Abstract
Enzymes involved in the last 2 steps of nicotinamide adenine dinucleotide (NAD) cofactor biosynthesis, which catalyze the adenylylation of the nicotinic acid mononucleotide (NaMN) precursor to nicotinic acid dinucleotide (NaAD) followed by its amidation to NAD, constitute promising drug targets for the development of new antibiotics. These enzymes, NaMN adenylyltransferase (gene nadD) and NAD synthetase (gene nadE), respectively, are indispensable and conserved in nearly all bacterial pathogens. However, a comparative genome analysis of Francisella tularensis allowed us to predict the existence of an alternative route of NAD synthesis in this category A priority pathogen, the causative agent of tularaemia. In this route, the amidation of NaMN to nicotinamide mononucleotide (NMN) occurs before the adenylylation reaction, which converts this alternative intermediate to the NAD cofactor. The first step is catalyzed by NMN synthetase, which was identified and characterized in this study. A crystal structure of this enzyme, a divergent member of the NadE family, was solved at 1.9-Å resolution in complex with reaction products, providing a rationale for its unusual substrate preference for NaMN over NaAD. The second step is performed by NMN adenylyltransferase of the NadM family. Here, we report validation of the predicted route (NaMN → NMN → NAD) in F. tularensis including mathematical modeling, in vitro reconstitution, and in vivo metabolite analysis in comparison with a canonical route (NaMN → NaAD → NAD) of NAD biosynthesis as represented by another deadly bacterial pathogen, Bacillus anthracis.
Keywords: genomic reconstruction, in vitro reconstitution, mathematical modeling, NAD intermediates, substrate preference
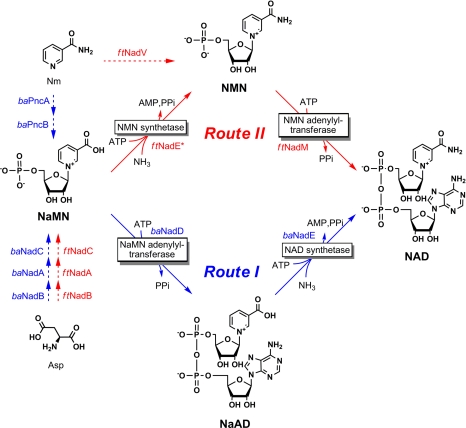
Biosynthesis of NAD, an indispensable and universal redox cofactor, has been the subject of extensive studies in a variety of species (1–3). Despite some variations in the early steps of the de novo and salvage pathways, the final biochemical transformations from the committed NaMN intermediate to NAD appear to be the same across all 3 domains of life. The 2 consecutive steps, referred to here as conventional route I, include the adenylylation of NaMN to NaAD followed by the ATP-dependent amidation of NaAD to NAD (Fig. 1). In bacteria, the first step is catalyzed by the enzyme NaMN adenylyltransferase of the NadD family. Genes encoding both enzymes are conserved and are essential in most pathogenic bacteria, and thus constitute prominent targets for the development of new antibiotics (4–8). The nearly universal character of biosynthetic route I is supported by the observed strict cooccurrence of nadD and nadE genes in the overwhelming majority of bacterial species with completely sequenced genomes [>750 genomes in the current version of NAD(P) biosynthesis subsystem in The SEED (9), also see SI Appendix, Table S1]. Route I is shared by both de novo synthesis from l-aspartate (genes nadB-nadA-nadC) and nicotinic acid salvage (genes pncA-pncB) pathways that converge at the common NaMN intermediate. Most bacteria have both pathways, as illustrated here for Bacillus anthracis (Fig. 1 and SI Appendix, Fig. S2), whereas some are strictly dependent on either de novo (e.g., Helicobacter pylori) or salvage (e.g., Streptococcus pneumoniae) pathways. The rarely observed absence of either pathway strongly correlates with the absence of both nadD and nadE genes. For example, Haemophilus influenzae overcomes this deficiency by the salvage of nicotinamide riboside (RNm) via NMN adenylyltransferase of the NadR family (10). In some intracellular pathogens with truncated genomes (such as Chlamydia spp), the loss of the entire NAD biosynthetic machinery is compensated for by the uptake of the intact cofactor from the host cells (11). A comparative genomic reconstruction of NAD biosynthesis within the collection of completely sequenced bacterial genomes integrated in The SEED allowed us to hypothesise that, in contrast to all previously studied species, the final 2 steps of NAD biosynthesis in Francisella tularensis occur in reverse order. In this pathogen, the amidation of NaMN to NMN appears to precede the adenylylation step converting NMN to NAD (Fig. 1). The enzymes involved in this pathway (here termed route II) were identified and characterized. The first reaction of route II is catalyzed by a divergent member of the NadE family endowed with NMN synthetase activity. Steady-state kinetic characteristics of this newly identified enzyme (here denoted ftNadE* to emphasize its distinction from other members of the NadE family with NAD synthetase activity) were established. The 3D structure of ftNadE* in complex with reaction products was solved at 1.9-Å resolution, revealing active site features that are likely responsible for its strong preference for NaMN vs. NaAD substrate. The in vivo NMN synthetase function of ftNadE* was verified by activity and metabolite measurements in crude extracts of the wild-type and ΔnadE* mutant of F. tularensis subsp novicida. The second step of route II is catalyzed by the NMN-preferring adenylyltransferase of the NadM family. This activity is encoded within the N-terminal domain of the ftNadM/Nudix bifunctional protein whose 3D structure and some enzymatic properties were reported in ref. 5. Here, we used a mixture of the purified recombinant enzymes ftNadE* and ftNadM for the experimental validation of the predicted pathway by its in vitro reconstitution with HPLC-based monitoring of the reaction products and intermediates. Similar experiments were performed using purified recombinant enzymes baNadD and baNadE from B. anthracis, representing a typical route I of NAD synthesis. The obtained results were consistent with a generalized mathematical model of the 2-step conversion of NaMN to NAD. In addition to establishing a precedent of this unexplored NAD biosynthetic route, this study provides new guidelines for targeting NAD biosynthesis in F. tularensis.
