Abstract
Protein kinase A (PKA)-dependent signaling cascades play an important role in mediating the effects of dopamine and other neurotransmitters in striatal medium spiny neurons. We have identified a prominent striatal PKA substrate as Rap1-GTPase activating protein (Rap1GAP), a negative regulator of Rap1 signaling. Although present throughout the brain, Rap1GAP is enriched in striatal medium spiny neurons and is phosphorylated by PKA at Ser-441 and Ser-499 in response to activation of D1 dopamine receptors. Phosphorylation of Rap1GAP is associated with inhibition of GAP activity, as demonstrated by increased Rap1 activity in striatal neurons. Phosphorylation of Rap1GAP is also associated with increased dendritic spine head size in cultured neurons. These findings suggest that phosphorylation of Rap1GAP by PKA plays an important role in striatal neurons by modulating Rap1 actions.
Keywords: dopamine, medium spiny neuron, cAMP, PKA, EPAC
Medium spiny neurons (MSNs) of the striatum are optimized for receiving information from cortex and thalamus via glutamatergic inputs and integrating them with contextual cues mediated by dopaminergic signaling. Thus, dopamine plays a critical role in the regulation of plasticity in MSNs and as a result controls aspects of associative learning including motor and reward learning and habit formation (1). Striatal dopamine also plays a major role in the process of drug addiction and in neurological disorders such as Parkinson's disease, depression, compulsive disorders, and schizophrenia. For this reason, a complete understanding of dopaminergic signaling in MSNs is an important goal for understanding striatal function and for developing future therapies.
Dopamine acts on MSNs in part through the D1/D5 subclass of dopamine receptors, which in turn stimulate the cAMP/protein kinase A (PKA) signaling pathway. Dopamine also acts via the D2 class of receptors to attenuate cAMP/PKA-dependent signaling. PKA-dependent phosphorylation of receptors, ion channels, transcription factors, and other proteins accounts for much of the structural and functional plasticity regulated by dopamine (2). Our previous studies that identified and characterized targets of the cAMP/PKA pathway have been critical for understanding the actions of dopamine in MSNs (3, 4). For example, the PKA substrate, dopamine and cAMP-regulated phosphoprotein, Mr 32 kDa (DARPP-32) has been extensively analyzed and found to amplify dopamine action in MSNs through inhibition of protein phosphatase-1 (PP1). In the present study, we have characterized a striatal-enriched PKA substrate, previously designated as cAMP-regulated phosphoprotein, 90 kDa (ARPP-90) (5, 6). The results obtained indicate that ARPP-90 is the same as Rap1-GTPase activating protein (Rap1GAP), a regulator of the Ras-like small G protein Rap1. Phosphorylation of Rap1GAP leads to its inhibition and to subsequent activation of Rap1. Phosphorylation of Rap1GAP is likely to play an important role in regulation of MSN function through the ability of the dopamine/cAMP/PKA signaling pathway to regulate Rap1 activity.
Results
Identification of a Major Striatal PKA Substrate as Rap1GAP.
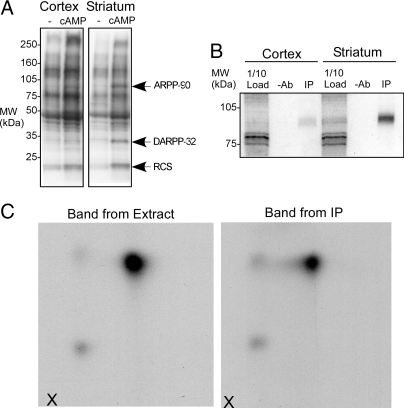
We compared the phosphorylation of proteins in soluble extracts from cortex or striatum following incubation without or with cAMP in the presence of [γ-32P]ATP. Autoradiography of proteins separated by SDS/PAGE revealed several protein bands incorporating more 32P label with cAMP treatment (Fig. 1A). Two of these proteins, the 21-kDa protein regulator of calmodulin signaling (RCS) and DARPP-32, were identified in previous studies using this approach (7–9). A third prominent band of ≈95 kDa is also enriched in striatum. This band, previously named ARPP-90, has remained unidentified (6). We therefore initiated efforts to identify this protein, using column chromatography and proteomic analysis.
Fig. 1.
Identification of ARPP-90 as Rap1GAP. (A) Proteins in low salt extracts from cortex and striatum were incubated with [γ-32P]ATP in the absence or presence of cAMP and samples analyzed by SDS/PAGE and autoradiography. The positions of ARPP-90, DARPP-32, and RCS are shown. (B) Proteins in extracts from cortex or striatum were incubated with [γ-32P]ATP in the presence of cAMP and subjected to immunoprecipitation without (−Ab) or with anti-Rap1GAP antibody (IP). Samples were analyzed by SDS/PAGE and autoradiography. Ten percent of the protein extract was analyzed. (C) Phospho-ARPP-90 bands cut from SDS/PAGE gels were subjected to digestion with trypsin and digests analyzed by 2-dimensional phosphopeptide mapping. Peptides were first separated by electrophoresis in the horizontal direction (origin is indicated by “X”) and chromatography in the vertical direction. Samples were from striatal samples before (Band from Extract) and after immunoprecipitation with anti-Rap1GAP antibody (Band from IP).
