Abstract
Large clusters of coexpressed tissue-specific genes are abundant on chromosomes of diverse species. The genes coordinately misexpressed in diverse diseases are also found in similar clusters, suggesting that evolutionarily conserved mechanisms regulate expression of large multigenic regions both in normal development and in its pathological disruptions. Studies on individual loci suggest that silent clusters of coregulated genes are embedded in repressed chromatin domains, often localized to the nuclear periphery. To test this model at the genome-wide scale, we studied transcriptional regulation of large testis-specific gene clusters in somatic tissues of Drosophila. These gene clusters showed a drastic paucity of known expressed transgene insertions, indicating that they indeed are embedded in repressed chromatin. Bioinformatics analysis suggested the major role for the B-type lamin, LamDmo, in repression of large testis-specific gene clusters, showing that in somatic cells as many as three-quarters of these clusters interact with LamDmo. Ablation of LamDmo by using mutants and RNAi led to detachment of testis-specific clusters from nuclear envelope and to their selective transcriptional up-regulation in somatic cells, thus providing the first direct evidence for involvement of the B-type lamin in tissue-specific gene repression. Finally, we found that transcriptional activation of the lamina-bound testis-specific gene cluster in male germ line is coupled with its translocation away from the nuclear envelope. Our studies, which directly link nuclear architecture with coordinated regulation of tissue-specific genes, advance understanding of the mechanisms underlying both normal cell differentiation and developmental disorders caused by lesions in the B-type lamins and interacting proteins.
Keywords: coexpressed genes, nuclear lamina
Previous studies by others and us have shown that many coexpressed tissue-specific genes are organized in large continuous clusters on chromosomes of diverse species (1–3), and similar clustering has been observed for the genes deregulated in diverse diseases (4–8). These findings imply that higher-order chromatin structure may be involved in regulation of extensive multigenic regions in both normal development and its pathological disruptions. In support of this suggestion, alterations in chromatin structure across large multigenic domains have been correlated to changes in gene expression (9–13). Repressed multigenic regions are frequently localized to nuclear periphery (14), and a number of studies linked derepression/activation of genetic loci with their translocation away from the nuclear envelope (15–21). These observations suggest that tethering of genetic loci to nuclear lamina causes their silencing. Recent reports of repression of integrated transgenes and adjacent endogenous genes upon their artificial recruitment to the nuclear envelope (21, 22) support this hypothesis; however, similar studies on different loci did not show repression (23, 24) indicating that silencing at nuclear envelope can be locus-specific. Therefore, although studies on individual loci have suggested that silent tissue-specific gene clusters can be embedded in multigenic chromatin domains repressed at nuclear lamina, it is not clear to what extent this model is applicable to all of the numerous tissue-specific gene clusters present in the genome. In addition, these studies stopped short of elucidating the molecular mechanism(s) that control gene repression within such clusters. In particular, the repressory role of the B-type lamins—the major components of the nuclear lamina (25)—has not been demonstrated.
The possible evolutionarily conserved role of the B-type lamins in developmental gene repression is suggested by interaction of these lamins with silent multigenic regions in both Drosophila and mammals (20, 26). This hypothesis is supported by the studies that link mutations in mammalian B-type lamins with developmental disorders such as autosomal dominant leukodystrophy, premature cell senescence, and lipodystrophy (27–29), and by observation of defects suggesting accelerated aging and neuromuscular degeneration in the Drosophila mutant for the B-type lamin LamDm0 (30). Moreover, even a wider spectrum of similar degenerative tissue-specific diseases has been associated with mutations in the nuclear envelope components interacting with both the B- and the A/C-type lamins, and in the A/C-type lamins themselves (31–38). Intriguingly, recent studies strongly imply that such disorders (collectively identified as laminopathies) involve impaired maintenance of stem cells and their aberrant differentiation (39, 40). Thus, accumulating evidence suggests that the nuclear lamina components, and in particular B-type lamins, regulate cell differentiation through repression of numerous multigenic tissue-specific chromatin domains at the nuclear envelope. Our studies presented here support this hypothesis by providing evidence that clusters of testis-specific genes, in general, represent genome regions silenced in somatic cells, and that at least three-quarters of these repressed gene clusters interact with the B-type lamin LamDm0. Further, we directly show that LamDm0 is required for somatic repression of testis-specific gene clusters at nuclear envelope, and that transcriptional activation of such cluster during male germ-line differentiation is coupled with its detachment from the nuclear lamina.
