Abstract
DNA double strand breaks (DSBs) initiate reversible cellular checkpoint and repair activities. Whereas many of the activating events at DSBs have recently been elucidated, the mechanisms used to terminate responses at these sites are largely undefined. Here we report a pathway required to reverse RNF8-Ubc13 dependent ubiquitination events on chromatin flanking DSBs. Inhibition of the Rap80-BRCC36 de-ubiquitinating enzyme complex partially restored DSB-associated ubiquitin levels following RNF8 knockdown or proteasome inhibition. Similarly, BRCC36 knockdown or expression of a BRCC36 de-ubiquitinating enzyme-inactive mutant rescued both 53BP1 recruitment to DSBs and ionizing radiation-induced γH2AX ubiquitination following RNF8 depletion, and mitigated ionizing radiation sensitivity resulting from RNF8 deficiency. Thus, concomitant and opposing RNF8-Ubc13 ubiquitin ligase and Rap80-BRCC36 ubiquitin hydrolysis activities are responsible for determining steady-state ubiquitin levels at DNA DSBs. These findings reveal a Rap80-BRCC36 dependent pathway that is required for appropriate DSB recruitment and repair responses.
Keywords: BRCA1, DNA damage, ubiquitin
Multiple, parallel signaling pathways converge upon DNA double-strand breaks (DSBs) to initiate temporal and spatial coordination of checkpoint and repair responses. These DNA damage response activities are mediated in part by accumulation of DNA repair proteins at both chromatin and non-nucleosomal DNA regions flanking DSBs (1–3). Repair factor targeting occurs within minutes of DSB induction, providing strong evidence that molecular recognition events rapidly develop at sites of DNA damage (1, 4, 5). Following the resolution of DNA damage, repair proteins dissociate from DSBs, thus alleviating cell cycle checkpoint responses and allowing resumption of cell proliferation. It is unclear, however, if DSB termination and activation pathways occur simultaneously in an equilibrium process during the earliest stages of repair, or if DNA damage response resolution occurs in a step-wise manner subsequent to the completion of DNA repair.
Some of the recruitment events at DSBs have recently been elucidated for the breast and ovarian cancer suppressor protein BRCA1 (Breast Cancer 1, early onset) and the checkpoint and repair protein 53BP1 (p53 Binding Protein 1) (6–12). Histone H2AX phosphorylation by the PI3K-like kinase members ATM (Ataxia Telangiectasia Mutated) and DNA-PKcs (DNA-Dependent Protein Kinase catalytic subunit) occurs at chromatin flanking DSBs to initiate a direct interaction between phosphorylated serine 139 of H2AX (γH2AX) and the MDC1 (Mediator of DNA Damage Checkpoint Protein 1) protein (13–15). PI3K-like kinase phosphorylation of MDC1 (13, 16) creates a binding site for the E3 ubiquitin ligase RNF8 (Ring Finger 8). In conjunction with the E2 enzyme Ubc13 (ubiquitin-conjugating 13), RNF8 ubiquitinates histones H2A and H2AX in a K63-linked manner to create a docking site for DNA repair proteins to accumulate at DSBs (6–8, 17). These DSB chromatin-associated K63-linked ubiquitin species are recognized by the tandem ubiquitin interacting motif (UIM) containing protein Rap80/UIMC1 (Receptor Associated Protein 80/Ubiquitin Interaction Motif Containing 1) as part of a biochemical complex that includes BRCA1 (9–11, 18). The mechanistic basis underlying RNF8-Ubc13-dependent DSB recognition by the 53BP1 protein is less clear. 53BP1 DSB localization depends upon recognition of di-methylated lysine residues on histone H4 (H4 K20-Me2) by the 53BP1 tudor domain (19, 20). It is possible that RNF8-Ubc13 substrate ubiquitination imparts changes to chromatin that facilitates access of 53BP1 to methylated histones at DSBs. A second possible scenario is that a 53BP1-associated ubiquitin binding protein mediates 53BP1 binding to RNF8-Ubc13 synthesized ubiquitinated structures at DSBs.
