Abstract
Recent calorimetric measurements of the solvation enthalpies of some dipeptide analogs confirm our earlier prediction that the principle of group additivity is not valid for the interaction of the peptide group with water. We examine the consequences for understanding the properties of peptide solvation. A major consequence is that the current value of the peptide-solvation enthalpy, which is a basic parameter in analyzing the energetics of protein folding, is seriously wrong. Electrostatic calculations of solvation-free energies provide an estimate of the size and nature of the error. Peptide hydrogen exchange rates provide an experimental approach for testing the accuracy of the solvation-free energies of peptide groups found by electrostatic calculations. These calculations emphasize that ignoring electrostatic interactions with neighboring NHCO groups should be a major source of error. Results in 1972 for peptide hydrogen exchange rates demonstrate that peptide-solvation-free energies are strongly affected by adjoining NHCO groups. In the past, the effect of adjoining peptide groups on the exchange rate of a peptide NH proton was treated as an inductive effect. The effect can be calculated, however, by an electrostatic model with fixed partial charges and a continuum solvent.
Keywords: group additivity, peptide solvation, peptide hydrogen exchange
In 2006, we pointed out (1) that group additivity is probably not valid for the interaction of the polar peptide group with water, and, consequently, the accepted value for the solvation enthalpy of the peptide group (−14.2 kcal/mol at 25 °C) (2, 3) is probably seriously wrong. Our argument about the probable breakdown of group additivity was based on electrostatic calculations of the solvation-free energies (esf values) of NHCO groups in short peptides. The calculated esf values depend strongly on the presence or absence of adjacent NHCO groups. If group additivity is valid, the esf value of a peptide group should be independent of the presence or absence of neighboring NHCO groups. The solvation enthalpies are expected to behave in a similar manner as the esf values because experimental values of solvation-free energies and enthalpies in simple amides differ only by small amounts (1, 4) and because the parameters for calculating esf are fixed by requiring that esf values agree with experimental solvation-free energies for a library of small polar molecules (5).
The solvation enthalpy of the peptide groups in an unfolded protein is the largest single factor in the energetics of protein folding, according to the standard analysis of folding energetics by Makhatadze and Privalov (refs. 2 and 3; and see also ref. 6). Data on the solvation enthalpies of the NHCO and CONH2 groups in dipeptide analogs have been published by Della Gatta and coworkers (7), and their results strongly suggest a failure of the group additivity assumption (7). Here, we compare the calorimetric results for solvation enthalpies (7) with predictions based on esf values calculated by using the DelPhi program (5). The present calculations employ numerical solution to the Poisson equation, using a continuum solvent and fixed, atom-centered partial charges. The background for DelPhi calculation of esf values is given in ref. 5.
The 4 dipeptide analogs for which solvation enthalpy results are available (7) contain 1 primary (CONH2) and 1 secondary (NHCO) amide group. The authors found that the solvation enthalpy of an NHCO group in these compounds is distinctly different from the values found in monoamides studied earlier (8). The authors treat the solvation enthalpies of the CONH2 and NHCO groups as being equal (6), and they conclude that group additivity is probably not valid for the interaction of the NHCO group with water. The standard analysis of protein folding energetics is based on the principle of group additivity (2, 3), and it is important to learn why group additivity should break down for the interaction of NHCO groups with water. Our article shows that a primary cause of the breakdown is the strong electrostatic interactions made by neighboring NHCO groups in peptides.
We point out that failure of group additivity for the interaction of the NHCO group with water, caused by electrostatic interaction with neighboring NHCO groups, is verified independently by a quite different experimental approach, based on data for the hydrogen exchange (HX) rates of peptide NH protons in short peptides. A strong effect of neighboring NHCO groups on HX rates was demonstrated experimentally by Molday et al. (9) in 1972. They found that the base-catalyzed HX rate of the peptide NH group increases almost 100-fold from 2.5 × 108 for N-methyl-acetamide (NMA) to 1.6 × 1010 for a tripeptide with NHCO groups on either side of the central NH group. Fogolari et al. (10) recognized that this result can be interpreted by calculating electrostatic free energies.
