Abstract
Direct control of microRNA (miRNA) expression by oncogenic and tumor suppressor networks results in frequent dysregulation of miRNAs in cancer cells and contributes to tumorigenesis. We previously demonstrated that activation of the c-Myc oncogenic transcription factor (Myc) broadly influences miRNA expression and in particular leads to widespread miRNA down-regulation. miRNA transcripts repressed by Myc include several with potent tumor suppressor activity such as miR-15a/16–1, miR-34a, and let-7 family members. In this study, we have investigated mechanisms downstream of Myc that contribute to miRNA repression. Consistent with transcriptional down-regulation, Myc activity results in the decreased abundance of multiple miRNA primary transcripts. Surprisingly, however, primary transcripts encoding several let-7 miRNAs are not reduced in the high Myc state, suggesting a posttranscriptional mechanism of repression. The Lin-28 and Lin-28B RNA binding proteins were recently demonstrated to negatively regulate let-7 biogenesis. We now show that Myc induces Lin-28B expression in multiple human and mouse tumor models. Chromatin immunoprecipitation and reporter assays reveal direct association of Myc with the Lin-28B promoter resulting in transcriptional transactivation. Moreover, we document that activation of Lin-28B is necessary and sufficient for Myc-mediated let-7 repression. Accordingly, Lin-28B loss-of-function significantly impairs Myc-dependent cellular proliferation. These findings highlight an important role for Lin-28B in Myc-driven cellular phenotypes and uncover an orchestration of transcriptional and posttranscriptional mechanisms in Myc-mediated reprogramming of miRNA expression.
In eukaryotic cells, small RNAs participate in diverse processes including the regulation of gene expression, control of chromatin structure, and suppression of transposons (1). Among the various families of small RNAs, microRNAs (miRNAs) have attracted considerable attention because of their increasingly recognized importance in development and disease (2). miRNAs are 18–24 nucleotide RNAs that regulate the translation and stability of target messenger RNAs (mRNAs). The biogenesis of miRNAs initiates with RNA polymerase II-mediated transcription of long primary transcripts known as pri-miRNAs, which are capped, polyadenylated, and usually spliced (3). Within these transcripts, a region containing the miRNA sequence forms a double stranded hairpin structure that is excised in the nucleus by the endonuclease Drosha and its obligate partner DGCR8. The liberated miRNA hairpin, referred to as the premiRNA, is exported to the cytoplasm where it undergoes further processing by the Dicer endonuclease to produce a short double-stranded RNA duplex. One strand of this duplex, destined to become the mature miRNA, associates with a member of the argonaute family of proteins, core components of a large assembly known as the RNA-induced silencing complex (RISC). The miRNA then guides RISC to sites of imperfect complementarity within mRNAs, leading to translational repression or accelerated turnover of the target.
Early studies of miRNA function in C. elegans and Drosophila revealed important roles for these RNAs in the regulation of cellular differentiation, proliferation, and death (4–6). Consistent with these activities, a large body of evidence has since documented that miRNA dysfunction contributes to cancer pathogenesis in humans. miRNA expression patterns in cancer cells are globally abnormal compared with normal tissues (7). These expression patterns are highly informative for tumor classification, staging, and prognosis. Moreover, specific miRNAs possess potent oncogenic and tumor suppressor activity, demonstrating that miRNA gain- and loss-of-function functionally contributes to tumorigenesis (8). For example, the miR-17–92 cluster, a group of 6 miRNAs cotranscribed from a locus on human chromosome 13, is frequently amplified in B lymphoma and other tumor types (9). Enforced expression of these miRNAs promotes tumorigenesis in animal models of hematopoietic and solid malignancies (10, 11). Conversely, the let-7 family of miRNAs are known to act as tumor suppressors. These miRNAs down-regulate oncogenes such as Ras, Myc, and HmgA2 and inhibit proliferation and tumorigenesis when ectopically expressed (12–20). Accordingly, multiple loci that encode let-7 miRNAs are frequently deleted in cancer cells (21).