The Rap80 tandem UIM domains have binding specificity for lysine 63-linked ubiquitin (K63-Ub) (11), enabling Rap80 to target BRCA1 to DSB-associated chromatin in a γH2AX-MDC1- and RNF8-Ubc13-dependent manner (6–8, 10, 11, 17). BRCA1 is a component of several other protein complexes, in addition to the BRCA1-Rap80 complex (21). It is likely that some of these Rap80-independent complexes are able to recognize non-chromatinized DSB regions (1, 22, 23). In addition, Rap80 directs a nearly stoichiometric binding partner, the BRCC36 de-ubiquitinating enzyme (DUB) to DSBs, raising the possibility that Rap80 simultaneously targets BRCA1 E3 ubiquitin ligase and BRCC36 DUB activities to DSBs for ubiquitin remodeling events (11, 24). In particular, Rap80-dependent BRCC36 delivery to RNF8-Ubc13 synthesized K63-Ub structures at DSBs may represent a mechanism to terminate DSB recruitment signals, preventing unabated DNA damage responses following completion of DSB repair.
Here we report a pathway that contributes to the termination of RNF8-Ubc13 dependent DSB associated ubiquitination events. The Rap80-BRCC36 complex is recruited to RNF8-Ubc13 synthesized K63-Ub structures at DSBs, where it is required to de-ubiquitinate γH2AX. Depletion of either Rap80 or BRCC36 increases DSB-associated ubiquitin levels and 53BP1 DSB targeting. Moreover, Rap80-BRCC36 inhibition antagonizes the effects of RNF8 knockdown with respect to DSB-associated ubiquitin and 53BP1, and cellular viability in response to ionizing radiation (IR). These data reveal that concomitant and opposing activities of RNF8-Ubc13-dependent K63-Ub synthesis and Rap80-BRCC36-dependent K63-Ub breakdown occur in an equilibrium process that regulates DSB ubiquitination events throughout the course of DNA repair.
Results
Rap80-BRCC36 DUB Activity and γH2AX Ubiquitination.
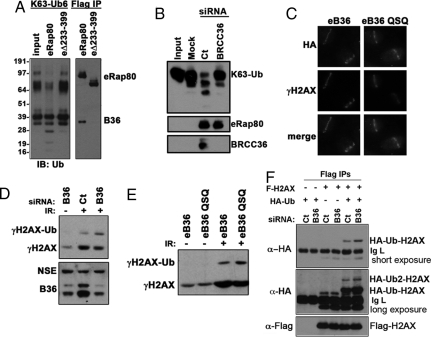
Rap80 is required for BRCA1 and BRCC36 localization to DSBs (11). We therefore hypothesized that BRCC36 would exert its DUB activities to influence DNA damage responses in the context of binding Rap80. BRCC36 has been reported to hydrolyze K63-Ub polymers (11). To determine if BRCC36 also had K63-Ub DUB activity in association with the Rap80 complex, affinity purified Rap80 species from HeLa nuclear extracts were incubated with recombinant K63-Ub chains and reactions analyzed for ubiquitin hydrolytic activity by immunoblot (IB). The wt Rap80 (eRap80) complex demonstrated K63-Ub DUB activity, whereas DUB activity was not readily detectable for the Rap80Δ233–399 complex that does not bind BRCC36 (Fig. 1A). Moreover, Rap80 complexes purified from HeLa S3 cells 48 h after transfection with BRCC36-specific siRNA demonstrated strongly reduced K63-Ub DUB activity (Fig. 1B). In addition, the Rap80 wt complex largely failed to hydrolyze K48-Ub chains in the time frame required for K63-Ub hydrolysis, demonstrating relative DUB specificity for K63-Ub compared with K48-Ub chains [supporting information (SI) Fig. S1].
Fig. 1.
