5. GAPDH protein levels were determined in liver cytosolic samples from mouse and MRs by Western blot analysis s using a specific GAPDH polyclonal rabbit anti-human GAPDH (Alamo Laboratories). The intensities of the bands were quantified by using ImageQuant version 5.0 (Molecular Dynamics). β-Actin was used as the loading control.
Supplementary Material
Acknowledgments.
This work was supported by the American Federation for Aging Research (V.I.P.), and National Institutes of Health/National Institute on Aging Grants K07 AG025063 04 (to A.C.), AG-022891 (to R.B.), AG025362 (to W.W.), AG23843 (to A.R.), and R37 AG26557 (to A.R. and A.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809620106/DCSupplemental.
References
- 1.Buffenstein R, Jarvis J U. The naked mole rat: A new record for the oldest living rodent. Sci Aging Knowledge Environ. 2002;2002:pe7. doi: 10.1126/sageke.2002.21.pe7. [DOI] [PubMed] [Google Scholar]
- 2.de Magalhaes JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J Gerontol A Biol Sci Med Sci. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: Insights from a successfully aging species. J Comp Physiol B. 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 4.O'Connor TP, Lee A, Jarvis JU, Buffenstein R. Prolonged longevity in naked mole-rats: Age-related changes in metabolism, body composition, and gastrointestinal function. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:835–842. doi: 10.1016/s1095-6433(02)00198-8. [DOI] [PubMed] [Google Scholar]
- 5.Csiszar A, et al. Vascular aging in the longest-living rodent, the naked mole-rat. Am J Physiol. 2007;293:H919–H927. doi: 10.1152/ajpheart.01287.2006. [DOI] [PubMed] [Google Scholar]
- 6.Hulbert AJ, Faulks SC, Buffenstein R. Oxidation-resistant membrane phospholipids can explain longevity differences among the longest-living rodents and similarly-sized mice. J Gerontol A Biol Sci Med Sci. 2006;61:1009–1018. doi: 10.1093/gerona/61.10.1009. [DOI] [PubMed] [Google Scholar]
- 7.Buffenstein R. The naked mole-rat: A new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- 8.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 9.Barja G, et al. Low mitochondrial free radical production per unit O2 consumption can explain the simultaneous presence of high longevity and high aerobic metabolic rate in birds. Free Radical Res. 1994;21:317–327. doi: 10.3109/10715769409056584. [DOI] [PubMed] [Google Scholar]
- 10.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 11.Droge W, Schipper HM. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell. 2007;6:361–370. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohal RS. Role of oxidative stress and protein oxidation in the aging process. Free Radical Biol Med. 2002;33:37–44. doi: 10.1016/s0891-5849(02)00856-0. [DOI] [PubMed] [Google Scholar]
- 13.Miwa S, Riyahi K, Partridge L, Brand MD. Lack of correlation between mitochondrial reactive oxygen species production and life span in Drosophila. Ann N Y Acad Sci. 2004;1019:388–391. doi: 10.1196/annals.1297.069. [DOI] [PubMed] [Google Scholar]
- 14.Andziak B, et al. High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell. 2006;5:463–471. doi: 10.1111/j.1474-9726.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- 15.Wilhelm Filho D, Althoff SL, Dafre AL, Boveris A. Antioxidant defenses, longevity, and ecophysiology of South American bats. Comp Biochem Physiol C Toxicol Pharmacol. 2007;146:214–220. doi: 10.1016/j.cbpc.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Van Remmen H, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 17.Labinskyy N, et al. Comparison of endothelial function, O2-* and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole-rat, and mice. Am J Physiol. 2006;291:H2698–H2704. doi: 10.1152/ajpheart.00534.2006. [DOI] [PubMed] [Google Scholar]