ARPP-90 was partially purified from rat brain extracts to a point where a single Coomassie-stained band could be resolved. Analysis of a tryptic digest of this protein band by liquid chromatography/mass spectrometry/mass spectrometry (LC/MS/MS) identified the protein as Rap1GAP (Rap1GAP1a) [see supporting information (SI) Fig. S1 for summary of peptide coverage]. To confirm this result, specific antibodies against Rap1GAP were used to immunoprecipitate proteins from striatal and cortical extracts incubated with cAMP and [γ-32P]ATP. A single 32P-labeled protein matching ARPP-90 was seen after immunoprecipitation with enrichment being found in the striatal sample (Fig. 1B). Tryptic phosphopeptide mapping was used to compare the ARPP-90 band from the striatal extract and the Rap1GAP band obtained following immunoprecipitation (Fig. 1C). The 2 peptide maps were identical, indicating that ARPP-90 is Rap1GAP.
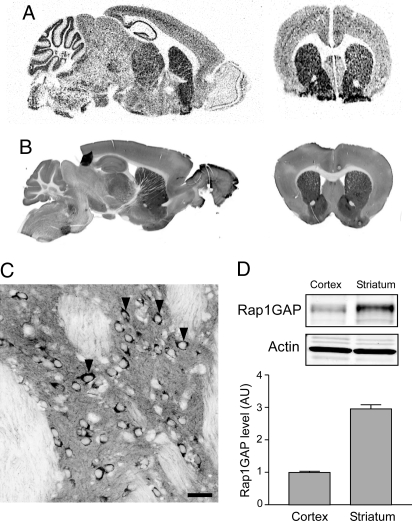
In situ hybridization and immunofluorescence labeling of fixed brain slices revealed that Rap1GAP mRNA and protein levels were enriched in the striatum (Fig. 2 A and B). Examination at higher magnification revealed that although present in neurons in most brain regions, many MSNs in the striatum were heavily labeled (Fig. 2C). Several areas of the di- and mesencephalon, the amygdala core, and cortex layer V also contained cells with high mRNA levels (Fig. S2). Immunoblot comparisons using an antibody against the N-terminal region of Rap1GAP revealed that the striatum contains ≈3.0-fold higher amounts of Rap1GAP protein compared to cortex (Fig. 2D).
Fig. 2.
Expression of Rap1GAP in mouse brain. Analysis of (A) Rap1GAP mRNA expression by in situ radiography and (B) Rap1GAP protein expression by immunoflorescence is shown. Similar sagittal and coronal sections shown that included the striatum were chosen for each method. (C) Confocal analysis of Rap1GAP protein expression in the dorsal striatum. Arrowheads indicate dark staining of cell bodies of MSNs. (Scale bar, 50 μm.) (D) Immunoblotting of Rap1GAP in cortex and striatum. Band intensities were measured and normalized first to actin (Lower panels) and then to cortex. Quantitation of results is shown in the bar graph (n = 12; error bars show mean ± SEM).
Identification of PKA Phosphorylation Sites on Rap1GAP.
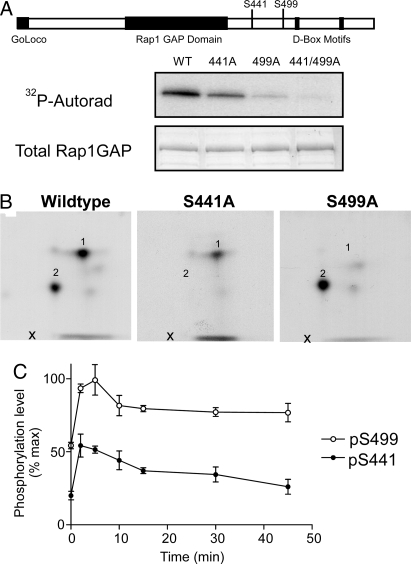
Previous in vitro studies of Rap1GAP identified 2 sites, at Ser-490 and Ser-499, that could be phosphorylated by PKA (10). Our phosphopeptide mapping analysis of Rap1GAP purified from transiently transfected HEK293 cells and phosphorylated by PKA revealed 2 major phosphopeptides. Mutation of Ser-499 to Ala removed only 1 of the observed phosphopeptides (Fig. 3 A and B). Mutagenesis of Ser-490 had no effect on phosphorylation of Rap1GAP and we found no evidence for phosphorylation of this site. However, mutation of other candidate PKA phosphorylation sites led to the identification of Ser-441 as the second major phosphorylated amino acid (Fig. 3 A and B and data not shown). Mutation of both sites resulted in a version of Rap1GAP that was not significantly phosphorylated by PKA in vitro (Fig. 3A). These 2 PKA phosphorylation sites on Rap1GAP are located in an area of unknown significance C-terminal to a well-characterized GAP domain (11).
Fig. 3.