Rap80-BRCC36 K63-DUB activity and DSB responses. (A) Equivalent amounts of Rap80 wt (eRap80) or Rap80Δ233–399 (eΔ233–399) were purified from HeLa nuclear extracts and incubated in DUB buffer with K63-linked hexa-ubiquitin (K63-Ub6). Higher molecular weight K63-Ub polymers are also present (Left, Top). IB was performed as indicated to detect input and K63-Ub hydrolysis products (Left), and ectopic Rap80 (eRap80) and BRCC36 (B36) (Right). (B) eRap80 complexes were purified from HeLa S3 cells at 48 h after control (Ct) or BRCC36 siRNA and DUB activity assays performed as in A. Mock represents DUB assay performed with Flag IP material from HeLa S3 cells that do not express eRap80. (C) Ectopic BRCC36 (eB36) and ectopic BRCC36 QSQ (eB36 QSQ) are recruited to laser-induced DSBs in U2OS cells 30 min after stripe induction. IF was performed against eBRCC36 with anti-HA antibody. (D) siRNA knockdown of BRCC36 versus control was performed in HeLa S3 cells and ubiquitinated forms of γH2AX analyzed by IB without IR or at 1 h after 10 Gy IR. Non-specific epitope of the BRCC36 antibody was used to indicate similar protein loading. (E) IB was performed for γH2AX in HeLa S3 cells expressing either eB36 wt or eB36 QSQ without IR or at 1 h after 10 Gy IR. (F) 293T cells were transfected with expression constructs for Flag-H2AX and HA-ubiquitin along with either control (Ct) or BRCC36 siRNA. Flag IP followed by IB was performed as indicated. Short and long exposures of the same IB are presented to reveal mono- and di-ubiquitinated Flag-H2AX. IgL indicates IgG light chain.
Both BRCC36 wt and a catalytically inactive mutant, BRCC36H122Q,H124Q (BRCC36 QSQ), bound Rap80 and BRCA1 (Fig. S1) and migrated to laser-induced DNA DSBs (Fig. 1C). Mutation of these active site histidine residues to glutamines disrupts the ability of JAMM domain-containing metallo-proteases such as BRCC36 to coordinate the active site zinc ion that directly participates in the catalytic hydrolysis of ubiquitin and ubiquitin-like moieties (25). Thus, BRCC36 wt and BRCC36 QSQ exhibit similar protein-protein interactions and sub-cellular localization in response to DNA damage, enabling an investigation of the role of BRCC36 K63-Ub DUB activity in the DNA damage response.
Identification of BRCC36 DUB targets is expected to provide mechanistic insights into ubiquitin signaling pathways relevant to DSB repair. Detailed kinetic studies have revealed that RNF8 is recruited to γH2AX-MDC1-containing stripes during the earliest stages of DNA DSB repair (8). Rap80 localization to DSBs occurs at slightly later time points, ostensibly by binding to RNF8-Ubc13-synthesized K63-Ub structures. This suggests a temporal sequence of events, occurring within minutes of DSB induction, leading to a step-wise recruitment of RNF8-Ubc13 followed by Rap80-BRCC36 to DSBs. Following the initial DSB accumulation events, RNF8-Ubc13 and Rap80-BRCC36 are predicted to be present at the same DSB regions, consistent with the hypothesis that both enzymatic complexes simultaneously regulate DSB-associated K63-Ub levels. γH2AX has recently been shown to be an RNF8-Ubc13 substrate; thus, ubiquitinated γH2AX (γH2AX-Ub) is a putative recognition element for the Rap80-BRCC36 complex (6, 8, 17). To determine if Rap80-BRCC36 targets ubiquitinated forms of γH2AX during DSB repair responses, cells were treated with control versus BRCC36 siRNA and histones purified by acid elution 1 h after 10 Gy IR. BRCC36 knockdown increased the ratio of ubiquitinated to non-ubiquitinated forms of endogenous γH2AX (Fig. 1D). In agreement with BRCC36-dependent γH2AX DUB activity, expression of BRCC36 QSQ stabilized γH2AX-Ub compared with cells expressing BRCC36 wt (Fig. 1E). Moreover, ectopic H2AX demonstrated increased ubiquitination in BRCC36 deficient cells compared with controls (Fig. 1F).
Rap80-BRCC36 Knockdown Restores DSB-Associated Ubiquitination During Proteasome Inhibition.