Results and Discussion
Paucity of the Marked Transposon Insertions in the Large Clusters of testes-Specific Genes: Evidence for Somatic Repression of Multigenic Chromatin Domains.
Recent studies (14) showed that transcriptionally active multigenic regions support expression of the transgenes integrated therein, whereas transgenes inserted in silent multigenic regions are suppressed. These findings indicated that integrated transgenes faithfully report the state of genome regions as permissive or repressive for gene expression. Data from our group and others suggest that clusters of testis-specific genes are embedded in repressed multigenic chromatin domains in somatic tissues (9, 41). If this model is correct for the majority of testis-specific gene clusters in the genome, the transgenes integrated in these clusters should be silenced in somatic cells. To test this prediction, we analyzed the distribution of the 39,894 known transgene insertions identified in saturating mutagenesis screens owing to expression of the transgene-based marker genes in somatic tissues (e.g., 42). Because these insertions mark genome regions permissive for somatic gene expression, the genome regions showing paucity of insertions are likely repressed in somatic cells.
Gene sets for this analysis were generated by using the database of Affymetrix data on gene expression in Drosophila tissues (43). Testis-specific genes were identified as showing consistently detectable expression in testes across the 4 replicate microarrays, and no consistent expression in any of the other tissues including adult brain, head, crop, gut, Malpighian tubule, ovaries, male accessory glands, thoracoabdominal ganglia, and thoracic and abdominal carcass, as well as larval Malpighian tubule and salivary gland. Analysis of chromosomal distribution of the testis-specific genes identified 74 large clusters of 3 or more genes (containing a total of 293 genes). In addition, we identified a set of 330 testis-specific genes present in small clusters of 2 genes, and a set of 827 testis-specific genes that are not clustered [supporting information (SI) Table S1, Table S2, and Table S3].
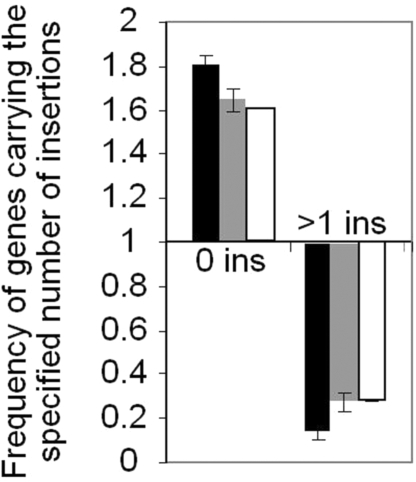
The frequency of transgene insertions in the testis-specific genes was compared with the average frequency across the entire genome. On average, 53% of genes in the Drosophila euchromatic genome have been “hit” by at least one transgene insertion, and 37% of genes have been hit more than once. The genes present in large testis-specific clusters did not contain insertions 1.8 times more frequently than the average (P < 0.0001, one-tailed z test), and the number of such clustered genes with multiple insertions was more than 7 times lower than the average (Fig. 1, black bars). This paucity of transposon insertions in clustered testis-specific genes is not related to the sizes of analyzed genes, because not only the frequency of insertions per gene, but also the frequency of insertions per kilobase was decreased (>3-fold) within the testis-specific gene clusters, as compared with the entire euchromatic genome. Finally, paucity of known transposon insertions is specifically related to the large gene clusters, because the testis-specific genes that are present in small clusters or not clustered at all showed a significantly lesser reduction in insertion frequency (P = 0.013 and P = 0.002, respectively, one-tailed z test) (Fig. 1, gray and white bars). These observations strongly suggest that large testis-specific gene clusters are embedded in the repressive genome regions in somatic tissues.
Fig. 1.
Paucity of the known marked transposon insertions in large testis-specific gene clusters. The frequency of genes with no known insertions or with multiple known insertions, as indicated on the horizontal axis, is shown. The average frequency of insertions per gene in the entire genome served as the reference. The following gene sets were analyzed: testis-specific genes found in large clusters of 3 or more genes (black bars), in small clusters of 2 genes (gray bars), or not clustered at all (open bars).