Phosphorylation of Rap1GAP by PKA in striatal slices. (A) Wild-type or mutant 6xHis-Rap1GAP [in which Ser-441 (S441A), Ser-499 (S499A), or both (S441/499A) were mutated to alanine] were purified from HEK293 cells using Ni-NTA Agarose (Qiagen) and phosphorylated on the beads by a PKA catalytic subunit in the presence of [γ-32P]ATP. Samples were analyzed by SDS/PAGE and autoradiography. Total Rap1GAP levels were analyzed using Coomassie stain. (B) Phospho-Rap1GAP bands were then subjected to tryptic digestion and analyzed by 2-dimensional phosphopeptide mapping as described in Fig. 1. Numbers indicate the location of major phosphopeptides. The positions of the phosphorylation sites are shown in the cartoon at the Top of the figure. In the absence of added PKA, there was a low level of phosphorylation of Rap1GAP that was distributed in some of the minor peptides but not in peptides 1 and 2. (C) Mouse neostriatal slices were treated for the indicated times with 10 μM SKF38393. Samples were then analyzed by SDS/PAGE and immunoblotting using antibodies specific for total Rap1GAP or for Rap1GAP phosphorylated at Ser-499 or Ser-441 (see immunoblots in Fig. S4 in SI). Rap1GAP phosphorylation was normalized to total Rap1GAP and also to the level of phosphorylation observed in the presence of the protein phosphatase inhibitor, okadaic acid (see Fig. S4; n = 4; error bars show mean ± SEM).
To further examine the regulation of phosphorylation at Ser-441 and Ser-499, phosphorylation-state specific antibodies were produced for each site (Fig. S3). These antibodies were then used to analyze Rap1GAP phosphorylation in ex vivo striatal slice preparations. Both sites were phosphorylated under basal conditions. Treatment of slices with the dopamine D1 agonist SKF38393 resulted in a rapid increase in phosphorylation of both Ser-441 and Ser-499, peaking after 5–10 min and followed by a slow decline (Fig. 3C).
Phosphorylation of Rap1GAP Controls Rap1 Activity.
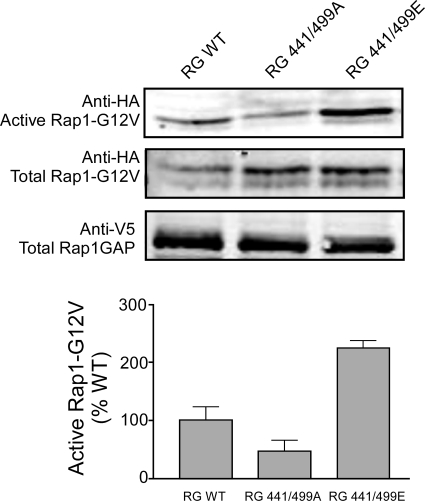
The role of Rap1GAP phosphorylation by PKA was analyzed using site-directed mutagenesis. Wild-type Rap1GAP, or mutants in which both phosphorylation sites were changed to alanine to mimic the dephospho-state, or to glutamate to mimic the phospho-state, were expressed in N2a cells (Fig. 4). To compensate for the increase in Rap1GAP activity that accompanies Rap1GAP overexpression, we coexpressed Rap1 with a Gly12Val mutation that greatly increases the affinity of Rap1 for GTP while remaining susceptible to hydrolysis and inactivation by GAPs (11–13). Using a RalGDS pull-down assay to measure Rap1 activity (14), we found that mutation of Ser-441 and Ser-499 to alanine resulted in a lower level of active Rap1. Conversely, mutation of both PKA sites to glutamate resulted in a higher level of active Rap1.
Fig. 4.
Mutation of Rap1GAP phosphorylation sites modulates Rap1 activation. Neuro-2a (N2a) cells in 10-cm plates were transiently transfected with HA-Rap1Gly12Val (5 μg) plus the indicated V5-Rap1GAP wild-type (WT) or mutant constructs (1 μg each), using Fugene6 (Roche). After 20 h, cells were serum starved for 24 h and then harvested, and Rap1 activity was measured using the RalGDS pull-down assay. The Upper panels show immunoblot analyses of active HA-Rap1Gly12Val-GTP (Rap1-GTP) and total levels of HA-Rap1Gly12Val (total Rap1) and V5-Rap1GAP using specific epitope-tag antibodies. Quantitation of Rap1-GTP levels was normalized to total levels of protein and expressed as a percentage of control. Significance for each group was determined by ANOVA, P < 0.001. Each bar represents the mean ± SEM (n = 6 from 2 experiments).
cAMP- and Ca2+-Dependent Regulation of Rap1GAP Phosphorylation and Rap1 Activity in Striatal Slices.
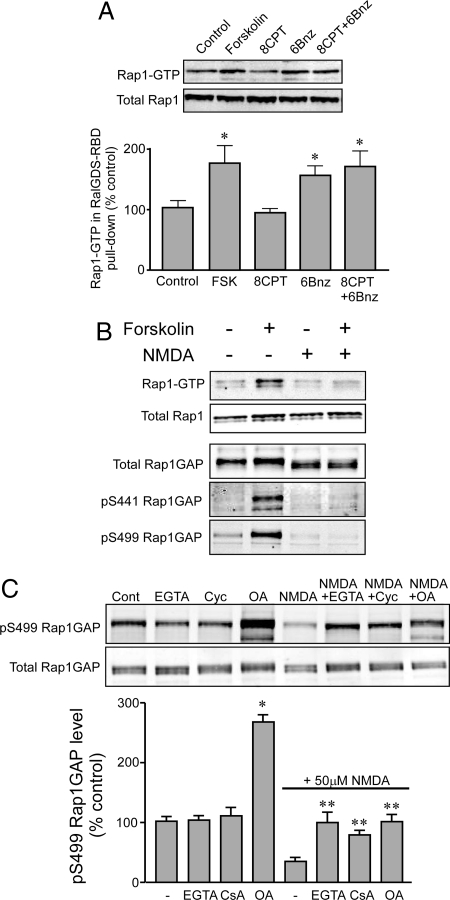
While cAMP is known to regulate Rap1 activity via direct activation of the Rap guanine nucleotide exchange factor (GEF), exchange protein factor directly activated by cyclic AMP (EPAC) (15, 16), our results suggested an alternative mechanism involving the regulation of Rap1GAP by PKA-dependent phosphorylation. We therefore examined the potential role of Rap1GAP phosphorylation in Rap1 activation, using striatal slices. cAMP production induced by forskolin was found to significantly increase the level of active Rap1-GTP after 5 min (Fig. 5A). This effect was also observed using the PKA-selective cAMP analog N6-Benzoyladenosine-cAMP (6Bnz) but not with the EPAC-selective analog 8-(4-Chlorophenylthio)-2′-O-methyl-cAMP (8-CPT-2Me-cAMP) (8CPT) under these conditions.