Solvation Enthalpy of an NHCO Group Depends Strongly on the Presence of Adjoining NHCO Groups
Earlier (1), we pointed out that the esf value of a specified NHCO group depends strongly on its residue position for NHCO groups close to either end of a peptide and also depends on the total number of NHCO groups if the peptide is very short. When the solvation enthalpy of a peptide is measured calorimetrically, the results give ΔHsolvo, the total solvation enthalpy for the whole peptide. To study whether or not group additivity can be used to analyze the results, we proceed as follows. First the contribution to Hsolvo from the alkyl groups present in the peptide is subtracted: The remainder is ΔHpol, the contribution to ΔHsolvo from the polar groups of the peptide. Then the dependence of (ΔHpol/n) on n is calculated, where n is the number of NHCO groups in the peptide. For the dipeptide analogs studied by Della Gatta and coworkers (7), it is also necessary to determine whether the solvation enthalpy of the CONH2 group equals that of the NHCO group. We first use esf calculations to investigate this question and give the predicted trend in (ΔHpol/n) with n for peptides larger than dipeptides.
Table 1 shows the total solvation enthalpy (ΔHpol) of the 2 polar groups (CONH and CONH2) in 4 dipeptide analogs (7). The contribution of the alkyl moiety has been subtracted (7) by using the experimental solvation enthalpy of the alkane that corresponds to the alkyl moiety. Both esf values and solvation enthalpy data indicate that the ΔHpol values should be similar for the NHCO and CONH2 groups (see below). Della Gatta and coworkers (7) found that ΔHpol/2 obtained for dipeptide analogs is distinctly different from ΔHpol for a single NHCO group in monoamides.
Table 1.
Values of esf and solvation enthalpy for polar groups in 4 dipeptide analogs (kcal/mol at 25 °C)
| Compound* | -ΔHpol† | -esf‡ |
|---|---|---|
| Ac-G-NH2 | 22.39 | 20.02 |
| Ac-A-NH2 | 21.58 | 19.53 |
| Ac-V-NH2 | 23.45 | 18.85 |
| Ac-L-NH2 | 21.89 | 19.00 |
*Ac-G-NH2 is a glycyl dipeptide analog (CH3CONHCH2CONH2); Ac-A-NH2, Ac-Val-NH2 and Ac-Leu-NH2 are the dipeptide analogs of Ala, Val and Leu, respectively.
†The total solvation enthalpy of the 2 polar groups (CONH and CONH2); data are from ref. 7.
‡The esf value is calculated for both polar groups (NHCO and CONH2). The esf value for the extended-β (−120°, 120°) conformation is given.
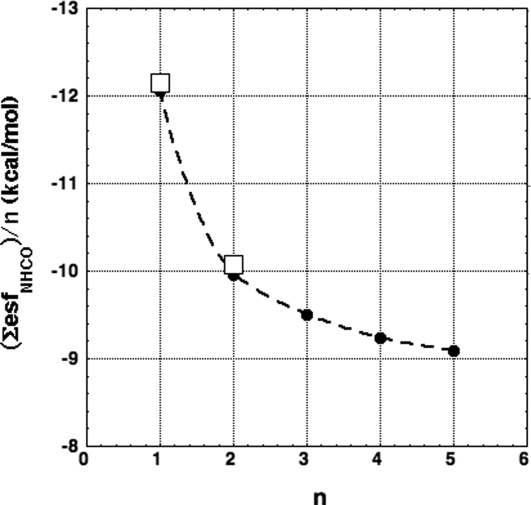
To compare experimental values of solvation enthalpy with esf values, we note that for simple monovalent anions and cations, the ratio of solvation enthalpy to free energy is 1.072 (1), and so we compare (solvation enthalpy/1.072) with esf. Fig. 1 shows the curve of (ΣesfNHCO)/n versus n for a set of short alanine peptides; n is the number of NHCO groups in a peptide and ΣesfNHCOis the total esf value for the peptide. Only the polar NH and CO groups of a peptide contribute to its esf value when the DelPhi algorithm and PARSE partial charges (5) are used to calculate esf. Fig. 1 includes values of [(ΔHpol/n)/1.072)] for a monoamide (NMA, plotted at n = 1) and for a dipeptide analog (alanine, plotted at n = 2). The agreement is satisfactory. Both the calculated esf values and the experimental solvation enthalpies indicate that group additivity fails for the solvation-free energy and enthalpy of the NHCO group.
