Abstract
We present a general strategy for identification of conformation-specific antibodies using phage display. Different covalent probes were used to trap caspase-1 into 2 alternative conformations, termed the on-form and the off-form. These conformation-trapped forms of the protease were used as antigens in alternating rounds of selection and antiselection for antibody antigen-binding fragments (Fabs) displayed on phage. After affinity maturation, 2 Fabs were isolated with KD values ranging from 2 to 5 nM, and each bound to their cognate conformer 20- to 500-fold more tightly than their noncognate conformer. Kinetic analysis of the Fabs indicated that binding was conformation dependent, and that the wild-type caspase-1 sits much closer to the off-form than the on-form. Bivalent IgG forms of the Fabs were used to localize the different states in cells and revealed the activated caspase-1 is concentrated in a central structure in the cytosol, similar to what has been described as the pyroptosome. These studies demonstrate a general strategy for producing conformation-selective antibodies and show their utility for probing the distribution of caspase-1 conformational states in vitro and in cells.
Keywords: allostery, caspase-1, phage display, protein conformational change
Protein allostery is a central means to regulate protein function in cells. Allostery is mediated through conformational selection upon binding of different small molecules, biopolymers, or metal ions or through posttranslational modifications. Structural methods give us high-resolution insight into the nature of these conformational transitions in vitro but have limited use for determining the equilibrium distribution of these states in solution or in cells. To expand the tools useful for trapping and analyzing conformational states in enzymes, both in solution and in cells, we developed a general strategy for 2-state selection of conformation-specific antibodies using phage display.
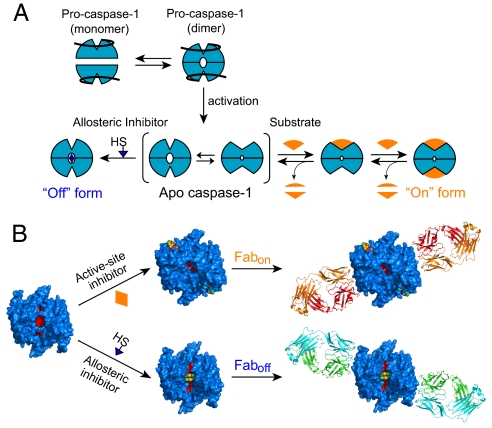
As a test case, we focused on caspase-1, an aspartate-specific thiol protease that is critical for processing of proinflammatory cytokines during the innate immune response (for review, see refs. 1–3). The enzyme is produced as an inactive proenzyme that exists primarily as a monomer in solution (4, 5). Upon innate immune stimuli, the proenzyme is believed to dimerize by binding to scaffolding proteins known collectively as the inflammasome. This triggers proteolytic autoactivation or transactivation, in which the propeptide and an intersubunit linker are cleaved (6, 7). Crystal structures of the mature protease with various small molecules bound show that it can exist in at least 2 conformations (8–10). When an active site inhibitor is bound, the enzyme appears to be in a catalytically competent form (called the on-form) (9). However, binding of covalent disulfide ligand to a central cavity ≈15 Å from the active site stabilizes the protease in an inactive state (called the off-form) (8). This allosterically inhibited state is virtually identical to the apo-form of the enzyme as seen in the crystal structures. The dimeric enzyme shows positive cooperativity [nhill = 1.5 (8)], and mutational studies reveal that only a small set of residues (a “hot-wire”) mediates the on-to-off transition between the allosteric and active sites (11). These data support a dynamic activation model for caspase-1 (Fig. 1A).
Fig. 1.
Model and labeling design. (A) Proposed model for the dynamic activation of caspase-1. It has been suggested that in cells procaspase-1 exists primarily as monomer. Upon binding to scaffolding proteins (NALPs, ASC, etc.), procaspase-1 dimerizes and undergoes proteolytic activation. Mature caspase-1 is in equilibrium between off- and on-conformations. Binding of ligands at the active or allosteric site can shift the equilibrium toward the on- or off-state. (B) Covalent labeling of apo-caspase-1. Irreversible active-site inhibitors or allosteric compounds were used to trap caspase-1 into a stable conformation for antibody selection.