Fig. 5.
Regulation of Rap1 in striatal slices. (A) Striatal slices were incubated for 5 min without treatment (control), or with 10 μM forskolin, 8CPT-2Me-cAMP, or 6Bnz-cAMP. Active Rap1-GTP levels were determined using a GST-RalGDS pull-down assay. Upper panels show immunoblots for Rap1-GTP pulled down by GST-RalGDS and for total Rap1 in the striatal slice lysates. The Lower bar graph shows quantitation of multiple experiments. Data were normalized to total Rap1 levels in each sample and then expressed as a percentage of the control value. Asterisks indicate results significantly different from control (1-way ANOVA with Tukey's post hoc test, n = 5, P < 0.05). (B) Striatal slices were incubated for 10 min without or with forskolin (10 μM), NMDA (50 μM), or forskolin plus NMDA (NMDA added 10 min before forskolin). Rap1 activity was measured by the RalGDS pull-down assay as in A. Rap1GAP phosphorylation was measured by immunoblotting with antibodies specific for total Rap1GAP or for Rap1GAP phosphorylated at Ser-499 or Ser-441. Quantitation of the Rap1 assay is shown in Fig. S5 in SI. (C) Striatal slices were pretreated with phosphatase inhibitors [5 mM EGTA, 10 μM Cyclosporin A (Cyc), 1 μM Okadaic Acid (OA)] for 30 min before stimulation with 50 μM NMDA for 10 min as indicated. Rap1GAP phosphorylation was measured by immunoblotting. Data for phospho-Ser-499 were normalized to total Rap1GAP levels in each sample and then to control. Data are means ± SEM, n = 4. *, P < 0.01 vs. control; **, P < 0.01 vs. NMDA alone.
Our previous studies of striatal phosphoproteins have highlighted a role for glutamate-dependent regulation of protein phosphatases PP2A and PP2B (17, 18). Pretreatment of striatal slices with NMDA efficiently blocked the forskolin-induced Rap1GAP phosphorylation at Ser-441 and Ser-499, as well as Rap1 activation (Fig. 5B and Fig. S5). NMDA treatment, in the absence or presence of forskolin, also led to faster migration of Rap1GAP on SDS/PAGE, suggesting the possible dephosphorylation of additional sites in the protein. Pretreatment of slices with a Ca2+ chelator, or with the PP2B inhibitor cyclosporin A, blocked the dephosphorylation at Ser-499 caused by NMDA (Fig. 5C). NMDA treatment also reduced phosphorylation of Ser-499 in the presence of okadaic acid, although notably okadaic acid increased basal phosphorylation ≈2-fold. Together these results suggest that both PP2A and PP2B can dephosphorylate Rap1GAP and that PP2B likely mediates the effect of NMDA receptor activation.
Rap1GAP contains a truncated N-terminal GoLoco domain that interacts with various membrane-bound Gα subunits. Activation of Gαz induces the translocation of Rap1GAP from the cytosol to form membrane complexes and inhibits Rap1 signaling in PC12 cells (19). We examined soluble and particulate fractions from forskolin- or NMDA-treated striatal slices by SDS/PAGE. No evidence for translocation of Rap1GAP was observed (data not shown). Previous in vitro studies have reported no effect of phosphorylation by PKA on Rap1GAP activity toward purified bacterial Rap1 (20). We also found no change in Rap1GAP activity in striatal slice lysates following prior forskolin treatment (data not shown).
Phosphorylation of Rap1GAP Controls Dendritic Spine Morphology.
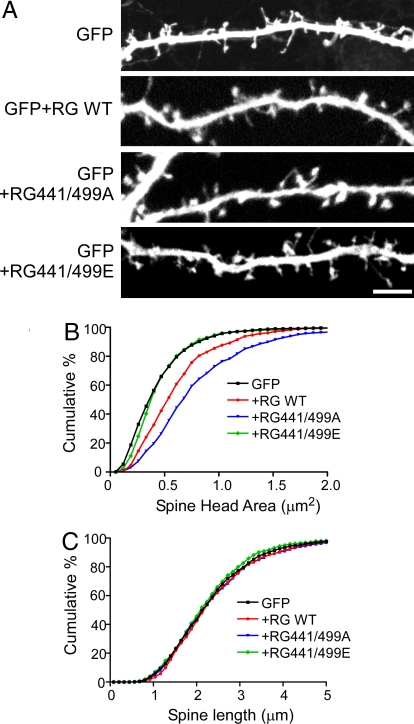
Neuronal Rap1 is known to regulate dendritic spine structure (21, 22). We therefore examined the ability of phosphorylation of Rap1GAP to influence this phenomenon. Mature hippocampal neuronal cultures from E18 rat embryos were transfected with GFP alone or GFP plus the wild-type or phosphorylation-site mutant Rap1GAP constructs (Fig. 6A). A significant increase in spine head area was found following expression of wild-type Rap1GAP, and a greater increase was found with expression of the Ser-441/499Ala mutant. In contrast, expression of the Ser-441/499Glu mutant had no significant effect on spine head area. None of the Rap1GAP constructs had any effect on spine length (Fig. 6C).