Fig. 1.
ΣesfNHCO is the total ESF value of the n NHCO groups (ΣesfNHCO/n) is shown as filled circles) for a set of short alanine peptides (extended-β conformation, −120°, 120°). The peptides are blocked with CH3CO- at the N terminus, -NHCH3 at the C terminus. Also shown are values of [(solvation enthalpy/n)/1.072)] (open squares) for the polar groups of N-methyl-acetamide and for the average of the 2 polar groups in the dipeptide analog of alanine.
Different Partial Charges on the CONH2 Group of Acetamide and the NHCO Group of NMA
Before concluding that these results confirm the failure of group additivity, it is necessary to consider how the ΔHpol values for the NHCO and CONH2 groups are related. The PARSE partial charges (5) for the NH of NMA are −0.40 (N) and 0.40 (H), whereas for the NH2 group of acetamide they are −0.78 (N) and 0.39 (for each H). Although these partial charge differences appear large, the esf values of NMA (−12.20 kcal/mol) and acetamide (−11.75 kcal/mol) are similar (Table 2 and ref. 5). Thus, the esf values suggest that the ΔHpol values of the NHCO and CONH2 groups in the dipeptide analogs should be similar; comparison of experimental ΔHpol values confirms this conclusion (see below).
Table 2.
Comparison between solvation-free energy and enthalpy for 3 monoamides (kcal/mol at 25 °C)
| Compound* | -ΔGsolvo† | -ΔHsolvo‡ | -ΔGpol§ | -ΔHpol¶ | esf‖ |
|---|---|---|---|---|---|
| Acetamide | 9.72 | 16.32 | 11.65 | 13.74 | 11.75 |
| NMA | 10.08 | 17.07 | 12.23 | 13.01 | 12.20 |
| DMA | 8.56 | 16.56 | 10.86 | 10.31 | — |
*NMA, N-methyl-acetamide; DMA, dimethyl-acetamide.
‡Solvation enthalpy of the entire amide: data from ref. 8.
§Solvation-free energy of the polar group: data from ref. 4.
¶Solvation enthalpy of the polar group: data from ref. 8.
‖Calculated solvation-free energy of the NHCO group (NMA, DMA) or CONH2 group (acetamide): data from ref. 4 and here. The esf of DMA has not been determined.
Surprisingly, there is another possible source of different ΔHpol values for the NHCO group of NMA compared with the CONH2 group of acetamide. The ΔHpol values for monoamides are different for various sets of monoamides (8), suggesting that the partial charges on atoms of the NHCO group are not fixed but instead vary among different classes of monoamides. ΔHpol values for acetamide derivatives are different from those of formamide derivatives and values for N-disubstituted acetamide derivatives are different from those of N-monosubstituted derivatives (8). Moreover, calculated esf values for monoamides and amines sometimes show disturbingly large discrepancies from experimental values of ΔGpol (11). The variation in ΔHpol values among various classes of monoamides can be a large effect: The measured difference in ΔHpol between dimethyl-formamide and N-butyl-acetamide is ≈4.5 kcal/mol (8).
A likely explanation is provided by quantum chemistry calculations made by Wiberg and coworkers (12, 13). Their results indicate that the partial charge on the amide N may vary among amides because there is a mobile equilibrium between planar and twisted forms of the amide and the equilibrium is affected by amide substituents (12, 13). The difference between the ΔHpol values of the NHCO group in NMA (−13.01 kcal/mol) and the CONH2 group of acetamide (−13.74 kcal/mol) (Table 2 and ref. 8) is probably caused chiefly by a difference in the twisted-planar equilibrium of NMA versus acetamide.