Multiple mechanisms contribute to aberrant miRNA expression in cancer cells. We and others have previously demonstrated that miRNAs are critical downstream components of key oncogenic and tumor suppressor signaling pathways (22, 23). Gain- or loss-of-function of these pathways in tumor cells directly influences miRNA expression, which consequently affects malignant cellular behavior. This is well illustrated by the program of miRNA expression controlled by the c-Myc oncogenic transcription factor (Myc), one of the most commonly activated oncoproteins in human cancer (24). Myc directly induces expression of the miR-17–92 cluster, which subsequently promotes proliferation, inhibits apoptosis, and promotes tumor angiogenesis (10, 11, 25, 26). Furthermore, Myc activation leads to widespread repression of many additional miRNA genes including several with known tumor suppressor activity such as members of the let-7 family, miR-15a/16–1, the miR-29 family, and miR-34a (27). Myc associates with the promoters of these miRNA genes, suggesting direct transcriptional repression. Enforced expression of several of these miRNAs potently suppresses tumorigenesis, supporting a critical role for miRNA repression in the Myc oncogenic program. Interestingly, widespread miRNA down-regulation has been observed in many human tumors (28, 29). Given the frequency of Myc hyperactivity in cancer, this oncoprotein likely contributes to this phenomenon.
In the present study, we have investigated mechanisms downstream of Myc, which contribute to miRNA repression. Consistent with transcriptional repression, we find that the abundance of multiple pri-miRNAs is reduced in the high Myc state. Unexpectedly, however, we observed that despite repression of mature let-7 family members upon Myc activation, the levels of multiple let-7 primary transcripts are not reduced under these conditions. This observation suggests posttranscriptional regulation of let-7 expression. Recently, the Lin-28 and Lin-28B RNA binding proteins were described as negative regulators of let-7 biogenesis (30–34). These proteins directly inhibit both Drosha- and Dicer-mediated processing of let-7 family members and accelerate decay of let-7 precursors. Here, we demonstrate that Lin-28B is induced by Myc and is necessary and sufficient for Myc-mediated repression of let-7 miRNAs. Accordingly, loss-of-function of Lin-28B impairs Myc-mediated proliferation. These findings provide insight into mechanisms that contribute to miRNA repression in cancer and highlight an important role for let-7 repression in Myc-driven cellular phenotypes.
Results
Posttranscriptional Repression of let-7 Family Members by Myc.
We previously demonstrated that activation of Myc in human and mouse models of B cell lymphoma leads to up-regulation of the miR-17–92 cluster and down-regulation of numerous miRNAs including multiple let-7 family members, miR-22, and miR-34a (26, 27). To investigate the mechanisms through which Myc regulates these miRNAs, we used quantitative real-time PCR (qPCR) to examine the abundance of their primary transcripts in P493-6 cells, a model of human B cell lymphoma with tetracycline (tet)-repressible Myc expression (35). Consistent with regulation at the transcriptional level, we observed induction of the primary transcript encoding the miR-17–92 cluster and repression of the primary transcripts encoding miR-34a and miR-22 in the high Myc state (Fig. 1A). In contrast, we unexpectedly found that the abundance of the let-7a-1/let-7f-1/let-7d and let-7g primary transcripts were not repressed under these conditions. We previously showed that the mature miRNAs derived from these transcripts are down-regulated under conditions of high Myc activity (27). qPCR further confirmed that mature let-7g is down-regulated by ≈2-fold by Myc in these cells (Fig. 1B). These findings demonstrate that Myc utilizes multiple mechanisms to repress miRNA expression and specifically down-regulates multiple let-7 family members through a posttranscriptional pathway.
Fig. 1.
Posttranscriptional repression of let-7 expression by Myc correlates with Lin-28B induction. (A and B) qPCR analysis of the abundance of pri-miRNAs (A) and mature let-7g (B) in P493-6 cells with high or low Myc expression. (C) RT-PCR analysis of Lin-28 and Lin-28B expression in P493-6 cells. DNA Plasmids bearing the ORF of Lin-28 or Lin-28B were used as templates for positive controls. (D) qPCR analysis of Lin-28B expression in P493-6 cells. (E) Western blot of Lin-28B abundance in P493-6 cells after removal of tet. Signals were normalized to α-tubulin levels before calculating fold changes. (F) qPCR analysis of Lin-28B expression in murine B cell lymphomas. Two independent tumors from MycON and MycOFF neoplasms were assayed. (G) qPCR analysis of Lin-28B expression in Ras-transformed p53-null colon carcinomas. Four independent tumors from Ras/Myc and Ras-only neoplasms were assayed. For all qPCR analyses, error bars represent standard deviations derived from 3 independent measurements.