Fig. 6.
Phosphorylation of Rap1GAP regulates dendritic spine morphology. (A) Primary hippocampal neurons were transfected at 18 days in vitro (DIV) with GFP alone or GFP together with the indicated wild-type and mutant Rap1GAP constructs. Spine morphology was examined at 20 DIV. Images show the GFP expression. (Scale bar, 10 μm.) (B) Cumulative frequency distribution of spine areas for neurons transfected with the Rap1GAP constructs. For each construct, 1,000–1,500 spines from 20–30 neurons combined from 5 independent experiments were used. (C) Cumulative frequency distribution of spine lengths for the same spines analyzed in B.
Discussion
Signaling through Rap1 has been the subject of intense study, as activation of this cascade is thought to play an important role in cell adhesion (23, 24), polarity (25), and secretion (26, 27). The mode, specificity, and localization of Rap1 action is determined by the presence of various upstream Rap1 GEFs and GAPs and downstream effector proteins. Rap1 activity can also be controlled by the second messengers, cAMP and Ca2+. Recent studies have highlighted the roles of the Rap GEFs, EPAC1 and EPAC2, which are directly activated by cAMP (16, 28). A number of studies also indicate that Rap1 can be regulated by cAMP via PKA (29, 30), perhaps through direct phosphorylation of Rap1 (31, 32). In the present study, we have identified Rap1GAP as a prominent phosphoprotein that is enriched in striatal MSNs and shown that phosphorylation by PKA is associated with inhibition of Rap1GAP activity. Increased phosphorylation of Rap1GAP by PKA is therefore coupled to activation of Rap1 and represents an alternative mode of regulation of Rap1 by cAMP.
Rap1GAP is found in many cell types but is highly expressed in the brain (33, 34). Our studies have shown that while Rap1GAP is enriched in MSNs of striatum, the protein is widely distributed in the brain. Regulation of Rap1GAP by phosphorylation is therefore likely to contribute to control of Rap1 in many parts of the brain. In neurons, Rap1 has been found to regulate both structural and functional processes underlying synaptic plasticity and consequently learning and memory. However, the precise role of Rap1 is unclear. For example, studies with transgenic mice expressing a dominant-negative form of Rap1 in brain have suggested a role for the protein in cAMP- and ERK-dependent gating of long-term potentiation (35). The ability of Rap1 to control ERK in brain is consistent with studies in PC12 cells, where cAMP-dependent Rap1 activation provides a long-lasting stimulation of ERK that is required for differentiation and growth (32). Phosphorylation of Rap1GAP by PKA might therefore contribute to the regulation of ERK observed in those studies. In contrast, Rap1 can act in opposition to Ras to control ERK activation (36, 37), and Rap1 has been found to mediate the NMDA-dependent removal of AMPA receptors during long-term depression, an effect that was opposite to that of Ras (38).
Rap1 has also been shown to influence dendritic spine morphology. Spine-associated Rap1GAP (SPAR) is localized in the postsynaptic density through its association with PSD-95, and expression of SPAR results in increased spine head size (21). Other studies have found that expression of active Rap1 in neurons leads to formation of thin spines with reduced levels of AMPA receptors, a process that involves the PDZ-containing protein AF-6 (22). Our studies of the effects of expression of Rap1GAP are consistent with these reports. Expression of wild-type Rap1GAP, which would reduce Rap1 activity, resulted in spines with larger heads. Compatible with the observation that Rap1GAP was phosphorylated to a significant level at Ser-441 and Ser-449 under basal conditions, expression of the Ser-441/499Ala mutant had a larger effect than the wild-type protein on spine head size. Expression of the Ser-441/499Glu mutant had no effect on spine head size, presumably reflecting the inactive state of the phosphoprotein mimic.
Mechanistically, it is unclear how phosphorylation of Rap1GAP by PKA influences the function of its GAP domain. Consistent with another study (20), we did not measure any effect of Rap1GAP phosphorylation using an in vitro Rap1 GAP assay. Perhaps posttranslational modifications lacking in the recombinant Rap1 used may be responsible for this observation. For example, Rap1 has been shown to be a direct substrate for PKA (39). Other proteins, such as Gαo, that interact independently with both Rap1GAP (40) and the PKA catalytic subunit (41), may also be required to observe the effect of phosphorylation. Additionally, gel shifts of Rap1GAP seen with NMDA treatment suggest the presence of additional unidentified Rap1GAP phosphorylation sites that may also participate in regulating its GAP activity (see also refs. 20 and 42). Previous studies have found that Rap1GAP is a substrate for several serine/threonine kinases including PKA, GSK3β, and cdc2 kinase (10, 42). Our studies identified 2 major PKA phosphorylation sites at Ser-441 and Ser-499. Ser-499 is contained within a strong canonical PKA phosphorylation motif, and although phosphorylated to a significant level under basal conditions, phosphorylation at this site occurred physiologically in response to a dopamine D1 receptor agonist. Dephosphorylation of Ser-499 was attributed to the phosphatases PP1/PP2A under basal conditions and to PP2B (calcineurin) in response to treatment with the glutamate receptor agonist, NMDA. The other identified PKA site, Ser-441, contains a low-homology PKA motif that was phosphorylated to lower levels than Ser-499 under basal conditions; however, its responsiveness to dopamine was similarly robust.