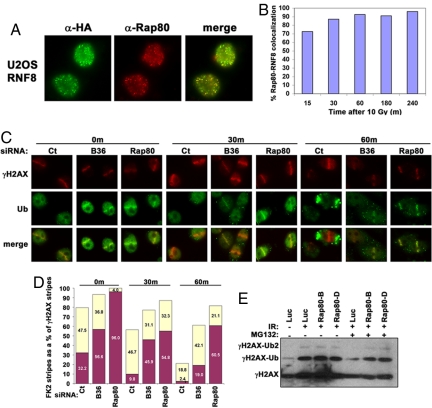
The preceding findings suggest a scenario whereby Rap80-BRCC36 DUB activity targets a subset of RNF8-Ubc13-dependent, K63-Ub-containing substrates in vivo during the DNA damage response. Consistent with this assertion, Rap80 and RNF8 exhibited extensive co-localization at IR-induced foci (IRIF) from 15 min through 4 h after 10 Gy IR (Fig. 2 A and B), indicating that each is simultaneously present at many of the same DSB regions throughout much of the DNA damage response. Both γH2AX ubiquitination and DSB targeting of several repair proteins are strongly reduced at 30 min following administration of the proteasome inhibitor MG132 (7, 8, 26, 27). Proteasome inhibition reduces nuclear ubiquitin concentrations by preventing degradation of poly-ubiquitinated proteins in the cytosol and regeneration of free ubiquitin pools. MG132 administration is therefore thought to limit the availability of ubiquitin for conjugation reactions by E3 ligases in the nucleus (28). The rapid loss of DSB ubiquitination and targeting events upon proteasome inhibition is consistent with an unidentified DUB activity being responsible for the short half-life of γH2AX-Ub species.
Fig. 2.
Rap80-BRCC36 regulates DSB associated ubiquitination. (A) eRNF8 and endogenous Rap80 co-localize at IRIF 4 h after 10 Gy IR. IF was performed with a mouse monoclonal antibody against the HA epitope to detect eRNF8 and a rabbit polyclonal antibody that was raised to Rap80 amino acids 233 to 710. (B) Quantification of eRNF8 and endogenous Rap80 at IRIF from 15 min to 4 h after 10 Gy IR. The experiment was performed in duplicate with greater than 200 cells examined for each time point. (C) Laser stripes were performed in HeLa cells 48 h after transfection with the indicated siRNA. MG132 (0.5 μM) was added 30 min after stripe induction. Cells were incubated at 37 °C for the indicated time in minutes following MG132 addition, and subsequently fixed and IF performed for conjugated ubiquitin and γH2AX. The 0 min time point indicates 60 min after stripe induction in the absence of MG132. This experiment was performed in duplicate with a minimum of 100 stripes analyzed for each time point. (D) Quantification of strong (purple) versus weakly detectable (yellow) ubiquitin (FK2) stripes as a percentage of the number of γH2AX stripes is displayed graphically. Total fluorescence intensity was calculated for each stripe to set an intensity threshold limit for both strong and weak stripes using a custom designed macro (Phase 3 Imaging) as described in Materials and Methods. (E) HeLa S3 cells expressing shRNA to luciferase (Luc), or either of two different Rap80 target sequences, were treated with 10 Gy IR, and DMSO or 0.5 μM MG132 was added 30 min later. Histones were acid-extracted 30 min after addition of drug and IB performed with an antibody to γH2AX. γH2AX, mono-ubiquitinated γH2AX (γ-H2AX-Ub), and di-ubiquitinated γH2AX (γ-H2AX-Ub2) are indicated.
To test the hypothesis that Rap80-BRCC36 contributes to this putative DSB-associated DUB activity, laser-induced DSBs were introduced in cells treated with Ct, Rap80, or BRCC36 siRNA. MG132 was administered 30 min following DSB induction and cells fixed at the indicated times after drug treatment (Fig. 2 C and D). MG132 treatment resulted in a barely detectable ubiquitin signal at γH2AX-positive stripes in control siRNA-treated cells by 30 min and a nearly complete absence of ubiquitin stripes at 60 min, whereas depletion of either Rap80 or BRCC36 produced a quantifiably stronger DSB ubiquitin signal at all time points, even in the absence of drug (Fig. 2 C and D). Additionally, ubiquitinated γH2AX was maintained 30 min after IR in MG132-treated HeLa cells expressing either of two different shRNAs targeted to Rap80, whereas it was strongly diminished in control shRNA cells (Fig. 2E).
Rap80-BRCC36 and RNF8-Ubc13 Play Opposing Roles in DSB Ubiquitination.