We wished to generate monoclonal antibodies to each of the on- and off-states to better understand the equilibrium distribution of these states in solution and in cells and to provide probes to localize these forms in cells. We trapped homogeneous forms of the on- and off-states of caspase-1 using the active site inhibitor (Ac-YVAD-cmk) to lock the on-state and compound 34 (1-methyl-3-trifluoromethyl-1H-thieno[2,3-c]pyrazole) to lock the off-state. These conformation-locked forms of caspase-1 were then used as antigens in sorting codon-restricted phage display libraries (12) to generate high-affinity antibody fragments (Fabs) that were selective for either the on- or the off-state. The combination of small molecules to trap different molecular conformations coupled with in vitro selection provides a general approach to produce multiple conformation- selective antibodies to biomolecules.
Results
Selection of Conformation-Specific Fabs Using Phage Display.
A major challenge to produce conformation-selective antibodies against caspase-1 is to have relatively homogenous preparations of each conformer. To lock caspase-1 in the off-state, we labeled the enzyme with a thiol-containing compound 34 that covalently bound to Cys-331 on the small subunit within the allosteric pocket (8). Similarly, an irreversible active-site inhibitor Ac-YVAD-cmk (CalBiochem) was used to trap the on-state (Fig. 1B). The stoichiometric labeling by the different covalent inhibitors to thiols in either the active site or allosteric site was confirmed by LC-MS (Fig. S1).
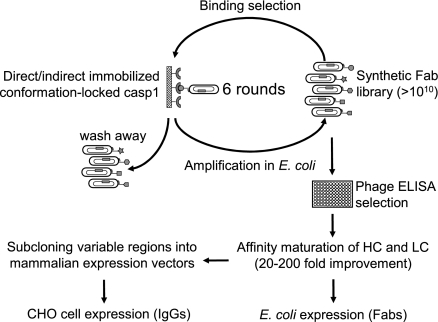
We used a phage-display strategy that allowed us to maintain the state of antigen throughout the selection process in vitro (Fig. 2). Two synthetic libraries were built on a single antibody fragment framework by introducing limited diversity into the 3 heavy chain CDRs (Complementarity-determining regions: CDR-H1, -H2, and -H3) and the third CDR of the light chain (CDR-L3) (13, 14). A Tyr/Ser focused library was used that explicitly placed Tyr and Ser at solvent accessible positions responsible for antigen recognition (14). A focused Tyr/Ser/Gly/Arg library added further chemical diversity by incorporating Gly and Arg to the binary Tyr/Ser background in CDR-H3 (25% Tyr, 25% Ser, 25% Gly, 25% Arg) (13). The combined naïve library pools contained >1010 unique clones.
Fig. 2.
Selection for conformation-selective antibodies. Six rounds of phage selection were performed for each target. Colonies were picked and tested by spot phage ELISA. Positive clones that showed both good affinity and selectivity were analyzed by DNA sequencing. HC and LC optimization were carried out for affinity maturation. Fabs and IgGs were expressed and purified in E. coli and mammalian cells, respectively. See Materials and Methods for details.
A total of 6 rounds of selection were performed for both target antigens. After the first 2 rounds, antiselection was enforced by preincubating the amplified phage pool with the off-target conformer. Ninety-six colonies were tested by spot phage ELISA after round 6, and clones showing <20-fold selectivity (based on OD450 for the target conformer/OD450 for the off-target conformer) were discarded. To exclude Fabs that bound to the active-site or allosteric inhibitors, a second ELISA screen was performed by using a different set of inhibitor-trapped caspase-1 (z-WEHD-fmk for trapping the on-form and compound 11 for trapping the off-form). Only Fabs that showed high selectivity and that bound independently of the specific inhibitors were sequenced. Affinities of Fabs on selected phage were measured by solution-competition phage ELISA (15, 16) (Table S1).
Two different affinity maturation strategies were undertaken to further improve the Fabs (Fig. 2). We focused first on optimizing Fabs for the on-form by partial randomization of all 3 CDR loops on the heavy chain (17). Four clones with affinities ranging from 50 to 110 nM were used as independent starting templates (Table S1). We found that a single amino acid change (M to T) at position 100c in the CDR-H3 resulted in the biggest improvement in affinity (>20-fold). The tightest binder (called Fabon) was chosen for expression and subsequent analysis (Table 1). The same strategy failed to improve the affinity of Fabs for the off-form. Therefore, we shifted our attention to the light-chain CDR loops by randomizing these sequences based on the natural diversity of human kappa light chain sequences in the Kabat database (18, 19). This resulted in >100-fold improvement of affinity for the best clone (called Faboff, Table 1).