Our findings provide a new mechanism whereby dopamine signaling can influence synaptic structure and function through the regulation of Rap1GAP phosphorylation and Rap1 inactivation. Rap1GAP is highly expressed in MSNs of striatum, where its phosphorylation and activity are controlled by dopamine acting at D1 receptors. MSNs also express high levels of the Rap1 GEF CalDAG-GEFI (43). These observations suggest that a unique system supporting Rap1 signaling exists in striatal neurons. Dopamine clearly has a key role in controlling signaling and plasticity in MSNs (1, 44–46). GABAergic MSNs, which represent ≈95% of all neurons in striatum, are composed of 2 intermingled subpopulations. One subpopulation of MSNs expresses high levels of dopamine D1 receptors, while the other expresses high levels of dopamine D2 receptors (1, 44–46). Recent studies using transgenic mice where different proteins have been selectively expressed in either subpopulation of MSNs have shown distinct patterns of phosphorylation of signaling molecules (47, 48), gene expression (49), and synaptic plasticity (50). Chronic exposure to psychostimulants has been found to increase dendritic spine density in MSNs in the dorsal and ventral striatum (51), and these drug-induced changes in spine morphology are more sustained in D1-containing MSNs (52). As a substrate for PKA whose phosphorylation is stimulated by activation of D1 receptors, Rap1GAP is likely to be differentially phosphorylated in D1 receptor-containing MSNs and this may contribute to the distinct patterns of signaling and plasticity observed in the 2 subpopulations of MSNs.
Methods
Materials and Reagents.
Antibodies to Rap1 and Rap1GAP (N-terminal, clone Y134) were obtained from Epitomics or Santa Cruz (Rap1GAP, sc-v19). Antibodies for V5 and HA epitopes, and β-actin, were from Abcam. 8CPT-2Me-cAMP and 6-Bnz cAMP were from Biolog Life Science Institute (Bremen, Germany). All other drugs were from Tocris Bioscience. The plasmids pGEX-RalGDS-RBD and pMT2-HA-Rap1Gly12Val were kind gifts from Johannes L. Bos (Utrecht University, The Netherlands). The Rap1GAP full-length human clone (pcDNA3.1-Rap1GAP-V5/HIS) was from the Invitrogen clone collection. Site-directed mutagenesis used the Quickchange method (Stratagene) to create alanine and glutamate mutations at amino acid residues Ser-441 and Ser-499 of pGS-Rap1GAP-V5/HIS.
Purification and Identification of ARPP-90.
ARPP-90 was purified using ion exchange chromatography and gel filtration as described in detail in SI Text. Protein sequencing was carried out using LC/MS/MS analysis (Rockefeller University DNA/Protein Technology Center). Two-dimensional phosphopeptide mapping was performed essentially as described (53) (see also SI for details).
Generation of Phospho-Specific Antibodies.
Antibodies recognizing phospho-Ser-441 and phospho-Ser-499 of Rap1GAP were generated against the phosphopeptides SRSQpSMDAMC and SRRSpSAIGIC, respectively. Each peptide (4 mg) was conjugated via its C-terminal cysteine to Limulus hemocyanin (LHC, Sigma), using m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester (sulfo-MBS, Sigma), and used to produce polyclonal antibodies in rabbits (Cocalico Biologicals). Antibodies were affinity purified on columns (SulfoLink gel, Pierce) coupled to the respective phosphopeptide.
RalGDS RBD Pull-Down Assay.
The RalGDS RBD pull-down assay was carried out essentially as described (14). After treatment, 2 frozen striatal slices were combined and sonicated in 1 mL TLB buffer [50 mM Tris (pH 7.4)/500 mM NaCl/1% Nonidet P-40/2.5 mM MgCl2/10 mM β-glycerophosphate/20 mM NaF/10% glycerol/1 mM PMSF/miniEDTA-free protease inhibitor (Roche)] also containing 20 μg/mL purified GST-RalGDS RBD. After centrifugation at 14,000 × g for 10 min at 4 °C, a 60-μL aliquot was removed for analysis of total protein level. The remaining extract was incubated with 10 μL MagneGST Glutathione particles (Promega) and incubated for 45 min with rotation. The particles were washed 3 times with TLB buffer and boiled 5 min in 20 μL sample buffer with an additional 100 mM DTT. Samples were analyzed by SDS/PAGE and immunoblotting.
In Situ Hybridization, Immunohistochemistry, Neostriatal Slice Preparation, Neuronal Culture, Transfection, and Immunostaining.