Binding Sites for the B-Type Lamin LamDm0 Are Frequently Associated with Large Testis-Specific Gene Clusters in Somatic Cells.
To identify candidate regulators of somatic repression of the testis-specific gene clusters, we analyzed enrichment of testis-specific genes with known binding targets for a number of proteins implicated in transcriptional regulation. The data on protein binding in somatic cells were obtained from the genome-wide surveys (44, 45). In our analysis, we considered genes as binding targets if they showed enrichment in protein binding survey equal to or above the arbitrarily set threshold. The results were similar for 3 different thresholds of 1.5-, 2.0-, and 2.5-fold enrichment (Table S4, Fig. S1); the representative analysis using a threshold of 2.0 is shown on Fig. 2. The average frequencies of binding targets across the euchromatic genome were used as the references.
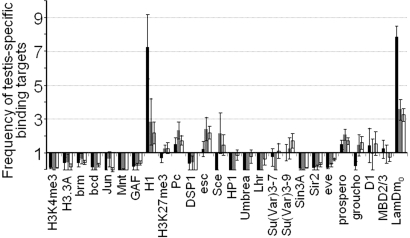
Fig. 2.
Selective enrichment of large testis-specific gene clusters with the LamDm0 and histone H1 binding targets. The frequencies of binding targets for the proteins indicated at the bottom were calculated among the testis-specific genes present in large clusters (black bars) or in small clusters (gray bars), or not clustered at all (open bars). The relative increase in binding target frequency is shown compared with the entire genome.
As expected, general markers of transcriptional activity such as H3K4-me3 and Histone 3.3A and transcriptional activators including Brm, Bcd, Jun, Mnt, and GAF are rarely associated with testis-specific genes in somatic tissues (Fig. 2). In contrast, binding targets of Histone H1, which has been implicated in transcriptional repression (46, 47), are strikingly enriched among the testis-specific genes present in large clusters. Although it remains to be determined whether H1 has an active role in somatic repression of these genes, or merely marks the silent loci as suggested by studies in yeast (48), it is possible that recruitment of this linker histone explains the “closing” of chromatin observed on testis-specific gene clusters in somatic cells (9, 41).
Among the other proteins, large testis-specific gene clusters show binding to the B-type lamin LamDm0 8 times more frequently than the average (P < 0.0001, one-tailed z test). As many as 56% of genes analyzed in these clusters interact with LamDm0, and among the 64 clusters in which at least one gene has been assayed for the LamDm0 binding, three-quarters (47) show such binding with the cutoff of 2.0. Testis-specific genes that are not clustered or are present in small clusters do not show such a drastic enrichment, and both these gene sets show significantly lower incidence of LamDm0-bound genes than large clusters (P < 0.0001 for both comparisons, one-tailed z test), suggesting that LamDm0 is explicitly involved in somatic repression of extensive multigenic testis-specific domains. Although lamins themselves probably do not regulate gene expression, they probably contribute to gene silencing at nuclear envelope by providing docking sites for diverse repressors (38, 49). Intriguingly, even though heterochromatin protein HP1 has been implicated in gene silencing at nuclear lamina (36, 50, 51), binding targets for HP1 and associated proteins Lhr, Umbrea, Su(Var)3-7, and Su(Var)3-9 (52–55) are not enriched among clustered testis-specific genes. Our analysis also did not show enrichment of this gene set with binding targets of Polycomb (Pc) and Pc group proteins DSP1, Esc, and Sce (56–58), or with the Pc-related trimethylation of H3-K27 (45), despite the previously suggested role for Pc in repression of testis-specific genes (59). Other analyzed repressors (Fig. 2) did not show strongly preferential binding to the clustered testis-specific genes as well. Even though a moderate (2- to 3-fold) enrichment of clustered genes with Groucho, D1, and Su(Var)3-9 binding targets was observed for the low-stringency enrichment cutoff of 1.5, these trends are not consistent at higher cutoff values (Fig. S1, Table S4) and therefore their significance is questionable. Thus, the repressory factors involved in silencing of large testis-specific gene clusters at nuclear envelope remain to be identified. Recent studies showing that silencing of lamina-tethered transgenes is linked to histone deacetylation (21, 22) suggest that gene repression at nuclear envelope role is mediated by HDACs, such as the lamina-associated enzymes HDAC1 and HDAC3 (36, 60). Further studies are required to elucidate the roles of these proteins in tissue-specific repression of lamina-associated multigenic domains.