- 18.Lambert AJ, et al. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- 19.Andziak B, O'Connor TP, Buffenstein R. Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech Ageing Dev. 2005;126:1206–1212. doi: 10.1016/j.mad.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5:525–532. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- 21.Salmon AB, Akha AA, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Gerontol A Biol Sci Med Sci. 2008;63:232–241. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung T, Bader N, Grune T. Oxidized proteins: Intracellular distribution and recognition by the proteasome. Arch Biochem Biophys. 2007;462:231–237. doi: 10.1016/j.abb.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Eaton P. Protein thiol oxidation in health and disease: Techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radical Biol Med. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 24.Thomas JA, Mallis RJ. Aging and oxidation of reactive protein sulfhydryls. Exp Gerontol. 2001;36:1519–1526. doi: 10.1016/s0531-5565(01)00137-1. [DOI] [PubMed] [Google Scholar]
- 25.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 26.Mannervik B, Axelsson K, Sundewall AC, Holmgren A. Relative contributions of thioltransferase-and thioredoxin-dependent systems in reduction of low-molecular-mass and protein disulphides. Biochem J. 1983;213:519–523. doi: 10.1042/bj2130519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez A, et al. Extracellular cysteine/cystine redox potential controls lung fibroblast proliferation and matrix expression through up-regulation of transforming growth factor-beta. Am J Physiol. 2007;293:L972–L981. doi: 10.1152/ajplung.00010.2007. [DOI] [PubMed] [Google Scholar]
- 28.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology. Free Radical Biol Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saibil HR. Chaperone machines in action. Curr Opin Struct Biol. 2008;18:35–42. doi: 10.1016/j.sbi.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Jiang C, Chang JY. Isomers of human α-synuclein stabilized by disulfide bonds exhibit distinct structural and aggregative properties. Biochemistry. 2007;46:602–609. doi: 10.1021/bi062068i. [DOI] [PubMed] [Google Scholar]
- 31.Perez VI, et al. Thioredoxin 2 haploinsufficiency in mice results in impaired mitochondrial function and increased oxidative stress. Free Radical Biol Med. 2008;44:882–892. doi: 10.1016/j.freeradbiomed.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Pierce AP, et al. Oxidation and structural perturbation of redox-sensitive enzymes in injured skeletal muscle. Free Radical Biol Med. 2007;43:1584–1593. doi: 10.1016/j.freeradbiomed.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Walss-Bass C, et al. Clozapine causes oxidation of proteins involved in energy metabolism: A possible mechanism for antipsychotic-induced metabolic alterations. Int J Neuropsychopharmacol. 2008;11:1097–1104. doi: 10.1017/S1461145708008882. [DOI] [PubMed] [Google Scholar]
- 34.Pierce A, et al. A novel approach for screening the proteome for changes in protein conformation. Biochemistry. 2006;45:3077–3085. doi: 10.1021/bi052031i. [DOI] [PubMed] [Google Scholar]
- 35.Kim JR, et al. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 36.Breusing N, Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem. 2008;389:203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- 37.Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 38.Conconi M, et al. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch Biochem Biophys. 1996;331:232–240. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson KD. Roles of ubiquitinylation in proteolysis and cellular regulation. Annu Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 40.Shang F, Nowell TR, Jr, Taylor A. Removal of oxidatively damaged proteins from lens cells by the ubiquitin-proteasome pathway. Exp Eye Res. 2001;73:229–238. doi: 10.1006/exer.2001.1029. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhuri AR, Khan IA, Luduena RF. Detection of disulfide bonds in bovine brain tubulin and their role in protein folding and microtubule assembly in vitro: A novel disulfide detection approach. Biochemistry. 2001;40:8834–8841. doi: 10.1021/bi0101603. [DOI] [PubMed] [Google Scholar]
- 42.Rafter GW, Chaykin S, Krebs EG. The action of glyceraldehyde-3-phosphate dehydrogenase on reduced diphosphopyridine nucleotide. J Biol Chem. 1954;208:799–811. [PubMed] [Google Scholar]
- 43.Harting J, Velick SF. Transfer reactions of acetyl phosphate catalyzed by glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1954;207:867–878. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.