Fig. 1.
Genomic reconstruction of NAD biosynthesis in F. tularensis and B. anthracis. In conventional route I, as occurs in B. anthracis (outlined in blue), NaMN is adenylated to NaAD by baNadD, which is then amidated to NAD by NAD synthetase baNadE. In route II postulated for F. tularensis (outlined in red), an NMN synthetase activity of ftNadE* amidates the NaMN precursor to NMN before its adenylylation by ftNadM. De novo biosynthesis of NaMN from aspartate (via NadB, NadA, and NadC enzymes), a common route for both species, and 2 alternative Nm salvage pathways, deamidating (via PncA and PncB enzymes as in B. anthracis) and nondeamidating (via NadV enzymes as in F. tularensis), are shown by dashed arrows with respective color coding.
Results
Genomic Reconstruction and Prediction of the Alternative Route of NAD Biosynthesis in F. tularensis.
We used a subsystems-based approach implemented in The SEED (9) for the genomic reconstruction and analysis of variations in NAD biosynthesis across the entire collection of completely sequenced bacterial genomes. The results of this analysis are available online in the respective subsystem (“NAD and NADP cofactor biosynthesis global” at www.nmpdr.org (12)) and are shown in the condensed form in SI Appendix, Table S1. Among other features, this analysis confirmed the nearly universal cooccurrence of NaMN adenylyltransferase (gene nadD) and NAD synthetase (gene nadE) that jointly comprise a classic 2-step conversion of NaMN to NAD via the NaAD intermediate (route I in Fig. 1). Moreover, the NadD enzyme is present in all bacterial pathogens that contain genes of NaMN de novo synthesis from l-aspartate (nadB, nadA, and nadC), with the singular exception of F. tularensis.
The presence of NAD de novo synthesis in F. tularensis inferred by genomic reconstruction (operon nadACB) is consistent with the known ability of this pathogen to grow on defined media in the absence of nicotinamide or any other NAD precursors (13). However, the nadD gene, which is required for the utilization of the produced NaMN by conventional route I, is missing in all 7 available genomes of F. tularensis strains and isolates (SI Appendix, Table S2 and Fig. S2). In this context, the presence of the nadE gene in F. tularensis genomes appears equally unusual, because its anticipated NAD synthetase activity would be obsolete without a supply of NaAD substrate, which is typically produced by the NadD enzyme. The absence of 1 of the 2 recognized drug targets (NadD) and an unclear physiological context of the second target (NadE) in F. tularensis, a high-priority pathogen, prompted us to further analyze this conundrum.
The presence of orthologs of nadV and nadM genes encoding nicotinamide phosphoribosyl transferase and NMN adenylyltransferase in the F. tularensis genome pointed to a possible nicotinamide (Nm) salvage/recycling pathway (Fig. 1). The existence of such a 2-step conversion (Nm → NMN → NAD) was described in some bacteria (14, 15) and mammals (16). Although the presence of this salvage pathway in F. tularensis was further confirmed in our study (see below), this finding alone could not explain the growth of F. tularensis in the absence of Nm, nor could it suggest a physiological role for the nadE gene. A dual role of the ftNadM enzyme as both NMN and NaMN adenylyltransferase could be considered among possible interpretations of the gapped de novo pathway. Indeed, a single enzyme with comparable activity toward both NaMN and NMN intermediates is involved in NAD synthesis in mammalian cells (17–19). However, a strong preference of ftNadM for NMN over NaMN substrate (≈400-fold based on the comparison of respective kcat/Km values, see Table 1) made this enzyme an unlikely candidate for the role of a missing NaMN adenylyltransferase in conventional route I (NaMN → NaAD → NAD) (5).
Table 1.