Myc-Mediated Induction of Lin-28B Expression.
Recently, the Lin-28 and Lin-28B RNA binding proteins were demonstrated to bind directly to let-7 stem loops and inhibit biogenesis of these miRNAs by blocking both Drosha- and Dicer-mediated cleavage and accelerating decay of let-7 precursors (30–34). We therefore speculated that induction of Lin-28 or Lin-28B by Myc could mediate repression of let-7 family members. To test this hypothesis, we first used RT-PCR to assay for expression of Lin-28 transcripts in P493-6 cells. Although Lin-28 expression was undetectable, Lin-28B was expressed in these cells specifically in the high Myc state (Fig. 1C). qPCR and Western blot analysis further documented that induction of Myc results in increased Lin-28B mRNA and protein abundance in this system (Fig. 1 D and E). To confirm that Lin-28B is induced by Myc in other settings, we examined 2 additional in vivo models with regulated Myc expression: a mouse B lymphoma model with estradiol-regulated Myc activity (36) and a mouse colon cancer model in which cells are transformed by Ras alone (low Myc) or by simultaneous expression of Ras and Myc (high Myc) (10). In tumor tissue from both model systems, activation of Myc resulted in elevated Lin-28B expression (Fig. 1 F and G).
Myc Directly Transactivates Lin-28B Expression.
To determine whether Lin-28B is a direct Myc target gene, we first performed chromatin immunoprecipitation (ChIP) to assess whether Myc associates with the genomic locus that encodes this protein. When transactivating target genes, Myc binds to enhancer (E)-box sequences, typically CACGTG or CATGTG (37). Within a 10-kb window centered on the transcriptional start site (TSS) of Lin-28B, 7 putative Myc binding sites conforming to these sequences are present on either strand, with 3 sites showing conservation between human and mouse (Fig. 2A, conserved sites in red). Real-time PCR amplicons were designed to assay for all 7 putative binding sites in ChIP samples. Clear evidence for Myc association with this locus was obtained with strongest binding occurring within a region containing a conserved E-box located within the first intron of Lin-28B (Fig. 2B, amplicon 3). Myc is known to frequently bind to sites in the first intron of its transcriptional target genes (37). Signals were dramatically reduced when Myc expression was inhibited by treatment with tet, demonstrating the specificity of these findings. No evidence was obtained for Myc binding to regions located further upstream or downstream of Lin-28B intron 1 (Fig. 2B, amplicons 1, 6, and 7). These data demonstrate that Myc interacts directly with the Lin-28B promoter.
Fig. 2.
Lin-28B is a direct Myc target gene. (A) Schematic representation of the genomic region near the transcription start site of Lin-28B. Putative Myc binding sites are indicated (CACGTG or CATGTG); those in red are conserved between human and mouse. The plot depicts evolutionary conservation and was generated from the University of California, Santa Cruz genome browser (human genome March 2006 assembly). qPCR amplicons are represented by numbered lines. (B) qPCR analysis of Myc chromatin immunoprecipitates. Fold enrichment represents signal obtained after Myc immunoprecipitation relative to signal obtained with irrelevant antibody. Error bars represent standard deviations derived from 3 independent measurements. (C) Activity of promoter reporter constructs in P493-6 cells with high or low Myc expression. Values shown represent relative firefly luciferase activity produced from each reporter construct normalized to renilla luciferase activity produced from a cotransfected control vector. Error bars represent standard deviations derived from 3 independent measurements. An internal ribosome entry site (IRES) upstream of luciferase was included in these vectors because several out-of-frame translation initiation codons are present in the cloned genomic fragments. E, potential Myc binding site (E-box).
To directly examine the ability of Myc to transactivate the Lin-28B promoter, a genomic fragment extending from 1.4 kb upstream to 1.4 kb downstream of the TSS was cloned upstream of a promoterless luciferase reporter cassette (Fig. 2C). This region contains the E-boxes downstream of the TSS, which exhibited Myc binding. This fragment was sufficient to drive Myc-dependent reporter activity when introduced into P493-6 cells (Fig. 2C, construct P1). Equivalent Myc-dependent reporter activity was conferred by a truncated fragment extending from 266 bp upstream to 1.4 kb downstream of the TSS that also contains the E-boxes (Fig. 2C, construct P2) whereas a fragment lacking the E-boxes did not exhibit transcriptional activity (Fig. 2C, construct P3). Taken together with the results demonstrating Myc-mediated up-regulation of Lin-28B expression and direct binding of Myc to the Lin-28B promoter, these data provide compelling evidence that Lin-28B is a direct Myc target gene.