Revised values of solvation enthalpies for monoamides, compared with ones we gave earlier in ref. 4, are given in Table 2. The values given for NMA, acetamide, and dimethyl-acetamide are different from our earlier values for 2 reasons. First, we earlier corrected for the alkyl moieties by assuming that the correction term is proportional to accessible surface area (ASA). Second, a standard state correction is needed for vapor to liquid transfer (16), and we made this correction (4). We now conclude that the procedure used by Della Gatta and coworkers (7, 8) to subtract the contribution of the alkyl moiety is preferable to our earlier procedure (4) because it is more direct and avoids the assumption that the alkyl correction is proportional to ASA. When the Della Gatta procedure for making the alkyl correction is used, the standard state correction drops out because it is subtracted together with the alkyl correction term, and the standard state correction has the same value (≈0.55 kcal/mol) (2) in both cases.
Consequently, the solvation enthalpies of the NHCO groups of monoamides shown here in Table 2 are somewhat larger (more negative) than our earlier values (4). The basic conclusion drawn earlier (4) from data like the results in Table 2 was that the experimental values of solvation-free energy and solvation enthalpy of the NHCO group in monoamides are closely similar. This conclusion still applies, but less strikingly. An ASA-dependent correction term is used here to obtain ΔGpol from ΔGsolvo because the PARSE parameters of DelPhi are calibrated by using an ASA-dependent correction term (5). Note that the alkyl correction term is always larger for the solvation enthalpy than for the solvation-free energy of the alkane.
In Table 2, the esf value of dimethyl-acetamide (DMA) cannot be computed by using known PARSE partial charges because they have not yet been determined for tertiary amides (5). However, because the experimental solvation-free energy of DMA is known (12), and the esf is required to be closely equal to the experimental ΔGpol value when the PARSE charges are assigned, the esf of dimethyl-acetamide should be close to the −10.86 kcal/mol given in Table 2 for ΔGpol. Earlier (4), we also gave values for ΔGpol and ΔHpol for propionamide, but by mistake, we gave the ΔHpol value for N,N-dimethyl-propionamide, taken from ref. 8, and the experimental value for propionamide is not available in ref. 8.
Hydrogen Exchange Rate as a Probe of Solvation-Free Energy in Peptides
There is a straightforward theory linking hydrogen exchange rate (kHX) in peptides with the electrostatic free energy of the peptide (10, 17). LogkHX for base-catalyzed exchange depends directly on ΔpKa, the difference in pKa between the catalyst OH−, with pKa 15.7, and the peptide NH group, with pKa 18.5, for a representative peptide group (17). (These are illustrative pKa values (17); the reader should be aware that their values depend on temperature and isotope effects (18) as well as on the neighboring residue effects discussed here.) The 2 reactants first form an encounter complex (AH + B− ⇔ A− + BH, AH = peptide, B− = OH−) in a fast, diffusion-controlled reaction whose second-order rate constant is kD. Then kHX is given by (17)
ΔpKa depends on the free-energy cost of ionizing the peptide NH group, which can be estimated by calculating ΔGel, the electrostatic free energy of the ionizing group (10), by using the finite difference method to solve the Poisson–Boltzmann equation for ΔGel. This is done for both the neutral and ionized forms of the peptide group of the peptide (pep) and of the reference compound, NMA.
The procedure used here is outlined in Methods and in refs. 5, 10, and 19. The partial charges of the solute molecule are placed on a grid running through a cavity in the solvent. ΔGel for the N atom at grid site i depends on the partial charge and the electrostatic potential at site i (5, 10, 19, 20) whose value depends on electrostatic interactions made between the NH or N− group with neighboring NHCO groups.