Table 1.
Sequences of Fab clones after affinity maturation
| CDR-H1 | CDR-H2 | CDR-H3 | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28 | 29 | 30 | 31 | 32 | 33 | 50 | 51 | 52 | a | 53 | 54 | 55 | 56 | 57 | 58 | 95 | 96 | 97 | 98 | 99 | 100 | a | b | c | |
| Fabon | N | F | S | Y | Y | S | S | I | S | S | Y | S | S | S | T | S | G | Y | Y | Y | I | G | T | ||
| Faboff | S | I | Y | S | S | S | S | I | S | P | Y | Y | G | Y | T | S | Y | S | S | Y | S | Y | Y | A | F |
| CDR-L1 | CDR-L2 | CDR-L3 | |||||||||||||||||||||||
| 28 | 29 | 30 | 31 | 32 | 33 | 50 | 51 | 52 | 53 | 54 | 55 | 91 | 92 | 93 | 94 | 95 | 96 | ||||||||
| Fabon | S | V | S | S | A | V | S | A | S | S | L | Y | S | Y | S | Y | P | S | |||||||
| Faboff | V | V | V | R | Y | L | L | A | S | N | L | A | S | S | A | F | P | L | |||||||
Only CDR sequences at positions that were randomized in the libraries are shown. The numbering is according to the nomenclature of Kabat et al. (34).
Biochemical Characterization of Conformation-Specific Antibodies.
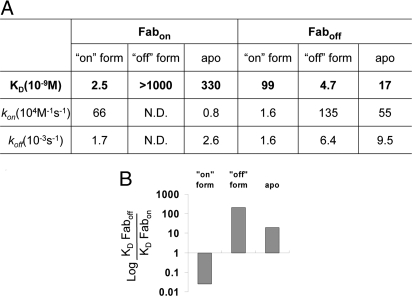
We expressed Fabon and Faboff in Escherichia coli and purified them by protein-A affinity chromatography. To characterize the binding affinity and selectivity of the Fabs, we tested their interaction against various caspase-1 conformers by surface plasmon resonance (SPR). As shown in Fig. 3A, both Fabs bind to their target conformers tightly with low-nM KD values that were similar to those estimated from solution-competition phage ELISA (12.9 ± 1.1 nM for Fabon-phage; 7.9 ± 0.5 nM for Faboff-phage). Fabon showed no binding to the off-state caspase-1 even at highest test concentration (1 μM) on BIAcore, whereas Faboff bound to the on-state caspase-1 at least 20-fold weaker than to its cognate form (99 vs. 4.7 nM, respectively). The high selectivity demonstrated by the Fabs was mostly contributed by the difference in kon not koff. This suggests these Fabs are selective for the specific conformation of caspase-1.
Fig. 3.
Fabs are conformation-selective. (A) SPR analysis of Fabs by BIAcore. Kinetic analysis was performed by immobilizing different forms of caspase-1 on the CM5 chip and injecting serial dilutions of Fabs. BIAevaluation software was used to fit the recorded sensograms and determine the kinetic parameters. (B) The relative difference in KD of Fabs binding to different forms of caspase-1 was calculated from data in A.
Given the sensitivity of the Fabs for different caspase-1 conformers, we were intrigued to investigate the conformational state of ligand-free caspase-1. Kinetic analysis using BIAcore showed that Faboff bound to apo caspase-1 with nearly 20-fold higher affinity than Fabon (17 vs. 330 nM, respectively; Fig. 3A). Again, most of the difference was in kon, not in koff. Comparing how well ligand-free caspase-1 bound to both Fabs with that of the locked-on and -off forms (Fig. 3B), we concluded that caspase-1 is in an equilibrium between the on- and off-states but that the dominant species is much closer to the off-state conformation.
Activation or Inhibition of Caspase-1 with Conformation-Specific Antibodies.