Standard methods were used for these routine procedures. Details are provided in SI. Images of dendritic segments were acquired with a Zeiss confocal microscope, using z-series stacks to fully encompass the entire segment. All healthy pyramidal cells that expressed the appropriate constructs at levels allowing clear visualization of dendritic spines were included in the analysis. Image analysis of the GFP channel for each neuron was performed using Metamorph software (Molecular Devices). For analysis of spine area, spines were thresholded to a similar level and carefully outlined. Statistical significance of cumulative frequency plots was determined using the Kolmogorov–Smirnov test.
Supplementary Material
Acknowledgments.
This work was supported by funding from the National Institutes of Health (DA10044 and MH74866 to A.C.N. and P.G. and DA11717 to A.C.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813263106/DCSupplemental.
References
- 1.Calabresi P, Picconi B, Tozzi A, Di Filippo M. Dopamine-mediated regulation of corticostriatal synaptic plasticity. Trends Neurosci. 2007;30:211–219. doi: 10.1016/j.tins.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Greengard P. The neurobiology of slow synaptic transmission. Science. 2001;294:1024–1030. doi: 10.1126/science.294.5544.1024. [DOI] [PubMed] [Google Scholar]
- 3.Fienberg AA, et al. DARPP-32: regulator of the efficacy of dopaminergic neurotransmission. Science. 1998;281:838–842. doi: 10.1126/science.281.5378.838. [DOI] [PubMed] [Google Scholar]
- 4.Rakhilin SV, et al. A network of control mediated by regulator of calcium/calmodulin-dependent signaling. Science. 2004;306:698–701. doi: 10.1126/science.1099961. [DOI] [PubMed] [Google Scholar]
- 5.Walaas SI, Nairn AC, Greengard P. Regional distribution of calcium- and cyclic adenosine 3′:5′-monophosphate-regulated protein phosphorylation systems in mammalian brain. II. Soluble systems. J Neurosci. 1983;3:302–311. doi: 10.1523/JNEUROSCI.03-02-00302.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walaas SI, Cala S, Greengard P. Localization of ARPP-90, a major 90 kiloDalton basal ganglion-enriched substrate for cyclic AMP-dependent protein kinase, in striatonigral neurons in the rat brain. Brain Res Mol Brain Res. 1989;5:149–157. doi: 10.1016/0169-328x(89)90006-5. [DOI] [PubMed] [Google Scholar]
- 7.Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci. 1984;4:84–98. doi: 10.1523/JNEUROSCI.04-01-00084.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmings HC, Jr, Greengard P. ARPP-21, a cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Purification and characterization of the protein from bovine caudate nucleus. J Neurosci. 1989;9:851–864. doi: 10.1523/JNEUROSCI.09-03-00851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walaas SI, Aswad DW, Greengard P. A dopamine- and cyclic AMP-regulated phosphoprotein enriched in dopamine-innervated brain regions. Nature. 1983;301:69–71. doi: 10.1038/301069a0. [DOI] [PubMed] [Google Scholar]
- 10.Rubinfeld B, et al. Localization of the rap1GAP catalytic domain and sites of phosphorylation by mutational analysis. Mol Cell Biol. 1992;12:4634–4642. doi: 10.1128/mcb.12.10.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkmann T, et al. Rap-specific GTPase activating protein follows an alternative mechanism. J Biol Chem. 2002;277:12525–12531. doi: 10.1074/jbc.M109176200. [DOI] [PubMed] [Google Scholar]
- 12.Maruta H, Holden J, Sizeland A, D'Abaco G. The residues of Ras and Rap proteins that determine their GAP specificities. J Biol Chem. 1991;266:11661–11668. [PubMed] [Google Scholar]
- 13.Scheffzek K, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 14.M'Rabet L, et al. Activation of the small GTPase rap1 in human neutrophils. Blood. 1998;92:2133–2140. [PubMed] [Google Scholar]
- 15.de Rooij J, et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 16.Bos JL. Epac proteins: multi-purpose cAMP targets. Trends Biochem Sci. 2006;31:680–686. doi: 10.1016/j.tibs.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Ahn JH, et al. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci USA. 2007;104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina I. Extrasynaptic NMDA receptors reshape gene ranks. Sci STKE. 2007;2007:pe23. doi: 10.1126/stke.3862007pe23. [DOI] [PubMed] [Google Scholar]
- 19.Meng J, Casey PJ. Activation of Gz attenuates Rap1-mediated differentiation of PC12 cells. J Biol Chem. 2002;277:43417–43424. doi: 10.1074/jbc.M204074200. [DOI] [PubMed] [Google Scholar]
- 20.Polakis P, Rubinfeld B, McCormick F. Phosphorylation of rap1GAP in vivo and by cAMP-dependent kinase and the cell cycle p34cdc2 kinase in vitro. J Biol Chem. 1992;267:10780–10785. [PubMed] [Google Scholar]
- 21.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 22.Xie Z, Huganir RL, Penzes P. Activity-dependent dendritic spine structural plasticity is regulated by small GTPase Rap1 and its target AF-6. Neuron. 2005;48:605–618. doi: 10.1016/j.neuron.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Retta SF, Balzac F, Avolio M. Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol. 2006;85:283–293. doi: 10.1016/j.ejcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 25.Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Dremier S, et al. Cyclic adenosine 3′,5′-monophosphate (cAMP)-dependent protein kinases, but not exchange proteins directly activated by cAMP (Epac), mediate thyrotropin/cAMP-dependent regulation of thyroid cells. Endocrinology. 2007;148:4612–4622. doi: 10.1210/en.2007-0540. [DOI] [PubMed] [Google Scholar]