Fogolari, et al. (10) made electrostatic calculations using the UHBD algorithm to interpret the HX rate data of Molday et al. (9) for various short peptides. Their procedure differs in several respects from the DelPhi procedure used by us, and we have repeated their calculations using DelPhi to determine whether equivalent results and conclusions are obtained. The basic differences between their UHBD calculations and ours are as follows. They assign small partial charges to the hydrocarbon groups, whereas partial charges of 0 are assigned when using the PARSE parameters of DelPhi (5). They use a dielectric constant (ε) of 4 for the peptide (10), whereas ε = 2 is normally used in DelPhi (5), and we investigate here values of ε = 2, 4, and 20. Finally, they use a closed van der Waals surface to describe the cavity surface for the solute molecule, whereas DelPhi uses the water-accessible surface (ASA) obtained by rolling a probe with radius 1.4 Å over the solute. The calculated values of logkHX depend substantially on backbone conformation (10), and they compared experimental results with calculated values for an assumed 70:30 mixture of extended-β (−120°, 120°) and αR (−60°, −40°) backbone conformations, for which they found good agreement with the experiment (10).
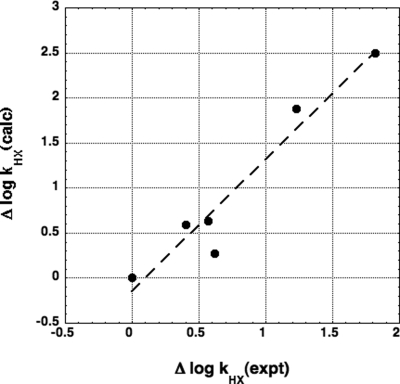
We made DelPhi calculations of logkHX for 3 backbone conformations (αR, −60°, −40°; β, −120°, 120°; and PII, −75°, 145°). The logkHX calculations for ε = 4 [the value they used (10)] are shown in Table 3 together with the experimental values of logkHX (9). As they also found (10), there are substantial differences between calculated values of logkHX for different backbone conformations. The average calculated value of logkHX for the 3 backbone conformations is plotted against the experimental value in Fig. 2: A good correlation is seen, with R = 0.964. A similarly good correlation is found when logkHX for 30% αR, 70% β is plotted (R = 0.968) for comparison with their results. Although individual values of logkHX (calculated) vary significantly between the results of ref. 10 and our own, the present results are broadly similar to the UHBD results found by Fogolari et al. (10). Both sets of results agree that electrostatic interactions among adjacent NHCO groups are responsible for the strong dependence of logkHX on neighboring peptide groups.
Table 3.
Experimental and calculated values of ΔlogkHX for short peptides with varying numbers of NHCO groups
| Compound* | Expt† | α | β | PII |
|---|---|---|---|---|
| 1 | 0.00 | 0.00 | 0.00 | 0.00 |
| 2 | 0.40 | 1.87 | 0.02 | −0.11 |
| 3L | 1.23 | 2.94 | 1.49 | 1.19 |
| 3R | 0.62 | 1.28 | −0.10 | −0.35 |
| 4L | 1.82 | 4.51 | 1.68 | 1.31 |
| 4R | 0.57 | 2.69 | −0.14 | −0.64 |
*The compounds are listed by the notation used in refs. 9 and 10. Compound 1 is CH3CONH*CH3, 2 is CH3CONHCH(CH3)CONH*CH3, 3 is CH3CONH*CH2CONH*CH3, 4 is CH3CONHCH(CH3)CONH*CH2CONH*CH3; L and R refer to the left and right NH* groups, and NH* refers to the NH group whose kHX is measured.
†The experimental value of ΔlogkHX is given, followed by calculated values for the αR (−60°, −40°), β (−120°, 120°), and PII (−75°, 145°) backbone conformations. The reference compound 1 (NMA) has the same geometry and pKa value throughout. The interior dielectric constant used in the calculations is ε = 4.
Fig. 2.
Comparison of experimental and calculated values of ΔlogkHX (slope = 1.46, R = 0.965). The experimental values are from ref. 9, the reference compound is N-methylacetamide, and the calculated values are the average for 3 backbone conformations (αR, β, PII) shown in Table 3.