Comparison of kinetic parameters of the enzymes involved in the last two steps of NAD synthesis in F. tularensis and B. anthracis
| Protein, substrate | Kinetic constants |
kcat/Km, s−1·M−1 | Subs. pref., fold | |
|---|---|---|---|---|
| kcat, s−1 | Km, mM | |||
| Amidation step | ||||
| ftNadE*, NaMN | 0.50 ± 0.02 | 0.20 ± 0.04 | 2,500 | ≈60† |
| ftNadE*, NaAD | 0.25 ± 0.02 | 5.82 ± 1.13 | 43 | |
| baNadE, NaMN | 0.004 ± 0.0002 | 1.18 ± 0.17 | 3 | ≈3,000‡ |
| baNadE, NaAD | 2.64 ± 0.06 | 0.29 ± 0.02 | 9,100 | |
| Adenylylation step | ||||
| ftNadM, NaMN | 0.16 ± 0.01 | 0.81 ± 0.27 | 200 | ≈400§ |
| ftNadM, NMN | 2.8 ± 0.1 | 0.034 ± 0.004 | 82,000 | |
| baNadD, NaMN | 25.60 ± 1.2 | 0.04 ± 0.01 | 640,000 | ≈45,000¶ |
| baNadD, NMN | 0.014 ± 0.001 | 0.94 ± 0.12 | 15 | |
The apparent values of Km (mM) and kcat (s−1) of ftNadE* and baNadE enzymes for amidation of both alternative substrates, NaMN and NaAD, at saturating concentrations of ATP (2 mM) and NH4Cl (4 mM), and of baNadD and ftNadM enzymes for adenylylation of NaMN and NMN at saturating concentration of ATP (2 mM) were determined as described in SI Appendix, SI Text. Errors represent standard deviation. Substrate preference (Subs. pref.) is expressed as a ratio of catalytic efficiencies (kcatapp/Kmapp) for the respective alternative substrates (see SI Appendix, SI Fig. 5 for kinetic rate plots).
†NaMN > NaAD;
‡NaAD > NaMN;
§NMN > NaMN;
¶NaMN > NMN.
Among other interpretations, the most compelling (albeit unconventional) was the hypothesis of the alternative pathway from NaMN to NAD with the opposite order of amidation and adenylylation steps (NaMN → NMN → NAD). This pathway (route II in Fig. 1) would proceed via the NMN (as opposed to the NaAD) intermediate, allowing the ftNadM enzyme to efficiently catalyze the NMN adenylyltransferase reaction at the second step of the pathway. An enzymatic activity inferred for the first step of the proposed pathway was termed NMN synthetase by analogy with the NAD synthetase reaction of NadE enzymes. Indeed, the postulated enzyme would catalyze essentially the same chemical transformation (ATP-dependent amidation of pyridine carboxylate) with the only feature differentiating NaMN and NAD substrates, the absence of an adenylyl moiety, being remote from the actual site of the reaction. Hence, it was tempting to suggest the product of the F. tularensis nadE gene a candidate for the proposed functional role of NMN synthetase (termed ftNadE*). Despite the straightforward chemical analogy, this alternative activity was previously never detected (20, 21), and, in most cases, this activity was not assayed for any of the characterized members of the NadE family (22–28). To test the bioinformatic predictions described above, we performed the kinetic and structural characterization of purified recombinant ftNadE* enzyme.
Enzymatic Characterization of ftNadE* Endowed with NMN Synthetase Activity.
The gene (FTL0685) encoding for ftNadE* from F. tularensis subsp. novicida strain U112 was cloned and overexpressed in E. coli with the N-terminal 6xHis tag. The recombinant ftNadE* protein was purified to homogeneity (SI Appendix, Fig. S3), using Ni-affinity chromatography, followed by gel filtration. The predicted NMN synthetase activity of ftNadE* was initially confirmed by a developed colorimetric coupled assay. It was then directly verified, kinetically characterized, and compared with a relatively low NAD synthetase activity also displayed by ftNadE*, using the HPLC-based protocol (SI Appendix, Fig. S4). The reaction requires conversion of 1 molecule of ATP to AMP and PPi per cycle of amidation of NaMN to NMN (as confirmed by HPLC, SI Appendix, Fig. S9). This is consistent with the known catalytic mechanism of all members of the ATP-dependent amidotransferase superfamily (29), including NAD synthetases of the NadE family (30). We have also demonstrated that ftNadE* can use only ammonia (NH3) but not glutamine (Gln) for the amidation of its substrate, consistently with the known properties of many bacterial NadE enzymes that do not contain an additional Gln-utilization (GAT) domain (22). The steady-state kinetic parameters of the ftNadE* enzyme shown in Table 1 revealed the enzyme's strong preference for the NaMN vs. the NaAD substrate. The conversion of NaMN to NMN with ATP and NH3 as cosubstrates is catalyzed by ftNadE* with ≈60-fold higher efficiency than the amidation of the canonical NaAD substrate, based on the ratio of the respective kcat/Km values. In contrast, we observed a >1,000-fold preference of NAD synthetase over NMN synthetase activity at saturating concentrations of each substrate for 2 other tested representatives of the NadE family from Corynebacterium glutamicum and Helicobacter pylori (SI Appendix, Table S3). Similar observations were made for the baNadE enzyme that was studied here in more detail (see below). All observed substrate preferences were fully consistent with the different physiological roles of ftNadE* in route II vs. NadE involved in conventional route I of NAD biosynthesis in other species, based on genomic reconstruction (SI Appendix, Table S1). Although some of these and other members of the NadE family were structurally and mechanistically characterized, the structural basis of their NaAD vs. NMN substrate preference was never addressed before, because the existence of the NMN synthetase activity in this family was previously unknown. To address this question, we used the comparative structural analysis as described below.
Structural Analysis of ftNadE* and Its Mechanistic Implications.