Lin-28B Loss-of-Function Impairs Myc-Mediated let-7 Repression and Proliferation.
To test whether Lin-28B is necessary for Myc-mediated repression of let-7 family members, we used 2 different short interfering RNAs (siRNAs) to inhibit Lin-28B expression in P493-6 cells in the high Myc state. Significant knockdown (≈70–80% reduction in Lin-28B mRNA abundance compared with negative control siRNA) was observed as early as 24h after siRNA delivery and reduced Lin-28B levels were maintained for at least 96h (Fig. 3A). Lin-28B knockdown completely reversed Myc-mediated repression of mature let-7g, documenting its essential role in this process (Fig. 3B). Furthermore, P493-6 cells with Lin-28B inhibition exhibited markedly impaired proliferation (Fig. 3C). Thus, Lin-28B provides functions that are critical for Myc-mediated cellular phenotypes including let-7 repression and enhanced proliferation.
Fig. 3.
Lin-28B knockdown reverses let-7 repression and slows proliferation. (A and B) P493-6 cells were grown the absence of tet (high Myc state) and transfected with 2 different siRNAs targeting Lin-28B or negative control siRNA. qPCR was used to measure Lin-28B (A) or mature let-7g abundance (B). (C) Proliferation of P493-6 cells after siRNA delivery. Error bars represent standard deviations of measurements of triplicate transfections. (D) Proliferation of P493-6 cells after transfection of siRNA and miRNA inhibitors. Error bars represent standard deviations of measurements of triplicate transfections. P value calculated by 2-tailed t test.
Let-7 family members are known to have potent anti-proliferative activity (13, 38). We therefore hypothesized that the impaired proliferation of P493-6 cells lacking Lin-28B function was a consequence of elevated levels of these miRNAs. To test this directly, we coadministered Lin-28B siRNA along with let-7 antisense inhibitors. Because our earlier studies documented that P493-6 cells express the let-7a-1/let-7f-1/let-7d, miR-99a/let-7c/miR-125b-2, and let-7g transcription units (27), we used a pool of inhibitors directed against let-7a, let-7c, let-7d, let-7f, and let-7g to simultaneously reduce activity of all family members expressed from these loci. Northern blot analysis confirmed a reduction in the abundance of let-7a, let-7d, and let-7g after administration of the inhibitors (Fig. S1). It was not possible to assess let-7c and let-7f knockdown because high levels of cross-hybridization preclude accurate measurement of these miRNAs by Northern blot analysis (27). The let-7 inhibitors had no effect on proliferation of cells transfected with control siRNA (Fig. 3D). In contrast, the let-7 inhibitors caused a moderate, but statistically-significant, increase in the rate of proliferation of cells transfected with Lin-28B siRNA. Therefore, activity of let-7 miRNAs contributes to the slow growth phenotype associated with Lin-28B knockdown. It is possible that residual function of let-7 family members in inhibitor-treated cells prevented a more complete rescue of the Lin-28B knockdown phenotype. However, these data are also consistent with the existence of additional let-7-independent functions of Lin-28B that are necessary for normal rates of proliferation.
Lin-28B Plays a Limited Role in Myc-Mediated miRNA Repression Beyond the let-7 Family.
We next investigated the broader role of Lin-28B in the regulation of Myc-responsive miRNAs and protein-coding genes. To examine the consequences of Myc-mediated induction of Lin-28B in isolation from the more widespread effects of Myc hyperactivity, P493-6 cells were infected with a retrovirus that stably expresses Lin-28B (MSCV-Lin-28B) or empty vector (MSCV-empty) as a control. Importantly, the level of enforced Lin-28B expression in these cells in the low Myc state (+tet) closely mirrored the endogenous level of expression that is induced in the high Myc state (−tet) (Fig. 4A). Tet-treated P493-6 cells with enforced expression of Lin-28B fully recapitulate Myc-mediated repression of let-7a, let-7d, and let-7g (Fig. 4B). Taken together with the results from the knockdown experiments (Fig. 3 A and B), these data demonstrate that Lin-28B is necessary and sufficient for Myc-mediated let-7 repression. As expected, miR-34a was not repressed by Lin-28B, consistent with our earlier findings suggesting transcriptional repression of this miRNA by Myc (Fig. 1A and 4B). Of note, enforced expression of Lin-28B in this cell line is not sufficient to drive Myc-independent proliferation and does not further enhance proliferation rates in the high Myc state (data not shown).