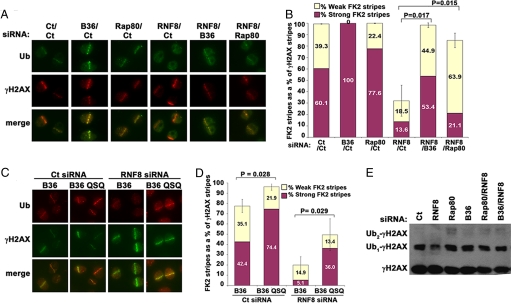
The aforementioned findings suggest that Rap80-BRCC36 may be responsible for degrading K63-Ub at DSBs. Such an activity is predicted to antagonize RNF8-Ubc13-dependent K63-Ub synthesis activities. To test this hypothesis, single and double siRNA knockdown experiments involving Rap80 or BRCC36 and RNF8 were performed in HeLa cells, followed by laser stripe induction and detection of conjugated ubiquitin at γH2AX-positive DSBs (Fig. 3A). Either Rap80 or BRCC36 knockdown alone reproducibly increased the DSB ubiquitin level compared with control-treated cells. Importantly, concomitant knockdown of Rap80 or BRCC36 in combination with RNF8 significantly rescued DSB-associated ubiquitin levels in HeLa cells as detected by the FK2 antibody (Fig. 3 A and B). Similar results were obtained by examining FK2-positive foci formation after IR in cells deficient for either BRCC36 or RNF8 alone or dual deficiency for RNF8 and BRCC36 (Fig. S2). Control experiments demonstrate no obvious influence of RNF8 on Mre11 IRIF (data not shown) as previously reported for NBS1 (8).
Fig. 3.
Rap80-BRCC36 DUB activity reverses RNF8-dependent ubiquitination events at DSBs. (A) HeLa cells were treated with equivalent amounts of siRNA as indicated and then subjected to laser-induced DSBs. Cells were fixed at 30 min following DSB induction and IF performed for γH2AX and conjugated ubiquitin. (B) Quantification of ubiquitin positive stripes (FK2) as performed in Fig. 2. Bars represent the average of at least 3 independent experiments and 150 to 200 stripes. Error bars indicate SEM. P values were calculated by the Student t test. (C) Laser stripes were performed in siRNA-treated U2OS cells that express either BRCC36 wt (B36) or BRCC36 mutant (B36 QSQ). IF for γH2AX and conjugated ubiquitin was performed as indicated. (D) Quantification of strong versus weakly detectable ubiquitin stripes as a percentage of total γH2AX stripes was performed as in Fig. 3B. The displayed figure is a compilation of 3 independent experiments, each done in duplicate. Error bars indicate SEM. P values were calculated by the Student t test. (E) Co-transfection of the indicated siRNA was performed and γH2AX ubiquitination (γH2AX-Ub and γH2AX-Ub2) assessed in HeLa cells 1 h after 10 Gy IR.
To confirm that Rap80-targeted BRCC36 DUB activity is directed to RNF8-Ubc13 dependent substrates, laser stripes were performed in eBRCC36 wt or eBRCC36 QSQ cells transfected with either control or RNF8 siRNA, and DSB-associated conjugated ubiquitin quantified at γH2AX-positive laser stripes. The ubiquitin signal was reproducibly stronger in the eBRCC36 QSQ cells versus cells expressing eBRCC36 wt. Additionally, BRCC36 DUB inhibition significantly rescued the ubiquitin signal at DSBs in RNF8-knockdown cells, whereas the DSB-associated ubiquitin signal in RNF8-depleted eBRCC36 wt cells was obviously reduced (Fig. 3 C and D). These results support the concept that Rap80-BRCC36 DUB activity is an important factor involved in reversing RNF8-Ubc13-dependent DSB ubiquitination.
To determine if Rap80 and BRCC36 depletion could maintain IR-induced γH2AX-Ub levels following RNF8 knockdown, transfection of equivalent amounts of either RNF8 plus control siRNA versus RNF8 plus Rap80 or RNF8 plus BRCC36 siRNA was performed and γH2AX-Ub levels examined by IB. Rap80 or BRCC36 knockdown in combination with RNF8 restored γH2AX-Ub at 1 h after 10 Gy IR in HeLa cells (Fig. 3E). Similar knockdown efficiency for RNF8 was achieved in combination with BRCC36 and Rap80 knockdown (Fig. S3). In sum, these results indicate that opposing ubiquitin synthesis and breakdown activities by RNF8-Ubc13 and Rap80-BRCC36, respectively, are determinants of steady-state DSB ubiquitin levels.
BRCC36 and RNF8 Play Opposing Roles for DSB Recruitment Events and Responses to IR.