LamDm0 Is Required for Somatic Repression of the Lamina-Bound Testis-Specific Gene Clusters.
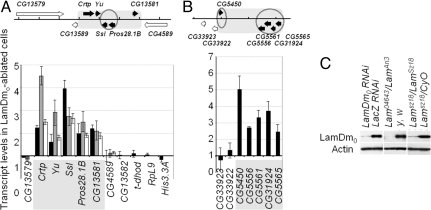
Because bioinformatics studies strongly suggested the major role for LamDm0 in somatic repression of the testis-specific gene clusters, we directly tested this hypothesis by analyzing gene expression in somatic cells with ablated LamDm0. We analyzed 2 representative LamDm0-bound testis-specific gene clusters. One cluster of 5 testis-specific genes in cytological region 60D1 was thoroughly characterized in our previous studies, which showed that this cluster is embedded in “closed” chromatin in somatic tissues (9). Another region 22A1 has been identified as a prominent lamina-bound domain in cultured somatic cells (20), and testis-specific expression of 5 genes clustered in this region was confirmed by the real-time RT-PCR (Fig. S2). Expression of the genes included in these 2 clusters was quantitatively studied in the first instar larvae homozygous for the null allele LamSz18 by the real-time RT-PCR. The heterozygous LamSz18/CyO,P{Ubi-GFP.S65T} siblings were identified by their GFP fluorescence and used as the reference. The first instar larvae essentially represented the samples of somatic tissues, because only a few primordial germ cells are present at this early stage of development. We found that the transcript levels for all 5 testis-specific genes in the 60D1 region are significantly higher (P < 0.01) in the LamSz18 homozygotes than in their siblings carrying the CyO,P{Ubi-GFP.S65T} chromosome which is “wild type” for the LamDm0 (Fig. 3A, black bars). Similarly, all 5 testis-specific genes in the 22A1 cluster showed strong up-regulation in the LamSz18 mutants (Fig. 3B). For comparison, we also analyzed the expression of the testis-specific transcript of the gene dhod. The testis-specific dhod promoter is juxtaposed to the ubiquitously active promoter of this gene (61), thus the dhod gene region is not repressed in somatic tissues. In addition, we analyzed the genes CG13579, CG4589, CG13582, RpL9, His-3.3A, CG33922, and CG33923, which are not testis-specific. Neither the testis-specific dhod transcript, nor the broadly expressed transcripts of the other analyzed genes were affected in the LamSz18 homozygotes (Fig. 3 A and B, black bars). These results indicated that loss of LamDm0 leads to selective derepression of testis-specific genes clustered in the 60D1 and 22A1 regions.
Fig. 3.
Ablation of LamDm0 leads to somatic derepression of the testis-specific gene clusters in the 60D1 (A) and 22A1 (B) regions. (Upper) Genome organization of the regions showing testis-specific genes (black) and broadly expressed (white) genes as arrows. Gene clusters are outlined as gray rectangles, and genes showing at least 2-fold enrichment with LamDm0 in somatic cells (20) are outlined by circles. Tick marks correspond to the 5-kb intervals. (Lower) Transcript levels in the homozygous LamSz18 first-instar larvae compared with their heterozygous siblings (black bars), in somatic tissues of the Lam04643/Lamari3 adult females compared with the y, w flies (gray bars), and in the LamDm0 RNAi-treated S2 cells compared with the LacZ RNAi-treated cells (open bars). The Actin5C transcript served as a template loading reference. (C) Western blot analysis confirms ablation of LamDm0 in RNAi-treated cells, in the Lam04643/Lamari3 females and in the LamSz18/LamSz18 larvae.