- 27.Li J, et al. Cyclic adenosine 5′-monophosphate-stimulated neurotensin secretion is mediated through Rap1 downstream of both Epac and protein kinase A signaling pathways. Mol Endocrinol. 2007;21:159–171. doi: 10.1210/me.2006-0340. [DOI] [PubMed] [Google Scholar]
- 28.Holz GG, Kang G, Harbeck M, Roe MW, Chepurny OG. Cell physiology of cAMP sensor Epac. J Physiol. 2006;577:5–15. doi: 10.1113/jphysiol.2006.119644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obara Y, Labudda K, Dillon TJ, Stork PJ. PKA phosphorylation of Src mediates Rap1 activation in NGF and cAMP signaling in PC12 cells. J Cell Sci. 2004;117:6085–6094. doi: 10.1242/jcs.01527. [DOI] [PubMed] [Google Scholar]
- 30.Zanassi P, et al. cAMP-dependent protein kinase induces cAMP-response element-binding protein phosphorylation via an intracellular calcium release/ERK-dependent pathway in striatal neurons. J Biol Chem. 2001;276:11487–11495. doi: 10.1074/jbc.M007631200. [DOI] [PubMed] [Google Scholar]
- 31.Hu CD, et al. Effect of phosphorylation on activities of Rap1A to interact with Raf-1 and to suppress Ras-dependent Raf-1 activation. J Biol Chem. 1999;274:48–51. doi: 10.1074/jbc.274.1.48. [DOI] [PubMed] [Google Scholar]
- 32.Vossler MR, et al. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 33.Rubinfeld B, et al. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–1042. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- 34.Kurachi H, et al. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2. Segregate expression profiles from a rap1GAP gene product. J Biol Chem. 1997;272:28081–28088. doi: 10.1074/jbc.272.44.28081. [DOI] [PubMed] [Google Scholar]
- 35.Morozov A, et al. Rap1 couples cAMP signaling to a distinct pool of p42/44MAPK regulating excitability, synaptic plasticity, learning, and memory. Neuron. 2003;39:309–325. doi: 10.1016/s0896-6273(03)00404-5. [DOI] [PubMed] [Google Scholar]
- 36.Stork PJ, Dillon TJ. Multiple roles of Rap1 in hematopoietic cells: complementary versus antagonistic functions. Blood. 2005;106:2952–2961. doi: 10.1182/blood-2005-03-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwartkruis FJ, Bos JL. Ras and Rap1: two highly related small GTPases with distinct function. Exp Cell Res. 1999;253:157–165. doi: 10.1006/excr.1999.4695. [DOI] [PubMed] [Google Scholar]
- 38.Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 39.Altschuler D, Lapetina EG. Mutational analysis of the cAMP-dependent protein kinase-mediated phosphorylation site of Rap1b. J Biol Chem. 1993;268:7527–7531. [PubMed] [Google Scholar]
- 40.Jordan JD, Carey KD, Stork PJ, Iyengar R. Modulation of rap activity by direct interaction of Galpha(o) with Rap1 GTPase-activating protein. J Biol Chem. 1999;274:21507–21510. doi: 10.1074/jbc.274.31.21507. [DOI] [PubMed] [Google Scholar]
- 41.Ghil S, et al. Compartmentalization of protein kinase A signaling by the heterotrimeric G protein Go. Proc Natl Acad Sci USA. 2006;103:19158–19163. doi: 10.1073/pnas.0609392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsygankova OM, Feshchenko E, Klein PS, Meinkoth JL. Thyroid-stimulating hormone/cAMP and glycogen synthase kinase 3beta elicit opposing effects on Rap1GAP stability. J Biol Chem. 2004;279:5501–5507. doi: 10.1074/jbc.M305824200. [DOI] [PubMed] [Google Scholar]
- 43.Kawasaki H, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 45.Surmeier DJ, Ding J, Day M, Wang Z, Shen W. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 2007;30:228–235. doi: 10.1016/j.tins.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Kreitzer AC, Malenka RC. Striatal plasticity and basal ganglia circuit function. Neuron. 2008;60:543–554. doi: 10.1016/j.neuron.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bateup HS, et al. Cell type-specific regulation of DARPP-32 phosphorylation by psychostimulant and antipsychotic drugs. Nat Neurosci. 2008;11:932–939. doi: 10.1038/nn.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertran-Gonzalez J, et al. Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. J Neurosci. 2008;28:5671–5685. doi: 10.1523/JNEUROSCI.1039-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen W, Flajolet M, Greengard P, Surmeier DJ. Dichotomous dopaminergic control of striatal synaptic plasticity. Science. 2008;321:848–851. doi: 10.1126/science.1160575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47(Suppl 1):33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 52.Lee KW, et al. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc Natl Acad Sci USA. 2006;103:3399–3404. doi: 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai Y, Nairn AC, Gorelick F, Greengard P. Ca2+/calmodulin-dependent protein kinase II: identification of autophosphorylation sites responsible for generation of Ca2+/calmodulin-independence. Proc Natl Acad Sci USA. 1987;84:5710–5714. doi: 10.1073/pnas.84.16.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.