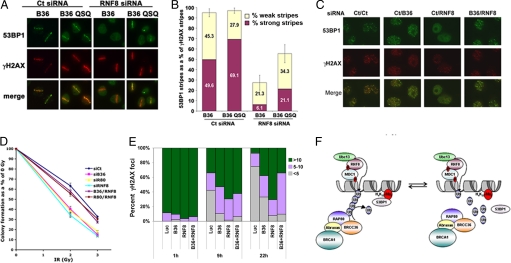
RNF8-Ubc13 ligase activity is required for 53BP1 and BRCA1 recruitment to chromatin adjacent to DSBs (6–8, 10, 26, 27). Therefore, increases in steady-state ubiquitin levels should provide additional docking sites for 53BP1 at DSBs. To directly test the influence of BRCC36 on RNF8-dependent 53BP1 DSB targeting, knockdown of RNF8 was performed in U2OS cells expressing either eBRCC36 wt or eBRCC36 QSQ mutant and DSB stripes examined for 53BP1 (Fig. 4A). Consistent with earlier results establishing that BRCC36 inhibition increases DSB ubiquitination, BRCC36 QSQ cells displayed a quantifiably stronger 53BP1 signal at DSBs. Conversely, RNF8 knockdown dramatically reduced 53BP1 stripe localization in eBRCC36 wt cells as previously reported (7, 8, 10), whereas co-expression of eBRCC36 QSQ resulted in a partial restoration of 53BP1 levels at DSBs (Fig. 4 A and B). Moreover, concomitant BRCC36 and RNF8 siRNA transfected cells displayed increased 53BP1 IRIF compared with RNF8/control siRNA treated cells, which largely failed to exhibit 53BP1 IRIF (Fig. 4C and Fig. S4).
Fig. 4.
BRCC36 deficiency restores 53BP1 DSB recruitment and DNA damage responses in RNF8-depleted cells. (A) Laser-induced DSBs were performed 48 h after control versus RNF8 knockdown in U2OS cells expressing either B36 wt or B36 QSQ. IF for 53BP1 and γH2AX was performed 30 min after stripe induction as indicated. Experiments were performed in triplicate with 150 to 200 stripes examined per group. (B) Quantification of strong versus weak 53BP1 stripes from A as a percentage of total γH2AX stripes. The displayed figure is a compilation of 3 independent experiments, with more than 100 stripes analyzed per sample. Error bars indicate SEM. (C) HeLa cells were co-transfected with siRNA to RNF8 and control, or RNF8 and BRCC36. Transfected cells were treated with 10 Gy IR at 48 h after transfection, and IF for 53BP1 and γH2AX was performed 5 h later. (D) Co-transfection of RNF8, Rap80 (R80), or BRCC36 siRNA with control siRNA or co-transfection of RNF8 siRNA with either Rap80 or BRCC36 siRNA was performed in HeLa cells, and the indicated doses of IR were administered 48 h later. Colony formation at 12 days after IR treatment was normalized as a percentage of colony formation at 0 Gy for each point. (E) γH2AX foci formation was quantified in HeLa cells at different times after 10 Gy IR in cells that were transfected with the indicated pairs of siRNA. The percentage of cells containing each number of γH2AX is displayed graphically and was derived from duplicate experiments with more than 200 cells counted each time. (F) Model for opposing regulation of poly-ubiquitination events at DSBs. Equilibrium synthesis and breakdown of poly-ubiquitin on common substrates by RNF8-Ubc13 and Rap80-BRCC36, respectively, is established at early stages of DSB repair. This dynamic equilibrium is predicted to determine steady-state levels of DSB-associated ubiquitin and DSB recruitment for BRCA1 and 53BP1 protein complexes.
Consistent with these findings, a functional interrelationship was observed between BRCC36 DUB and RNF8 ligase activities with respect to viability responses to IR (Fig. 4D). siRNA-mediated depletion of RNF8, BRCC36, or Rap80 resulted in reduced viability in response to 2 and 3 Gy IR compared with control siRNA-treated HeLa cells. However, double knockdown of RNF8 in combination with either BRCC36 or Rap80 largely rescued viability in response to IR, revealing that concomitant reduction in these opposing pathways can restore cellular sensitivity to IR (Fig. 4D). Similar interactions between E3 and DUB activity with respect to viability after IR were observed following knockdown of RNF8 in eBRCC36 wt versus eBRCC36 QSQ-expressing cells (Fig. S5). Viability responses to IR correlated with the kinetics of γH2AX disappearance, suggesting partial correction of DSB repair efficiency upon simultaneous reductions of RNF8 and BRCC36 proteins (Fig. 4E). RNF8 or BRCC36 knockdown increased the average cellular γH2AX foci number at each time point after IR, whereas depletion of both BRCC36 and RNF8 partially reduced γH2AX foci number compared with RNF8 deficiency in combination with control siRNA at 22 h after IR.