We confirmed our observations by analyzing gene expression in the 60D1 region in somatic tissues in which LamDm0 was ablated by using a different combination of the alleles, including the null allele Lam04643 and the hypomorphic allele Lamari3. All testis-specific genes in the 60D1 cluster were significantly (P < 0.01) up-regulated in the gonadectomized adult Lam04643/Lamari3 females, as compared with the gonadectomized y, w females that are “wild type” for the LamDm0. At the same time, the broadly expressed genes CG13579 and CG4589 were not affected (Fig. 3A, gray bars). The effect of LamDm0 ablation on up-regulation of the testis-specific 60D1 cluster was further corroborated in the RNAi studies on the cultured somatic S2 cells. In the cells treated with the LamDm0 dsRNA, the testis-specific genes in this cluster showed significant (P < 0.01) up-regulation, in contrast to the flanking broadly expressed gene CG4589 (Fig. 3A, open bars). Efficient ablation of LamDm0 in the RNAi-treated cells, as well as in the mutant somatic tissues, was confirmed by Western blot analysis (Fig. 3C).
Our observations showed that LamDm0 is required for somatic repression of testis-specific gene clusters. The derepression of testis-specific gene clusters upon ablation of LamDm0 in somatic cells was selective because a number of other genes not included in such clusters were not affected. This effect was different from the global down-regulation of RNA polymerase activity previously reported in the Lamin B1-ablated mammalian cells (62). Such difference is probably due to the high mortality caused by the Lamin B1 ablation (62), whereas in our experiments the reduced levels of LamDm0 were physiologically acceptable as evidenced by the survival of the Lam04643/Lamari3 mutants through adulthood. Thus, our studies, which took advantage of Drosophila that can tolerate severe changes in the B-type lamin dosage, provided unique direct experimental evidence that this major component of nuclear lamina is involved in repression of tissue-specific multigenic domains.
Transcriptional Up-Regulation of the Lamina-Bound Testis-Specific Clusters Is Coupled with Their Detachment from the Nuclear Envelope.
We hypothesized that, in addition to the somatic silencing of testis-specific gene clusters, LamDm0 is also required for attachment of these clusters to the nuclear envelope. To test this hypothesis, we determined intranuclear positions of the 60D1 and 22A1 regions in the interphase nuclei of cultured S2 cells in which LamDm0 was ablated by RNAi. Fluorescence in situ hybridization (FISH) combined with immunostaining for LamDm0 confirmed RNAi-induced ablation of LamDm0, and showed approximately 2-fold decrease in frequency of the 60D1 and 22A1 loci bound to lamina (Fig. 4 A and B and Fig. S3). Next, we analyzed association of the 60D1 gene cluster with nuclear envelope during male germ-line differentiation. The whole-mount testes dissected from the third instar larvae were analyzed for intranuclear localization of the 60D1 region by FISH combined with immunostaining for LamDm0. Morphologically, a group of small cells is located at the end of testis (lower left on Fig. 4C) and includes spermatogonia, cyst cells and stem cells, in which the 60D1 locus is silent (9). In these cells, the 60D1 region is associated with nuclear lamina in 76% of nuclei (Fig. 4D), similarly to the cultured somatic S2 cells (Fig. 4B). On the contrary, in spermatocytes (identified by characteristic large nuclei, Fig. 4C, upper right corner) in which testis-specific genes in the 60D1 region are expressed (9), this region is found at the nuclear envelope in only 6% of the nuclei (Fig. 4D). Thus, transcriptional activation of the testis-specific gene cluster in male germ line is coupled to its dissociation from the nuclear envelope. Similarly, detachment from the nuclear lamina has been associated with transcriptional activation of other lamina-bound loci both in Drosophila and in mammals (20, 21). These observations strongly suggest a model in which gene repression is controlled in a cell type-specific manner through regulated tethering of chromatin to the nuclear lamina. Further dissection of the mechanisms that mediate repression of lamina-bound multigenic regions and control localization of these regions at nuclear envelope will provide new insights into coordinated regulation of tissue-specific genes, thus advancing understanding of cell differentiation both in normal development and in disease.
Fig. 4.