Before a realistic comparison can be made between calculated and experimental values of logkHX, the actual conformations of the peptides must be determined, and the appropriate value of ε for use in these calculations must be evaluated. The slope of the line in Fig. 2 is 1.47, but the difference between this value and the expected value of 1.00 is not important until the backbone conformations of the peptides can be determined. The slope of the plot shown in Fig. 2 depends strongly on the value used for ε: The slope is 3.18 for ε = 2, 1.46 for ε = 4, and 0.26 for ε = 20. Whether use of the closed van der Waals surface or the ASA surface gives better results remains to be determined.
The standard literature explanation for the dependence of logkHX on neighboring NHCO groups invokes a through-bonds inductive effect (21, 22). Our results show, as was evident earlier (10), that the effect can be described instead as a through-space electrostatic effect.
Wider Consequences of Electrostatic Solvation of Peptide Groups
Our earlier studies of esf-dependent properties of peptides were hampered by our inability to make direct comparisons between esf values and experimental data (1, 4, 23–25). The basic problem is that most quantities we have studied are affected by other properties in addition to solvation, and these other properties are not yet known quantitatively. In this article, we report some progress with this problem. Solvation enthalpy should be directly related to solvation-free energy for highly polar molecules (1, 4), at least in simple cases. Moreover, the effect of adjoining NHCO groups on the HX rate of a given NH group is both a large effect (>2 kcal/mol in Fig. 2) and one for which it is easy to obtain accurate experimental data. The required experimental data have been present in the literature since 1972 (9). There is an intriguing twist to the problem because the data have long between interpreted as an inductive (through-bond) effect (21, 22), whereas the esf calculations are made for fixed partial charges, a point recognized earlier (10). Fig. 1 indicates that the effect of adjoining NHCO groups on the average solvation enthalpy of the peptide groups in a short peptide can be even larger than shown in Fig. 2 for HX rates.
A different esf-dependent effect that we studied earlier (23, 24) is that solvation strongly affects preferences for different backbone conformations, and the effect is amino acid-specific. The dependence of backbone preference on solvation may explain why alanine favors the polyproline II (PII) conformation in the coil library, whereas valine favors extended-β (23, 24). The esf dependence of backbone preference is in the size range 0.5–1 kcal/mol (23, 24).
A related esf-dependent effect is the neighboring-residue effect (24). Peptide solvation depends on the access of water to a peptide group because access is limited by neighboring side chains. A neighboring-residue effect was detected experimentally in Dobson's laboratory (26) by peptide NMR measurements of the 3JHNα coupling constant, which is directly related to the φ backbone angle. These workers divided neighboring residues into 2 groups according to the size of the effect: L (large effect, aromatic and β-branched residues, FHITVWY) and S (small effect, other amino acids). The same 2 groups of L and S residues (with G and P excluded) were found in esf calculations of a neighboring-residue effect in the coil library (24). The existence of a neighboring-residue effect has been confirmed by other types of studies (27, 28). The average esf difference caused by having L versus S neighbors is 0.2 kcal/mol (24), the smallest of the 3 effects discussed here.
The effect of adjoining NHCO groups is related to, but different from, the electrostatic effect on peptide backbone conformation considered by Flory (29) and by Avbelj and Moult (30). They point out that electrostatic interactions among adjoining NHCO groups should be a major determinant of peptide backbone conformational preferences because dipoles of adjacent peptide groups are antiparallel (favorable) in the extended-β conformation (−120°, 120°), whereas they are parallel in the αR conformation (−60°, −40°). Electrostatic interactions in the αR conformation are unfavorable unless peptide H-bonds are formed and the parallel dipoles are joined end-to-end. These same electrostatic considerations arise when discussing solvation of the peptide group, but there are 2 important differences. The esf is always negative (favorable), whereas the local electrostatic interaction energy can be either favorable or unfavorable. Second, the size of the esf varies among amino acid residues at fixed φ,ψ values (4, 20, 23, 24) whereas the local electrostatic interaction energy is fixed.