Fig. 4.
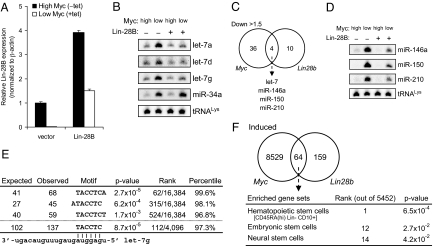
Enforced Lin-28B expression fully recapitulates Myc-mediated let-7 repression but has limited effects on other Myc-repressed miRNAs. (A) qPCR analysis of Lin-28B expression in P493-6 cells infected with MSCV-empty or MSCV-Lin-28B. (B) Northern blot analysis of miRNAs in retrovirally-infected cells. We used the conditions described in ref. 27 that prevent cross-hybridization between let-7 family members. tRNALys served as a loading control. (C) Comparison of Myc-repressed and Lin-28B-repressed miRNAs as assayed by microarray. Individual let-7 family members are not distinguishable on the array because of cross-hybridization and are therefore treated as 1 group. (D) Northern blot validation of miRNA microarray results. (E) Motifs complementary to the let-7 seed region are enriched in the 3′ UTRs of Lin-28B-up-regulated transcripts. Expected and observed counts for the corresponding motifs in up-regulated 3′ UTRs are shown. Rank and percentile columns indicate the relative enrichment of a given motif compared with all 16,384 possible heptamer motifs (above dotted line) or all 4,096 possible hexamer motifs (below dotted line). (F) Comparison of Myc-induced and Lin-28B-induced mRNAs as assayed by microarray. Gene sets characteristic of multiple types of stem cells are enriched in the transcripts up-regulated by both proteins. P values were calculated by the Molecular Signatures Database available as part of the Gene Set Enrichment Analysis toolset (www.broad.mit.edu/gsea/msigdb/annotate.jsp) and are not corrected for multiple hypothesis testing. The rank of enrichment of each gene set is shown relative to all 5,452 genes sets tested.
We next used a custom microarray to determine whether Lin-28B contributes to Myc-mediated repression of additional miRNAs (Table S1). Comparison of miRNAs repressed >1.5-fold after Myc-activation (MSCV-empty −tet vs. MSCV-empty +tet) to miRNAs repressed by Lin-28B (MSCV-Lin-28B +tet vs. MSCV-empty +tet) revealed only 3 miRNAs other than let-7 family members shared between the conditions (miR-146a, miR-150, and miR-210; Fig. 4C). Northern blot analysis validated these findings but also demonstrated that enforced Lin-28B expression only partially recapitulates Myc-mediated repression of these miRNAs (Fig. 4D). Thus, Lin-28B plays a limited role in the widespread program of Myc-mediated miRNA repression beyond regulating let-7 family members.
To examine the broader impact of Lin-28B-mediated repression of let-7 on gene expression, we performed Affymetrix microarray analysis. 538 genes with annotated 3′ UTRs were measurably up-regulated (>1.2-fold) in cells with enforced Lin-28B expression. Within this set of 3′ UTRs, heptamer and hexamer motifs that contain matches to the let-7 seed sequence were significantly enriched (Fig. 4E). Thus, Lin-28B-mediated repression of this family of miRNAs has a measurable impact on gene expression in these cells. We did not observe enrichment for matches to the seed sequences of the additional miRNAs repressed by both Myc and Lin-28B (miR-146a, miR-150, and miR-210) further supporting the conclusion that regulation of let-7 miRNAs is the major function of Lin-28B in the Myc program of miRNA repression.
Discussion
Lin-28 was first described as an important regulator of early developmental timing in C. elegans (39). The mammalian orthologue of Lin-28 also plays a role in early development and is highly abundant in embryonic stem (ES) cells and during embryogenesis after which its expression decreases (40). An additional Lin-28 homolog, Lin-28B, was identified as a highly expressed gene in hepatocellular carcinoma cells (41). Recently, Lin-28 and Lin-28B were demonstrated to function as negative regulators of let-7 biogenesis (30–34). These proteins are members of a highly conserved family of orthologous RNA binding proteins that contain a cold shock domain and dual zinc fingers. Through their ability to directly interact with the loop region of let-7 hairpins, Lin-28 proteins accelerate turnover of let-7 precursors and prevent both Drosha- and Dicer-mediated let-7 processing.