Expression of the LamDm0-bound testis-specific loci is coupled to their detachment from the nuclear envelope. (A) FISH shows localization of the testis-specific 60D1 locus within 0.4 μm from the nuclear envelope revealed by immunostaining for LamDm0, indicating direct contact between the locus and the envelope in cultured S2 cells treated with the control LacZ dsRNA. In the cells treated with the LamDm0 dsRNA, LamDm0 is ablated and the 60D1 locus is detached from the nuclear envelope (revealed by immunostaining for dLBR). (B) Quantitative analysis shows that LamDm0 dsRNA treatment decreases the frequency with which the testis-specific 60D1 and 22A1 loci are found in contact with nuclear envelope, as compared with the control LacZ dsRNA treatment (gray versus black bars). (C and D) The 60D1 locus is found in contact with nuclear envelope in the majority of small cells including spermatogonia, cyst cells, and stem cells located at the testis tip (C, lower left; D, black bar) but not in the larger spermatocytes (C, upper right; D, gray bar).
Methods
Detailed descriptions of procedures are included in SI Methods and Table S5.
Drosophila melanogaster Stocks.
y, w; Lamsz18 (63) and Lam04643 (64) mutants and the stock containing the CyO balancer chromosome carrying the P{Ubi-GFP.S65T} expression transgene were obtained from the Bloomington Drosophila Stock Center at Indiana University, and the LamAri3 mutant (65) was kindly provided by J. Fischer (University of Texas, Austin).
Cell Culture and RNAi.
Schneider-2 (S2) cells were grown in Schneider's Drosophila medium supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin at 26 °C. To obtain dsRNAs, coding sequences of Escherichia coli lacZ gene (≈650 bp) and D. melanogaster LamDm0 gene (≈800 bp) were amplified from the pCaSpeR-βgal (67) and J14–3A (63) plasmids, and transcribed in vitro by using T7 RNA polymerase (Promega). Treatment of cells with dsRNA was performed by using a previously described protocol (66).
Reverse Transcription and Quantitative PCR.
Total RNA was obtained by extraction with the TRIzol reagent (Invitrogen), and contaminating DNA was removed by DNase I treatment. Real-time RT-PCR assays were performed on cDNA synthesized with the oligo(dT) primer, using SYBR Green chemistry (Applied Biosystems) and the Mx3000P hardware (Stratagene). Acrylamide gel-based assay used cDNA synthesized with the random hexamer primer; reactions were run in the presence of 33P-dATP and reaction products were quantified with PhosphorImager Storm-820 (Molecular Dynamics).
Western Blot Analysis.
Proteins were extracted with 8 M urea, 1 M Tris·HCl, pH 7.0. LamDm0 was detected with the monoclonal murine antibody ADL67 (68), and β-actin, with the polyclonal murine antibody (Abcam).
Immuno-FISH.
FISH probe for the 60D1 region was prepared by using the ≈35 kb cosmid k9 (9). The probe for the 22A1 region was generated by PCR amplification of 4 tiling ≈5-kb fragments representing a ≈20 kb genome segment. Digoxigenin-labeled DNA probes were hybridized with cells or whole-mount larval testes as described in ref. 69 with some modifications, and immunostained by using rabbit polyclonal antibody against LamDm0 (no. 836) (70) or guinea pig polyclonal anti-dLBR antibody (71), and anti-DIG-FITC-conjugated antibody (Roche). Secondary Alexa 546- or Alexa 647-conjugated antibodies were from Invitrogen.
Image Analysis.
Three-dimensional image stacks were recorded with a confocal Zeiss LSM510 Meta microscope. Images were processed and analyzed by using IMARIS 5.0.1 software (Bitplane AG) to measure the distance between the FISH signal and the reconstructed nuclear surface. The distance of 0.4 μm or less was interpreted as localization of FISH signal at nuclear envelope, because both the nuclear lamina and the FISH signal had a visual width of ≈0.4 μm.
Analysis of Transposon Insertions and Protein Binding.
Gene spans included transcribed regions and, in addition, the 500-bp 5′-UTR segments to account for the proximal promoter regions. Coordinates of the gene spans, and of the 39,894 known transgenes insertions were obtained from the Drosophila genome database (www.flybase.net). Genes were analyzed for the number of insertions within the gene span, using custom-made software. Raw gene microarray data on binding of diverse chromatin proteins were obtained from the survey of de Wit et al. (44) and publications referenced therein.