We crystallized and solved the 3-dimensional structure of ftNadE* complexed with the products of ATP hydrolysis, AMP and pyrophosphate (PPi), and Mg ions at 1.9-Å resolution (Fig. 2A and SI Appendix, Fig. S6). A comparison with a recently published 3D structure of baNadE (1.28-Å average Cα RMSD) reveals a similar dimer organization (Fig. 2A), in which each subunit contributes to a partially shared active site (28). Most of the active site residues that provide key interactions with ATP, Mg ions, and ammonia (as originally mapped in the most detailed structure-function analysis of bsNadE (31)) are conserved in ftNadE*. Modeling of the NaMN substrate in the active site of ftNadE* suggests that it is likely to occupy the same position as the nicotinosyl moiety of NaAD in baNadE complex (Fig. 2B). The structural conservation within the catalytic site suggests that NMN synthetase shares key mechanistic aspects with other NAD synthetases, including activation of the pyridine carboxyl-group by transient adenylylation and further displacement of the AMP-moiety by ammonia leading to the formation of a carboxamido-group (30). This is in agreement with the stoichiometric ATP-to-AMP conversion observed in the course of the ftNadE*-driven NMN synthetase reaction (as described above).
Fig. 2.
Three-dimentional structure of NMN synthetase from F. tularensis (ftNadE*) and a comparison with B. anthracis NAD synthetase (baNadE) structure. (A) Overall structure of ftNadE* (cyan and green subunits) homodimer is shown with bound AMP, PPi, and catalytic metal ions. (B and C) Superposition of ftNadE* (cyan) and baNadE (magenta) is illustrated by a detailed view of the catalytic sites (B) and substrate binding pockets (C). The arrow points to the nicotinosyl carboxyl group that is adenylated in the baNadE complex and that would be amidated in the second step of the reaction.
A detailed comparison of ftNadE* and baNadE substrate binding pockets revealed structural differences that are likely to be responsible for the distinct substrate preferences of these enzymes. In the putative nicotinosyl binding site of ftNadE*, a replacement of baNadE residues Gly-149 and Arg-252 by Gln and Trp, respectively, together with the significant movements in the α9 helix (≈3.5 Å) and the α5 helix (≈1.0 Å) generate a tighter substrate binding pocket that would be more favorable for the binding of the mononucleotide substrate. A replacement of baNadE residue Val-255 by Arg in ftNadE* would provide additional interactions with a free phosphoryl group of NaMN (Fig. 2C). In the putative adenosyl binding site, a bulky side-chain of Tyr replacing baNadE Thr-37 in the ftNadE* structure would block the adenosyl moiety of NaAD (Fig. 2C). Additionally, a favourable H-bond between baNadE Tyr-33 and the adenine ring of NaAD would be lost in ftNadE, in which this residue is replaced by Ser. Finally, the side-chain orientation of the baNadE His-258 residue, which is conserved in both enzymes, is dramatically different between the 2 structures. In baNadE this residue forms favorable stacking interactions with the NaAD adenine ring, whereas in the ftNadE structure it adopts a different rotamer conformation. The latter is stabilized by an H-bond with the main-chain carbonyl of Tyr-27 and by a stacking interaction with the Tyr-27 side-chain. Therefore, ftNadE* His-239 would clash with the adenosyl moiety of NaAD, while presenting no steric hindrance for binding of the NaMN substrate. One may notice that the structural differences observed in the substrate binding pocket of ftNadE* are relatively subtle. Nevertheless, altogether they lead to a substantial shift in the observed substrate preference of this enzyme, from dinucleotide (typical for other NadE enzymes) to mononucleotide, as reflected in an ≈30-fold difference in the apparent Km values (5.8 mM for NaAD compared with 0.2 mM for NaMN).
The structural basis of the substrate preference for NMN over NaMN displayed by ftNadM, the second enzyme of proposed route II of NAD synthesis (Fig. 1), is discussed in ref. 5. In Table 1, we provide the steady-state kinetic parameters of ftNadM, revealing an ≈400-fold discrimination of adenylylation of NMN vs. NaMN (based on the ratio of the apparent kcat/Km values). A combination of the observed substrate preferences of both enzymes, ftNadE* and ftNadM, strongly suggests a preference for route II vs. route I for NAD synthesis in F. tularensis. For the comparison of this pathway with the conventional route I, we obtained the reciprocal kinetic parameters for baNadE and baNadD enzymes as described below.
Enzymatic Characterization of baNadE and baNadD.
The 2 recombinant enzymes from B. anthracis were cloned with His6-tag, expressed and purified as detailed in SI Appendix, SI Text and Fig. S3. The results of their steady-state kinetic analysis are shown in Table 1 and SI Appendix, Fig. S5. The NAD synthetase baNadE displayed >3,000-fold preference for NaAD over NaMN, revealing not only an opposite substrate preference compared with ftNadE* but also a 50-fold stronger selectivity toward its preferred substrate. The steady-state kinetic parameters of baNadD revealed even stronger substrate selectivity typical for NaMN adenylyltransferase of the NadD family. The observed preference for NaMN over NMN of baNadD corresponds to an ≈100-fold increase in selectivity compared with the inverse preference of ftNadM. An extremely high substrate selectivity of both enzymes, baNadE and baNadD, indicated that NAD synthesis in B. anthracis would follow route I exclusively (without any appreciable contribution from route II). These results were used for mathematical modeling studies as described in the next section.
Mathematical Modeling of NAD Biosynthetic Routes I and II.