Discussion
Equilibrium signaling reactions exist for nearly every cellular process, providing a regulatory mechanism to maintain cellular and organismal homeostasis. The ubiquitin proteasome system represents a prominent example of such regulation, in which proteasome-dependent protein degradation is impaired by deletion of either proteasome-associated E3 ligase or DUB activities, but restored upon concomitant deletion of both, supporting the idea that ubiquitin remodeling activities are critical to proteasome function (29). De-ubiquitination of the Fanconi Anemia D2 protein, FANCD2, by the USP1 DUB represents a second example in which DUB activity is necessary to terminate DNA damage-inducible ubiquitination events (30). Failure to either ubiquitinate FANCD2 or to de-ubiquitinate FANCD2-Ub confers sensitivity to DNA cross-linking agents (31, 32).
The data presented here reveal the existence of a Rap80-BRCC36 directed pathway to control equilibrium ubiquitination levels on chromatin during DSB repair (Fig. 4F). This model predicts that RNF8-Ubc13-synthesized K63-Ub structures attract Rap80-BRCC36 DUB complexes to terminate DSB-associated ubiquitin targeting signals. It is supported by several key findings: (i) RNF8-Ubc13 and Rap80-BRCC36 co-localize at DSBs throughout the course of DSB repair; (ii) knockdown of either Rap80 or BRCC36, or BRCC36 DUB inhibition, increases DSB-associated conjugated ubiquitin and 53BP1 DSB targeting; and (iii) dual inhibition of Rap80-BRCC36 and RNF8 partially restores DSB-associated ubiquitination, γH2AX-Ub levels, and viability responses to IR. Thus, activation (i.e., RNF8-Ubc13-dependent) and termination (i.e., Rap80-BRCC36-dependent) reactions occur together during early stages of DNA repair and are necessary for long-term viability in response to IR.
Although BRCC36 DUB activity contributes to viability responses following IR, the mechanism by which it does so is still incompletely understood. Reduced viability in response to IR in BRCC36 mutant cells could be caused by a failure to terminate RNF8-Ubc13-dependent DNA damage responses centered on γH2AX and perhaps other substrates at DSBs. Evidence does exist that increased γH2AX signaling may result in IR sensitivity. The PP2A phosphatase dephosphorylates γH2AX, and indeed, PP2A deficiency renders cells IR-sensitive (33). Additionally, sustained levels of mono-ubiquitinated FANCD2 in USP1-null cells confers reduced viability after DNA damage (31). Thus, opposing activation and termination responses on γH2AX and other proteins involved in DNA repair appear to be crucial to maintain viability after DNA damage. Consistent with this hypothesis, simultaneous depletion of Rap80-BRCC36 and RNF8 mitigates the IR supersensitivity observed upon knockdown of either Rap80-BRCC36 or RNF8 alone, reduces the persistence of γH2AX foci, and partially restores G2 checkpoint responses (Fig. 4 D and E and Fig. S6).
The balance of forward and reverse signaling reactions determine the outcome of a wide range of cellular processes ranging from metabolism to cellular proliferation and differentiation. Our findings provide additional support that equilibrium signaling involving ubiquitin metabolism at DSBs is also a critical determinant of DNA damage responses. It is possible that other DUBs, such as histone H2A DUBs USP3 and USP16 (34, 35), are also involved in negative regulation of RNF8-Ubc13-dependent events at DSBs. Indeed, BRCC36 knockdown does not completely rescue DSB-associated ubiquitin levels following proteasome inhibition or RNF8 knockdown, suggesting additional mechanisms could also be involved in removing ubiquitin at DSBs. However, unlike BRCC36, neither USP3 nor USP16 were found to localize to γH2AX-positive DSBs (Fig. S7), suggesting that they may not directly participate in an equilibrium mechanism to reverse RNF8-dependent events at DSBs. In addition, whereas Ubc13 has been reported to strongly influence DSB recruitment for Rad51, NBS1, and BRCA1 (17), RNF8 appears to control a more limited subset of repair factors at DSBs (7, 8), suggesting that Ubc13 may pair with other E3 ligases to control DSB recruitment. Therefore, these ubiquitination/de-ubiquitination pathways may not be completely linear.