We have shown previously that in human and mouse models of B cell lymphoma, induction of the Myc oncoprotein leads to widespread repression of miRNA expression, including members of the let-7 family (27). We now demonstrate that induction of Lin-28B fully accounts for the ability of Myc to repress let-7 miRNAs in a model B lymphoma cell line. Both loss- and gain-of-function experiments demonstrate that Lin-28B is necessary and sufficient for suppression of let-7 expression after Myc activation. Our earlier experiments indicated that Myc associates directly with the let-7g proximal promoter and a conserved element upstream of the let-7a-1/let-7f-1/let-7d cluster. Nevertheless, in light of the results described here, it is apparent that Myc binding to these loci does not result in transcriptional repression. Genome-wide ChIP analyses have shown that only a subset of Myc-bound loci exhibit expression changes in response to Myc activation (37), which likely explains this apparent discrepancy. Importantly, we have documented a reduction in the abundance of pri-miRNAs encoding other repressed miRNAs whose promoters are bound by Myc such as miR-22 and miR-34a, consistent with a mechanism that does involve direct transcriptional down-regulation. These results demonstrate that Myc utilizes multiple effector pathways, both transcriptional and posttranscriptional, to elicit broad miRNA repression.
Although Lin-28B plays a highly specific role in miRNA repression downstream of Myc, induction of this protein, and the resulting down-regulation of let-7 miRNAs, can strongly influence cellular behavior. We have shown that Lin-28B loss-of-function significantly impairs Myc-driven proliferation in a manner that is at least partly dependent on a resulting up-regulation of let-7 miRNAs. Consistent with this result, a large body of evidence has documented potent anti-proliferative and tumor suppressor activity for the let-7 family. Eight distinct loci in the human genome encode let-7 homologs and 4 of these loci are known to be within intervals that are frequently deleted in human cancers (21). Moreover, low let-7 expression in tumors of lung cancer patients portends poor prognosis (42, 43). In keeping with these observations, enforced expression of let-7 inhibits cellular proliferation in vitro and suppresses tumorigenesis in vivo (12, 13, 16). These effects are attributable to the ability of let-7 to down-regulate a program of oncogenes including Ras, HmgA2, and even Myc itself (14, 15, 17–20). Therefore, our findings establish a Myc-Lin-28B-let-7 regulatory circuit that may act in a positively reinforcing manner and likely contributes to Myc-mediated oncogenesis.
Notably, expression of Myc along with 3 other factors is sufficient to induce the reprogramming of somatic cells into a pluripotent state (44). Lin-28 can substitute for Myc in the formation of these induced pluripotent stem (iPS) cells (45). It is therefore tempting to speculate that the induction of Lin-28 or Lin-28B is a key downstream effect of Myc activity in iPS cell generation. Consistent with this notion, Myc has been documented to associate with the Lin-28 promoter in embryonic stem (ES) cells (46). Although Myc binding to the Lin-28B promoter was not assayed in this earlier study, our data raise the possibility that Myc activates Lin-28B in ES cells as well. Further support for a role for Lin-28B downstream of Myc in iPS cell generation is evident in the gene expression data obtained in the course of our study. A strict statistical algorithm (see methods) was used to identify genes regulated by Myc and Lin-28B in P493-6 cells (Table S2 and Table S3). Based on these stringent criteria, 64 genes are induced by both Myc activation and enforced Lin-28B expression (Fig. 4F). Gene set enrichment analysis (47) reveals overlap between these genes and genes expressed highly in ES cells, neural stem cells, and specific subsets of hematopoietic stem cells [CD45RA(hi) Lin- CD10+ cells]. Although future studies are necessary to evaluate the functional role of Lin-28B and let-7 repression in Myc-mediated iPS cell generation, these findings suggest that the Myc-Lin-28B-let-7 pathway described here impacts cellular behavior and identity beyond the context of cancer cell biology.
Materials and Methods
Cell Culture.