The verified existence of NMN synthetase activity completes a symmetry of possible biochemical transformations generating NAD cofactor from the nearly universal NaMN precursor (Fig. 1). Whereas both reactions, adenylylation and amidation, are required to accomplish this transformation, the order of these 2 reactions (or, in general, the relative contribution of route I vs. route II) in any organism would depend on the relative catalytic efficiencies and substrate preferences of the respective enzymes. A general kinetic model of this system was developed and applied to simulate NaMN-to-NAD transformations for F. tularensis in comparison with B. anthracis (Fig. 3 A and B). The steady-state kinetic parameters identified for individual purified recombinant enzymes (Table 1) were used for the transient time-course and steady-state simulations in the 2 binary mixtures ftNadE*/ftNadM and baNadD/baNadE. Both types of simulations suggested that in F. tularensis the contribution of route II to the overall flux from NaMN to NAD in optimal conditions (defined by the ratio of the 2 enzymatic activities) may reach 100% (Fig. 3A). Although some contribution of route I and accumulation of the NaAD intermediate is possible in suboptimal conditions (e.g., if [ftNadE*] ≪ [ftNadM]), this situation is unlikely as it would lead to a very low overall flux through the pathway. In the B. anthracis system, no contribution of route II and no appreciable accumulation of the off-pathway NMN intermediate are predicted by the simulations (Fig. 3B), even at a suboptimal ratio of the baNadD/baNadE enzymes. The more robust character of route I is a result of the more stringent substrate preferences of both B. anthracis enzymes compared with their counterparts from F. tularensis, as discussed above (Table 1). In addition to the important physiological implications, these simulations provided us with guidelines for the design of in vitro pathway reconstitution experiments (such as enzyme ratios and incubation times) aimed at the detection of pathway intermediates (see below).
Fig. 3.
Mathematical modeling, in vitro reconstitution, and in vivo assessment of the conventional (NaMN → NaAD → NAD, route I) and novel (NaMN → NMN → NAD, route II) pathways in F. tularensis and B. anthracis. (A and B) Examples of a simulated transient time-course of NaMN to NAD transformation for 2 binary mixtures: ftNadE*/ftNadM (A) and baNadD/baNadE (B). The details of model assumptions, boundary conditions, and additional results are provided in SI Appendix, SI Text. (C and D) In vitro reconstitution of a 2-step NaMN-to-NAD conversion by the same binary mixtures of pure recombinant enzymes: ftNadE*/ftNadM (C) and baNadD/baNadE (D), monitored by HPLC analysis at different time points. (E) HPLC-based assessment of the in vivo levels of NMN and NaAD intermediates: NaAD but not NMN is present in the crude extract from B. anthracis cells grown in rich media, whereas the extract of F. tularensis grown in rich media contains substantial amounts of NMN and a barely detectable level of NaAD. The NMN level is ≈2-fold higher in the wild-type strain compared with the nadE* knockout mutant of F. tularensis (ΔnadE*) grown in defined media (Chamberlain) supplemented with 200 μM nicotinamide. (F) The ftNadE* activity is required for the growth of F. tularensis in defined media in the absence of nicotinamide.
In Vitro Reconstitution of NAD Biosynthetic Routes I and II.
We used mixtures of purified recombinant enzymes ftNadE*/ftNadM and baNadD/baNadE for the in vitro reconstitution of respective pathways. A progression of NaMN-to-NAD conversion was initiated by adding the respective enzymes to the standard substrate mixture (1 mM NaMN, 2 mM ATP, and 4 mM NH4Cl) and was monitored by HPLC analysis of aliquots taken at several time points (Fig. 3 C and D, and SI Appendix, Fig. S12). Both consecutive reactions are reversible, and both generate PPi as one of the products. To simplify the analysis and to make the reaction conditions similar to those used in mathematical simulations, we added inorganic pyrophosphatase to all reaction mixtures. This addition made both steps of either pathway technically irreversible, which indeed led to almost 100% conversion of NaMN to NAD accompanied by the generation of equimolar amounts of AMP. Despite this similarity, the key differences between the 2 systems were in the nature of the transient intermediates, NMN for the ftNadE*/ftNadM mixture vs. NaAD for the baNadD/baNadE mixture. Overall, the observed dynamics of substrate consumption and formation of the intermediate and final products were in line with mathematical simulations. This observation is supportive of both the generalized mathematical model and the assertion of alternative pathways in the compared species.
In Vivo Assessment of NAD Biosynthesis in B. anthracis and F. tularensis.