The 3 esf-dependent effects summarized above have wider consequences. The backbone preference effect should be a major determinant of α-helix (4) and β-structure (25) propensities. The neighboring-residue effect demonstrates that Flory's “isolated pair” proposal (29) cannot be valid as a general principle. The isolated-pair hypothesis argues that the φ,ψ angles of 1 residue in an unfolded protein should be independent of adjoining residues but the neighboring-residue effect shows they are dependent. The effect of adjoining NHCO groups demonstrates that the principle of group additivity does not apply when solvation of the peptide group is considered. Because of its large size, and because the principle of group additivity has been the cornerstone of earlier experimental analyses of the energetics of protein folding (2, 3), the effect of adjoining NHCO groups poses a major problem for understanding the energetics of folding. Development of a new analysis will not be easy.
Methods
Calculation of esf Values.
The esf value is the transfer free energy from the gas phase to aqueous solution. The steps in the calculation when using DelPhi, and the rationale for these steps, are given in ref. 5. We obtained the DelPhi algorithm from the Department of Biochemistry, Columbia University, 630 West 168th Street, New York, NY 10032. A grid with 101 × 101 × 101 points and a grid spacing of 0.4 Å was used, and 1 focusing step with a spacing of 1 Å was made. The image charge method of calculating ΔGel was used. The pH is 7.0, and the ionic strength is 0. For the geometry of the amino acid residues, a new residue library was made from 3,628 high-resolution protein structures in the Protein Data Bank (resolution <2 Å, r-factor <0.2) and used to give the bond angles and distances.
Calculation of ΔlogkHX Values.
This calculation is based on the free-energy cost of ionizing the peptide NH group and is calculated according to equation 7 of ref. 19. Values of ΔGel are calculated for the neutral and ionized forms of the peptide NH group, and the difference is taken. A charge of −1.00 is placed on the N atom, as in ref. 10. The charge and the electrostatic potential at the N atom are used to calculate ΔGel. The ionic strength used in the calculations is 0.5.
Acknowledgments.
We thank Joe Dannenberg and George Makhatadze for calling to our attention papers by Wiberg et al. (12, 13) and Della Gatta et al. (7), respectively, and we thank George Rose for discussion.
Footnotes
The authors declare no conflict of interest.
References
- 1.Avbelj F, Baldwin RL. Limited validity of group additivity for the folding energetics of the peptide group. Proteins. 2006;63:283–289. doi: 10.1002/prot.20756. [DOI] [PubMed] [Google Scholar]
- 2.Makhatadze GI, Privalov PL. Contribution of hydration to protein folding thermodynamics. I. The enthalpy of hydration. J Mol Biol. 1993;232:639–659. doi: 10.1006/jmbi.1993.1416. [DOI] [PubMed] [Google Scholar]
- 3.Makhatadze GI, Privalov PL. Energetics of protein structure. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 4.Avbelj F, Luo P, Baldwin RL. Energetics of the interaction between water and the helical peptide group and its role in determining helix propensities. Proc Natl Acad Sci USA. 2000;97:10786–10791. doi: 10.1073/pnas.200343197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 6.Baldwin RL. Energetics of protein folding. J Mol Biol. 2007;371:283–301. doi: 10.1016/j.jmb.2007.05.078. [DOI] [PubMed] [Google Scholar]
- 7.Della Gatta G, Usacheva T, Badea E, Palecz B, Ichim D. Thermodynamics of solvation of some small peptides in water at T = 298.15 K. J Chem Thermodyn. 2006;38:1054–1061. [Google Scholar]
- 8.Della Gatta G, Barone G, Elia V. Enthalpies of solvation for N-alkylamides in water and in carbon tetrachloride at 25 °C. J Solution Chem. 1986;15:157–167. [Google Scholar]