BRCC36 is a specialized protease that, in principle, could be subject to small molecule inhibition. Indeed specific inhibitors have been developed for metallo-proteases involved in other biological processes (36). Thus, one could envisage potential in vivo strategies to pharmacologically manipulate DSB recruitment events and viability responses to DNA-damaging agents by modulating BRCC36 DUB activity.
Materials and Methods
IR Sensitivity.
Cells were transfected with the indicated siRNAs using Oligofectamine (Invitrogen). Cells were trypsinized and seeded at low density in 60 mm dishes (400 cells per dish) 24 h after transfection. Cells were irradiated the next day at the indicated doses, and incubated for 10 to 12 d. Resultant colonies were stained with Giemsa stain and counted. Colony formation was normalized for each siRNA-transfected population as a percentage of colonies formed at 0 Gy.
Immunoprecipitation.
Flag immunoprecipitation was performed in NETN buffer (10 mM TrisCl, pH 7.4, 1 mM EDTA, 0.05% Nonidet P-40, and protease inhibitors) containing 150 mM NaCl using anti-Flag M2 agarose beads (Sigma) and eluted with Flag peptide.
Laser-Generated DNA DSBs.
Laser-induced DNA DSBs and immunofluorescence (IF) was performed as previously described (5, 21) using a PALM MicroBeam laser microdissection system (Carl Zeiss MicroImaging).
Quantification of Stripes.
Following IF, 8-bit gray-scale pictures were taken of all stripes with a QImaging RETIGA-SRV camera coupled to a Nikon Eclipse 80i microscope using ImagePro 6.2. It should be noted that all images were captured at identical exposure times that were carefully selected so as to avoid saturation at any individual stripe. To discern strong versus weak stripes, a fluorescence intensity threshold was set for images based on a black and white intensity scale ranging between 0 and 255. Generally, threshold ranges fell between 100 and 255. The specific range for an experiment was selected before analysis to include strong intensity stripes within the threshold range while excluding apparent weaker stripes. Thus, strong stripes were identified as those falling within the selected intensity range, and weak stripes were identified as those that were clearly visible above background but did not fall within the threshold range.
siRNA and shRNA Constructs.
siRNA oligonucleotide duplexes were purchased from Dharmacon and Invitrogen. Transfections were performed in 6-well tissue culture dishes according to manufacturer's instructions using one of the following transfection reagents: Oligofectamine (Invitrogen), Hiperfect (Qiagen), or Lipofectamine RNAiMax (Invitrogen). siRNA sequences are listed in SI Methods.
Supplementary Material
Acknowledgments.
We thank T. Messick, N. Shanbhag, and T. Huang for critical discussion of the data, and B. Luo and H. Lodish for the retroviral shRNA expression vector. This study was supported by National Cancer Institute K08 award 1K08CA106597–01, the Sidney Kimmel Foundation Scholar Award, the Mary Kay Ash Charitable Foundation Cancer Research Grant, funds from the Abramson Family Cancer Research Institute (to R.A.G.), and National Institutes of Health training Grant T32-GM-007229 (to K.A.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.M.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807485106/DCSupplemental.
References
- 1.Bekker-Jensen S, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 3.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–260. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- 6.Huen MS, et al. RNF8 Transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 11.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 14.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 15.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 17.Zhao GY, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 18.Yan J, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 20.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117:305–317. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 25.Ambroggio XI, Rees DC, Deshaies RJ. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and Fanconi anemia pathway activation. Cancer Res. 2007;67:7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 27.Murakawa Y, et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 28.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 30.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Oestergaard VH, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 33.Chowdhury D, et al. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Joo HY, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 35.Nicassio F, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 36.Hu J, Van den Steen PE, Sang QX, Opdenakker G. Matrix metalloproteinase inhibitors as therapy for inflammatory and vascular diseases. Nat Rev Drug Discov. 2007;6:480–498. doi: 10.1038/nrd2308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.