To assess the physiological relevance of the proposed mechanism, we compared the NAD-related intermediates by HPLC in the extracts of B. anthracis and F. tularensis obtained in the log-phase of growth in rich media (SI Appendix, Fig. S13). A substantial level of NaAD intermediate was detected in B. anthracis, whereas the NMN level was below the detection threshold (Fig. 3E), in agreement with the model. In contrast, the level of NaAD intermediate was insignificant compared with NMN in F. tularensis (Fig. 3E). According to the steady-state simulations, some amounts of NaAD may accumulate in F. tularensis (generated by the residual NaMN adenylyltransferase activity of ftNadM) despite the negligible contribution of route I to the overall NAD biosynthetic flux. On the other hand, the NMN intermediate generated by ftNadE* within the major route II flux is not expected to accumulate in large quantities as it would be rapidly consumed by the ftNadM enzyme. However, the actual level of NMN in F. tularensis grown in the presence of nicotinamide could be somewhat higher because of the additional supply of NMN via ftNadV activity that was not accounted for by the described model (Fig. 1). Indeed, the observed level of NMN was >2-fold higher in the wild-type F. tularensis compared with the nadE* mutant strain when both were grown in the defined media supplemented with 200 μM nicotinamide (Fig. 3E). This observation supports the proposed physiological activities of ftNadE* and ftNadV supports the fact that both enzymes actually contribute to NAD biosynthesis in F. tularensis. The in vivo NMN synthetase function of ftNadE* was additionally confirmed by the detection of the respective activity in the crude extract of the wild type but not in the nadE* mutant of F. tularensis (SI Appendix, Fig. S14). The observation that the nadE* mutant could grow normally in the presence but not in the absence of Nm (Fig. 3F) confirmed the existence of the ftNadE*-independent NadV-NadM route of Nm salvage (Fig. 1) in F. tularensis. The observed Nm auxotrophy of this mutant was fully suppressed by the introduction of the ftNadE*-coding gene on a plasmid. Overall, the observations listed above provided us with sufficient proof of the physiological relevance of the newly identified NMN synthetase activity of the ftNadE* enzyme. In contrast to the predicted and observed conditional essentiality of the nadE* gene, the gene nadM encoding NMN adenylyltransferase should be essential under any growth conditions. This expectation is consistent with the biased distribution of the transposon insertions within the nadM gene that are present in all 3 viable nadM mutants from a transposon mutant library of F. tularensis subsp. novicida U112 (32). None of the mapped insertions would disrupt the N-terminal NMN adenylyltransferase domain, as in all 3 cases they occurred in the C-terminal ADPR pyrophosphatase domain, which is not involved in NAD biosynthesis and is expected to be fully dispensable.
Discussion
A subsystems-based approach to genomic annotation and metabolic reconstruction allows us to efficiently expand our knowledge of biosynthetic pathways of key metabolites, such as vitamins and cofactors, across the rapidly growing collection of completely sequenced diverse genomes (9). This approach enables the accurate projection of the known genes and pathways from model organisms to others, cataloguing of pathway variants (33), and establishment of feasible metabolic scenarios (34). Among various implications of these efforts is the identification and critical assessment of drug targets for the development of new anti-infectives (8). The comparative genomic approach also allows us to reveal gaps (e.g., missing genes) and inconsistencies (e.g., genes out of context) in the existing knowledge and generate conjectures to reconcile the revealed problems (35).
This study provided the evidence of the existence of NMN synthetase enzymatic activity, and the first proof of functional heterogeneity within the NadE family. This prompted us to characterize the ftNadE* enzyme in detail, including the steady-state kinetics and 3D structural analysis. The key mechanistic and structural features of ftNadE*—such as ATP dependence, the ability to use NH3 (but not Gln) as a N-source, the conservation of the dimeric structure, the overall fold, and many aspects of the active site topology—are quite similar to those of a typical single-domain bacterial NAD synthetase, such as baNadE. However, the detailed structural comparison of these enzymes revealed several elements in the ftNadE* substrate-binding site that are likely responsible for its altered substrate preference. Among them are the residues likely contributing to the preferential binding of the mononucleotide (Q133, W233, R236) and a decreased affinity for the dinucleotide substrate (H233, Y27). Remarkably, the observed relatively subtle structural changes lead to a rather dramatic shift in substrate specificity: an ≈60-fold preference of NaMN over NaAD in ftNadE*, compared with an ≈3,000-fold inverse preference of NaAD over NaMN in baNadE.
Of the 2 classes of biochemical transformations involved in the last steps of NAD biosynthesis (Fig. 1), variations in the substrate preference of respective enzymes have not been previously anticipated for the amidation step but are widely recognized for the adenylyltransferase reaction (36).
A combination of the observed substrate preference of both F. tularensis enzymes, ftNadE* for NaMN over NaAD and ftNadM for NMN over NaMN seem to be sufficient for reversing the usual order of biosynthetic steps toward the hypothesized route II (Fig. 1).
The difference between routes I and II was further analyzed using a generalized mathematical model of the 2-step NaMN-to-NAD conversion that accounts for all 4 possible reactions. The goal of the transient time-course simulations was to assess the reaction conditions for both pairs of enzymes, baNadD/baNadE and ftNadE*/ftNadM (enzyme concentrations and time points), that would allow us to detect the pathway intermediates. The simulations were simplified by forcing irreversibility of anabolic reactions by addition of inorganic pyrophosphatase to all reaction mixtures used for the in vitro pathway reconstitution. In the nearly optimal conditions, the only intermediates that could be detected were NaAD for conventional route I and NMN for proposed route II (Fig. 3 A–D). The detection of the respective intermediates in the extracts from B. anthracis and F. tularensis (Fig. 3E) is consistent with the steady-state simulations, suggesting that the respective routes would provide the major contribution to the actual NAD biosynthetic flux in each of these organisms.