Supplementary Material
Acknowledgments.
We thank Paul A. Fisher for the rabbit no. 836 and murine ADL67 anti-LamDm0 antibodies, Georg Krohne for the guinea pig anti-dLBR antibody, Janice A. Fischer for the LamAri3 flies, and Mark A. Krasnow for the LamDm0cDNA clone J14-3A. This work was supported by the Russian Foundation for Basic Research Grants 07-04-00695-a and 07-04-01467-a, the Grant to Scientific Schools from Russian Ministry of Science 3464.2008.4, and by Grant for Molecular and Cellular Biology from Russian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811933106/DCSupplemental.
References
- 1.Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 2.Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 3.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 4.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 5.Novak P, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 6.Staub E, et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol Cancer. 2006;5:37. doi: 10.1186/1476-4598-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buness A, et al. Identification of aberrant chromosomal regions from gene expression microarray studies applied to human breast cancer. Bioinformatics. 2007;23:2273–2280. doi: 10.1093/bioinformatics/btm340. [DOI] [PubMed] [Google Scholar]
- 8.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat. 2009;113:239–249. doi: 10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- 9.Kalmykova AI, Nurminsky DI, Ryzhov DV, Shevelyov YY. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 2005;33:1435–1444. doi: 10.1093/nar/gki281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Razin SV, Farrell CM, Recillas-Targa F. Genomic domains and regulatory elements operating at the domain level. Int Rev Cytol. 2003;226:63–125. doi: 10.1016/s0074-7696(03)01002-7. [DOI] [PubMed] [Google Scholar]
- 11.Dillon N. Gene regulation and large-scale chromatin organization in the nucleus. Chromos Res. 2006;14:117–126. doi: 10.1007/s10577-006-1027-8. [DOI] [PubMed] [Google Scholar]
- 12.Hansen JC. Linking genome structure and function through specific histone acetylation. ACS Chem Biol. 2006;1:69–72. doi: 10.1021/cb6000894. [DOI] [PubMed] [Google Scholar]
- 13.Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- 14.Gierman HJ, et al. Domain-wide regulation of gene expression in the human genome. Genome Res. 2007;17:1286–1295. doi: 10.1101/gr.6276007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francastel C, Magis W, Groudine M. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc Natl Acad Sci USA. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 17.Zink D, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, et al. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 2006;20:784–794. doi: 10.1101/gad.1392006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams RR, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 20.Pickersgill H, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 21.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 22.Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 25.Gerace L, Comeau C, Benson M. Organization and modulation of nuclear lamina structure. J Cell Sci Suppl. 1984;1:137–160. doi: 10.1242/jcs.1984.supplement_1.10. [DOI] [PubMed] [Google Scholar]
- 26.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 27.Padiath QS, et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- 28.Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegele RA, et al. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am J Hum Genet. 2006;79:383–389. doi: 10.1086/505885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muñoz-Alarcón A, et al. Characterization of lamin mutation phenotypes in Drosophila and comparison to human laminopathies. PLoS ONE. 2007;2(6):e532. doi: 10.1371/journal.pone.0000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann K, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly) Nat Genet. 2002;31:410–414. doi: 10.1038/ng925. [DOI] [PubMed] [Google Scholar]
- 32.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2002;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 33.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterham HR, et al. Autosomal recessive HEM/Greenberg skeletal dysplasia is caused by 3-beta-hydroxysterol delta(14)-reductase deficiency due to mutations in the lamin B receptor gene. Am J Hum Genet. 2003;72:1013–1017. doi: 10.1086/373938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manilal S, Nguyen TM, Sewry CA, Morris GE. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum Mol Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- 36.Holaska JM, Wilson KL. An emerin “proteome”: Purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattout A, Dechat T, Adam SA, Goldman RD, Gruenbaum Y. Nuclear lamins, diseases and aging. Curr Opin Cell Biol. 2006;3:335–341. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Shaklai S, Amariglio N, Rechavi G, Simon AJ. Gene silencing at the nuclear periphery. FEBS J. 2007;274:1383–1392. doi: 10.1111/j.1742-4658.2007.05697.x. [DOI] [PubMed] [Google Scholar]