- 9.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 10.Fogolari F, Esposito G, Viglino P, Briggs JM, McCammon JA. pKa shift effects on backbone amide base-catalyzed hydrogen exchange rates in peptides. J Am Chem Soc. 1998;120:3735–3738. [Google Scholar]
- 11.Tannor DJ, et al. Accurate first principles calculation of molecular charge distributions and solvation energies from ab initio quantum mechanics and continuum dielectric theory. J Am Chem Soc. 1994;116:11875–11882. [Google Scholar]
- 12.Wiberg KB, Breneman CM. Resonance interactions in acyclic systems. 3. Formamide internal rotation revisited. Charge and energy redistribution along the C-N rotational pathway. J Am Chem Soc. 1992;114:831–840. [Google Scholar]
- 13.Wiberg KB, Hadad CM, Rablen PR, Cioslowski J. Substituent effects. 4. Nature of substituent effects at carbonyl groups. J Am Chem Soc. 1992;114:8644–8865. [Google Scholar]
- 14.Wolfenden RV. Interaction of the peptide bond with solvent water: A vapor phase analysis. Biochemistry. 1978;17:201–204. doi: 10.1021/bi00594a030. [DOI] [PubMed] [Google Scholar]
- 15.Wolfenden RV, Cullis PM, Southgate CCF. Water, protein folding and the genetic code. Science. 1979;206:575–577. doi: 10.1126/science.493962. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Naim A, Marcus Y. Solvation thermodynamics of nonionic solutes. J Chem Phys. 1984;81:2016–2027. [Google Scholar]
- 17.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1984;16:521–6554. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 18.Connelly GP, Bai Y, Jeng M-F, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 19.Antosewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 20.Avbelj F. Amino acid conformational preferences and solvation of polar backbone atoms in peptides and proteins. J Mol Biol. 2000;300:1335–1359. doi: 10.1006/jmbi.2000.3901. [DOI] [PubMed] [Google Scholar]
- 21.Sheinblatt M. Determination of an acidity scale for peptide hydrogens from nuclear magnetic resonance kinetic studies. J Am Chem Soc. 1970;92:2505–2509. doi: 10.1021/ja00711a048. [DOI] [PubMed] [Google Scholar]
- 22.Molday RS, Kallen RG. Substituent effects on amide hydrogen exchange rates in aqueous solution. J Am Chem Soc. 1972;94:6739–6745. [Google Scholar]
- 23.Avbelj F, Baldwin RL. Role of backbone solvation and electrostatics in generating preferred backbone conformations: distributions of phi. Proc Natl Acad Sci USA. 2003;100:5742–5747. doi: 10.1073/pnas.1031522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avbelj F, Baldwin RL. Origin of the neighboring residue effect on peptide backbone conformation. Proc Natl Acad Sci USA. 2004;101:10967–10972. doi: 10.1073/pnas.0404050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avbelj F, Baldwin RL. Role of backbone solvation in determining thermodynamic β propensities of the amino acids. Proc Natl Acad Sci USA. 2002;99:1309–1313. doi: 10.1073/pnas.032665499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penkett CJ, et al. NMR analysis of main-chain conformational preferences in an unfolded fibronectin-binding protein. J Mol Biol. 1997;274:152–159. doi: 10.1006/jmbi.1997.1369. [DOI] [PubMed] [Google Scholar]
- 27.Keskin O, Yuret D, Gursoy A, Turkay M, Erman B. Relationships between amino acid sequence and backbone torsion angle preferences. Proteins. 2004;55:992–998. doi: 10.1002/prot.20100. [DOI] [PubMed] [Google Scholar]
- 28.Jha AK, et al. Helix, sheet and polyproline II frequencies and strong nearest neighbor effects in a restricted coil library. Biochemistry. 2005;44:9691–9702. doi: 10.1021/bi0474822. [DOI] [PubMed] [Google Scholar]
- 29.Flory PJ. Statistical Mechanics of Chain Molecules. New York: Wiley Interscience; 1969. [Google Scholar]
- 30.Avbelj F, Moult J. Role of electrostatic screening in determining protein main chain conformational preferences. Biochemistry. 1995;34:755–764. doi: 10.1021/bi00003a008. [DOI] [PubMed] [Google Scholar]