The genomic reconstruction of NAD biosynthesis in F. tularensis suggested that, in addition to the de novo pathway, this organism may generate NAD by conversion of Nam to NMN with nadV followed by NadM activity. This pathway bypasses the need of ftNadE*, which is consistent with the observed Nm auxotrophy of a viable nadE* knockout mutant (Fig. 3F). The observed decrease of the overall NMN level (Fig. 3E) and the lack of any measurable NMN synthetase activity in this mutant (SI Appendix, Fig. S14) are consistent with the proposed physiological role of ftNadE*.
Comparative genomic reconstruction of NAD metabolism suggests that 2 other members of the same divergent branch of NadE family from Mannhemimia succinoproducens and Actinobacillus succinogenes may be additional species that harbor a version of route II (see SI Appendix, Table S1, Fig. S2 and SI Text for the details of this analysis and its evolutionary implications).
In summary, by combining a comparative genomic approach with in vitro and in vivo experiments we have established that the nadE gene of F. tularensis encodes an enzyme, NMN synthetase. The comparative analysis of the ftNadE* enzyme 3D structure revealed features contributing to its unusual substrate preference for NaMN over NaAD. This enzyme is involved in the first step of the alternative pathway, converting NaMN to NAD via the NMN intermediate. Although the ftNadE* enzyme is dispensable in F. tularensis because of the existence of the ftNadE*-independent salvage of Nm, the second enzyme of the pathway, NMN adenylyltransferase ftNadM, is absolutely required for NAD biogenesis. Therefore, the ftNadM enzyme likely constitutes a potential target for the development of therapies against this deadly pathogen.
Methods
Bioinformatic Tools.
A set of subsystems-based genomic annotations and metabolic reconstruction tools implemented in The SEED (9) were used to explore NAD metabolic machinery in bacteria (SI Appendix, Table S1 and Table S2). We used the PROMALS server (http://prodata.swmed.edu/promals/promals.php) to construct the multiple protein alignment (Fig. S7) and PHYLIP software version 3.62 to build the phylogenetic tree of representative members of the bacterial NadE family by the maximum-likelihood method (Fig. S8).
Gene Cloning, Protein Overexpression, and Purification.
Proteins (ftNadM, ftNadE, baNadD and baNadE) were purified using standard protocols as described in SI Appendix, SI Text.
Structural Analysis of ftNadE*.
Crystallographic analyses followed established methods as detailed in SI Appendix together with statistics (SI Appendix, Table S4), electron density map of the bound ligand (SI Appendix, Fig. S6a), and stereoview of catalytic site (SI Appendix, Fig. S6b).
Enzyme Assays and Steady-State Kinetic Analysis.
Enzymatic characterization of purified proteins was performed using a series of specific assays, including continuous and discontinuous coupled enzyme assays and direct HPLC analysis, as described in detail in SI Appendix, SI Text together with initial rates plots (SI Appendix, Fig. S5).
Mathematical Modeling.
Simulation of transient kinetics and steady-state fluxes of routes I and II for binary mixtures of ftNadE*/ftNadM and baNadD/baNadE enzymes were performed using Matlab as detailed in SI Appendix, SI Text.
In Vivo Assessment of NAD Biosynthetic Intermediates.
Cellular levels of NMN and NaAD intermediates were determined by combining cellular extraction of pyridine nucleotides with enzymatic treatment as described in detail in SI Appendix, SI Text.
Supplementary Material
Acknowledgments.
We thank Xiaoqing Li for assistance in gene cloning; Dr. Y. Igarashi for help with sequence clustering; Ross Overbeek, Svetlana Gerdes, and other members of The SEED development team at the Fellowship for Interpretation of Genomes for the help with genome analysis; and David Scott for critical reading of the manuscript. This work was supported by National Institute of Allergy and Infectious Diseases Grant 5 R01 AI059146-02 (to A.L.O.) and National Institutes of Health Grant P01 AI57986 (to K.E.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors for the ftNadE* complex with AMP, PPi, and metal ions have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3FIU).
This article contains supporting information online at www.pnas.org/cgi/content/full/0811718106/DCSupplemental.