- 39.Espada J, et al. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J Cell Biol. 2008;181:27–35. doi: 10.1083/jcb.200801096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer JA, McCarrey JR, Djakiew D, Krawetz A. Differentiation: The selective potentiation of chromatin domains. Development (Cambridge, U.K.) 1998;125:4749–4755. doi: 10.1242/dev.125.23.4749. [DOI] [PubMed] [Google Scholar]
- 42.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 43.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 44.de Wit E, Braunschweig U, Greil F, Bussemaker HJ, van Steensel B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 2008;4(3):e1000045. doi: 10.1371/journal.pgen.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 46.Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K, et al. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schäfer G, McEvoy CR, Patterton HG. The Saccharomyces cerevisiae linker histone Hho1p is essential for chromatin compaction in stationary phase and is displaced by transcription. Proc Natl Acad Sci USA. 2008;105:14838–14843. doi: 10.1073/pnas.0806337105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: Integrating isolated clues. J Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- 50.Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope innermembrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;25:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 51.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delattre M, Spierer A, Tonka CH, Spierer P. The genomic silencing of position-effect variegation in Drosophila melanogaster: Interaction between the heterochromatin-associated proteins Su(var)3–7 and HP1. J Cell Sci. 2000;23:4253–4261. doi: 10.1242/jcs.113.23.4253. [DOI] [PubMed] [Google Scholar]
- 53.Schotta G, et al. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukai M, et al. MAMO, a maternal BTB/POZ-Zn-finger protein enriched in germline progenitors is required for the production of functional eggs in Drosophila. Mech Dev. 2007;124:570–583. doi: 10.1016/j.mod.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Brideau N, Barbash D. Investigating the interactions between the hybrid incompatibility protein LHR and heterochromatic proteins HP1 and HP6. A Dros Res Conf. 2008;49:706A. [Google Scholar]
- 56.Dejardin J, et al. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature. 2005;434:533–538. doi: 10.1038/nature03386. [DOI] [PubMed] [Google Scholar]
- 57.Fritsch C, Beuchle D, Muller J. Molecular and genetic analysis of the Polycomb group gene Sex combs extra/Ring in Drosophila. Mech Dev. 2003;120:949–954. doi: 10.1016/s0925-4773(03)00083-2. [DOI] [PubMed] [Google Scholar]
- 58.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- 60.Somech R, et al. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 61.Yang J, Porter L, Rawls J. Expression of the dihydroorotate dehydrogenase gene, dhod, during spermatogenesis in Drosophila melanogaster. Mol Gen Genet. 1995;246:334–341. doi: 10.1007/BF00288606. [DOI] [PubMed] [Google Scholar]
- 62.Tang CW, et al. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cell Sci. 2008;121:1014–1024. doi: 10.1242/jcs.020982. [DOI] [PubMed] [Google Scholar]
- 63.Guillemin K, Williams T, Krasnow MA. A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat Cell Biol. 2001;3:848–851. doi: 10.1038/ncb0901-848. [DOI] [PubMed] [Google Scholar]
- 64.Spradling AC, et al. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of the vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patterson K, et al. The functions of klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clemens JC, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 68.Stuurman N, Maus N, Fisher PA. Interphase phosphorylation of the Drosophila nuclear lamin: Site-mapping using a monoclonal antibody. J Cell Sci. 1995;108:3137–3144. doi: 10.1242/jcs.108.9.3137. [DOI] [PubMed] [Google Scholar]
- 69.Gemkow MJ, Verveer PJ, Arndt-Jovin DJ. Homologous association of the Bithorax-Complex during embryogenesis: Consequences for transvection in Drosophila melanogaster. Development (Cambridge, U.K.) 1998;125:4541–4552. doi: 10.1242/dev.125.22.4541. [DOI] [PubMed] [Google Scholar]
- 70.Osouda S, et al. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev Biol. 2005;284:219–232. doi: 10.1016/j.ydbio.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 71.Wagner N, Weber D, Seitz S, Krohne G. The lamin B receptor of Drosophila melanogaster. J Cell Sci. 2004;117:2015–2028. doi: 10.1242/jcs.01052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.