P493-6 cells (from D. Eick) were cultured in RPMI medium 1640 media supplemented with 10% FBS (FBS), penicillin, and streptomycin. To repress Myc expression, cells were grown in the presence of 0.1 μg/mL tetracycline (Sigma) for 72 h. Murine lymphoma cells and Ras-transformed p53-null colonocytes with high and low Myc were obtained as described (10, 36).
RT-PCR and qPCR.
Total RNA was isolated using TRIzol (Invitrogen) and further purified using RNeasy columns (Qiagen) with DNase I digestion. Reverse-transcription was performed using the SuperScript First-strand Synthesis System (Invitrogen) with random hexamers before PCR amplification. Quantitative PCR was performed using an ABI 7900 Sequence Detection System with the SYBR Green PCR core reagent kit (Applied Biosystems). Mature let-7g was quantified using a predesigned TaqMan MicroRNA Assay (Applied Biosystems). Eukaryotic 18S rRNA endogenous control (Applied Biosystems) was used as an internal standard. Primer sequences are provided in Table S4.
Lin-28B and let-7 Knockdown.
P493-6 cells were grown to ≈5 × 105/mL at the time of transfection. siRNAs targeting human Lin-28B (sequences provided in Table S4), nontargeting negative control siRNA, human let-7 inhibitors, and negative control inhibitor were purchased from Dharmacon. Before transfection, let-7 inhibitor was prepared by mixing an equal amount of let-7a-1, let-7c, let-7d, let-7f-1, and let-7g inhibitors. Transfection of siRNA and miRNA inhibitors was performed using an Amaxa Nucleofection device using Kit V according to the manufacturer's protocol. Briefly, 2 μM siRNA with or without 2 μM miRNA inhibitor were transfected into 2.5 × 106 P493-6 cells. After transfection, cells were harvested every 24 h for the indicated assays. Cell proliferation was monitored with the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.).
Retroviral Expression of Lin-28B.
Lin-28B was amplified from P493-6 cDNA and cloned into the XhoI site of the retroviral vector MSCV-PIG (48). Primer sequences are provided in Table S4. After transfection of Phoenix packaging cells (G. Nolan, Stanford University, Stanford, CA), retroviral supernatants were collected, filtered, and added to recipient P493-6 cells for 8 h in the presence of 2 μg/mL polybrene. Two days after infection, puromycin was added to the media at 2 μg/mL, and cell populations were selected for 2 weeks.
Northern Blot Analysis.
Northern blot analysis was performed as described in ref. 26 using Ultrahyb-Oligo (Ambion) and oligonucleotide probes perfectly complementary to the mature miRNA sequences. For let-7 miRNAs, optimized wash temperatures were used to prevent cross-hybridization, as described in ref. 27. Additional methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Eric Moss (University of Medicine and Dentistry of New Jersey, Stratford, NJ) for Lin-28B antibody and Chunfa Jie for assistance with microarray data analysis. J.T.M. is a Rita Allen Foundation Scholar and a Leukemia and Lymphoma Society Scholar. This work was supported by the Sol Goldman Center for Pancreatic Cancer Research and the Lustgarten Foundation for Pancreatic Cancer Research (J.T.M.) and National Institutes of Health Grants R01CA120185 (to J.T.M.) and R01CA122334 and R01CA102709 (to A.T.-T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data described in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14302).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808300106/DCSupplemental.
References
- 1.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 2.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 4.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA miR-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 10.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 13.Johnson CD, et al. The let-7 MicroRNA Represses Cell Proliferation Pathways in Human Cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 14.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Koscianska E, et al. Prediction and preliminary validation of oncogene regulation by miRNAs. BMC Mol Biol. 2007;8:79. doi: 10.1186/1471-2199-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 18.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SM, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 25.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 26.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 27.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaur A, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piskounova E, et al. Determinants of MicroRNA Processing Inhibition by the Developmentally Regulated RNA-binding Protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 32.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 33.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Pajic A, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 36.Yu D, Dews M, Park A, Tobias JW, Thomas-Tikhonenko A. Inactivation of Myc in murine two-hit B lymphomas causes dormancy with elevated levels of interleukin 10 receptor and CD20: Implications for Adjuvant Therapies. Cancer Res. 2005;65:5454–5461. doi: 10.1158/0008-5472.CAN-04-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 40.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 41.Guo Y, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 43.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 46.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.