References
- 1.Begley TP, Kinsland C, Mehl RA, Osterman A, Dorrestein P. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam Horm. 2001;61:103–119. doi: 10.1016/s0083-6729(01)61003-3. [DOI] [PubMed] [Google Scholar]
- 2.Foster JW, Moat AG. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980;44:83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S. Enzymology of NAD+ synthesis. Adv Enzymol Relat Areas Mol Biol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff HI, et al. Biosynthesis and recycling of nicotinamide cofactors in mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J Biol Chem. 2008;283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang N, et al. Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: Structure and function in bacterial NAD metabolism. Structure. 2008;16:196–209. doi: 10.1016/j.str.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdes SY, et al. From genetic footprinting to antimicrobial drug targets: Examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jedrzejas MJ. The structure and function of novel proteins of Bacillus anthracis and other spore-forming bacteria: Development of novel prophylactic and therapeutic agents. Crit Rev Biochem Mol Biol. 2002;37:339–373. doi: 10.1080/10409230290771537. [DOI] [PubMed] [Google Scholar]
- 8.Osterman AL, Begley TP. A subsystems-based approach to the identification of drug targets in bacterial pathogens. Prog Drug Res. 2007;131:133–170. doi: 10.1007/978-3-7643-7567-6_6. [DOI] [PubMed] [Google Scholar]
- 9.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurnasov OV, et al. Ribosylnicotinamide Kinase Domain of NadR Protein: Identification and Implications in NAD Biosynthesis. J Bacteriol. 2002;184:6906–6917. doi: 10.1128/JB.184.24.6906-6917.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haferkamp I, et al. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature. 2004;432:622–625. doi: 10.1038/nature03131. [DOI] [PubMed] [Google Scholar]
- 12.McNeil LK, et al. The National Microbial Pathogen Database Resource (NMPDR): A genomics platform based on subsystem annotation. Nucleic Acids Res. 2007;35:D347–D353. doi: 10.1093/nar/gkl947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson P, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes S, et al. Essential genes on metabolic maps. Curr Opin Biotechnol. 2006;17:448–456. doi: 10.1016/j.copbio.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Martin PR, Shea RJ, Mulks MH. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol. 2001;183:1168–1174. doi: 10.1128/JB.183.4.1168-1174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rongvaux A, et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Magni G, et al. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, et al. Structure of human nicotinamide/nicotinic acid mononucleotide adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin. J Biol Chem. 2002;277:13148–13154. doi: 10.1074/jbc.M111469200. [DOI] [PubMed] [Google Scholar]
- 19.Sorci L, et al. Initial-rate kinetics of human NMN-adenylyltransferases: Substrate and metal ion specificity, inhibition by products and multisubstrate analogues, and isozyme contributions to NAD+ biosynthesis. Biochemistry. 2007;46:4912–4922. doi: 10.1021/bi6023379. [DOI] [PubMed] [Google Scholar]
- 20.Yi CK, Dietrich LS. Purification and properties of yeast nicotinamide adenine dinucleotide synthetase. J Biol Chem. 1972;247:4794–4802. [PubMed] [Google Scholar]
- 21.Wagner R, Wagner KG. The pyridine-nucleotide cycle in tobacco Enzyme activities for the de-novo synthesis of NAD. Planta V. 1985;165:532–537. doi: 10.1007/BF00398100. [DOI] [PubMed] [Google Scholar]
- 22.Bieganowski P, Pace HC, Brenner C. Eukaryotic NAD+ synthetase Qns1 contains an essential, obligate intramolecular thiol glutamine amidotransferase domain related to nitrilase. J Biol Chem. 2003;278:33049–33055. doi: 10.1074/jbc.M302257200. [DOI] [PubMed] [Google Scholar]
- 23.Cantoni R, Branzoni M, Labo M, Rizzi M, Riccardi G. The MTCY428.08 gene of Mycobacterium tuberculosis codes for NAD+ synthetase. J Bacteriol. 1998;180:3218–3221. doi: 10.1128/jb.180.12.3218-3221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara N, et al. Molecular identification of human glutamine- and ammonia-dependent NAD synthetases. Carbon-nitrogen hydrolase domain confers glutamine dependency. J Biol Chem. 2003;278:10914–10921. doi: 10.1074/jbc.M209203200. [DOI] [PubMed] [Google Scholar]
- 25.Hughes KT, Ladika D, Roth JR, Olivera BM. An indispensable gene for NAD biosynthesis in Salmonella typhimurium. J Bacteriol. 1983;155:213–221. doi: 10.1128/jb.155.1.213-221.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nessi C, Albertini AM, Speranza ML, Galizzi A. The outB gene of Bacillus subtilis codes for NAD synthetase. J Biol Chem. 1995;270:6181–6185. doi: 10.1074/jbc.270.11.6181. [DOI] [PubMed] [Google Scholar]
- 27.Willison JC, Tissot G. The Escherichia coli efg gene and the Rhodobacter capsulatus adgA gene code for NH3-dependent NAD synthetase. J Bacteriol. 1994;176:3400–3402. doi: 10.1128/jb.176.11.3400-3402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald HM, et al. Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD+ synthetase from Bacillus anthracis. Acta Crystallogr D. 2007;63:891–905. doi: 10.1107/S0907444907029769. [DOI] [PubMed] [Google Scholar]
- 29.Zalkin H. The amidotransferases. Adv Enzymol Relat Areas Mol Biol. 1993;66:203–309. doi: 10.1002/9780470123126.ch5. [DOI] [PubMed] [Google Scholar]
- 30.Zalkin H. NAD synthetase. Methods Enzymol. 1985;113:297–302. doi: 10.1016/s0076-6879(85)13042-9. [DOI] [PubMed] [Google Scholar]
- 31.Rizzi M, et al. Crystal structure of NH3-dependent NAD+ synthetase from Bacillus subtilis. EMBO J. 1996;15:5125–5134. [PMC free article] [PubMed] [Google Scholar]
- 32.Gallagher LA, et al. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci USA. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Y, Osterman A, Overbeek R, Godzik A. Automatic detection of subsystem/pathway variants in genome analysis. Bioinformatics. 2005;21:478–486. doi: 10.1093/bioinformatics/bti1052. [DOI] [PubMed] [Google Scholar]
- 34.DeJongh M, et al. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139. doi: 10.1186/1471-2105-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osterman A, Overbeek R. Missing genes in metabolic pathways: A comparative genomics approach. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 36.Magni G, et al. Structure and function of nicotinamide mononucleotide adenylyltransferase. Curr Med Chem. 2004;11:873–885. doi: 10.2174/0929867043455666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.