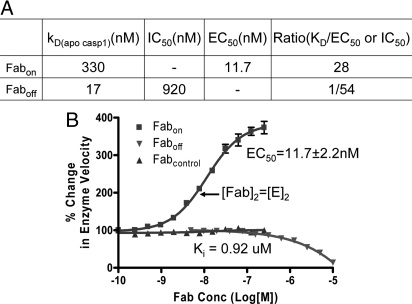
We explored the possibility of affecting caspase-1 activity by stabilizing each of the 2 conformational states upon binding to the 2 different Fabs. As seen in Fig. 4A, Fabon increased the catalytic activity of caspase-1 by over 3-fold in a dose-dependent manner. The EC50 value (11.7 ± 2.2 nM) was close to its KD for the fully locked on-state conformer (2.5 nM, Fig. 3A). In contrast, Faboff functioned as an inhibitor, which prevented substrate binding by trapping the free enzyme in the off-state. The EC50 and IC50 for activation and inhibition were 28-fold lower or 54-fold higher than their respective KDs for binding of the apo-caspase-1 to Fabon or Faboff (Fig. 4B). This is consistent with previous studies showing that the enzyme exhibits positive cooperativity (8, 11), where substrate drives the enzyme from the off- to the on-form (Fig. 1A).
Fig. 4.
Activation or inhibition of caspase-1 activity by Fabs. (A) Caspase-1 (5 nM) was preincubated with serial dilution of Fabs before assaying with fluorescent substrate Ac-WEHD-fmk at 100 μM concentration. The relative change in enzyme velocity was calculated by dividing the enzyme velocity at any given Fab concentration with the average velocity of enzyme in the absence of Fab. Nonlinear regression was used to fit the curve and calculate EC50 and Ki. (B) The ratios of KD(apo caspase-1) for each Fab to the corresponding EC50 or IC50 were calculated from data in A and Fig. 3A.
Effects on Binding to Procaspase-1 in Vitro and in Cells.
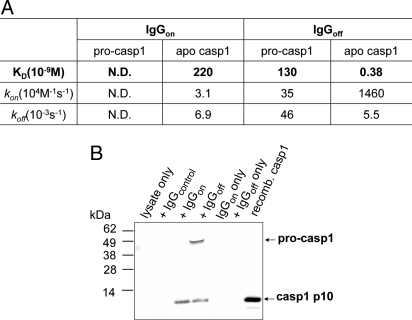
Previous biochemical studies have suggested that procaspase-1 exists as a monomer (4, 5). To determine the binding of the conformation-specific antibodies to procaspase-1, we expressed and purified a recombinant form of procaspase-1 in which the CARD domain was deleted, called p32 (residues 120–404). SPR analysis showed that neither monomeric Fab was able to bind to p32 (Fig. S2). We converted Fabs into full-length human dimeric IgGs and measured their binding to p32 and to ligand-free caspase-1. Both IgGs bound to ligand-free caspase-1 like their Fab counterparts; however, the binding affinity of IgGoff was at least 500-fold stronger than IgGon (Fig. 5A). The dimeric IgGoff was able to bind to p32 with an affinity of 130 nM, possibly by driving the dimerization of p32. Even though we cannot exclude the possibility that the binding epitope of IgGs might include the cleavage site, which is present in the mature form but not in the pro form, it is less likely because both antigens (on- and off-form caspase-1) we used in the selection had the same cleavage site present. That IgGon could not bind but IgGoff suggests p32 exists in a state much closer to the off-state.
Fig. 5.
IgGoff binds to procaspase-1. (A) SPR analysis of IgGs by BIAcore. Kinetic analysis was performed by immobilizing procaspase-1 or apo caspase-1 on the CM5 chip and injecting serial dilutions of IgGs. BIAevaluation software was used to fit the recorded sensograms and determine the kinetic parameters. (B) Immunopreciptation of procaspase-1 and apo caspase-1 from THP-1 lysates by IgGs. Monoclonal anti-caspase-1-p10 antibody (Calbiochem) that recognizes both procaspase-1 and caspase-1 p10 subunit was used in Western blot analysis.
To test whether IgGoff recognizes full-length procaspase-1 in cells, we carried out immunoprecipitation experiments in cell extracts from human THP-1 cells that have high levels of endogenous procaspase-1. As depicted in Fig. 5B, both IgGon and IgGoff pulled down caspase-1, whereas only the latter was able to immunoprecipitate full-length procaspase-1. These results align with the binding data above and further suggest that the IgGs are useful tools for studying the specific conformations of caspase-1 in cells.
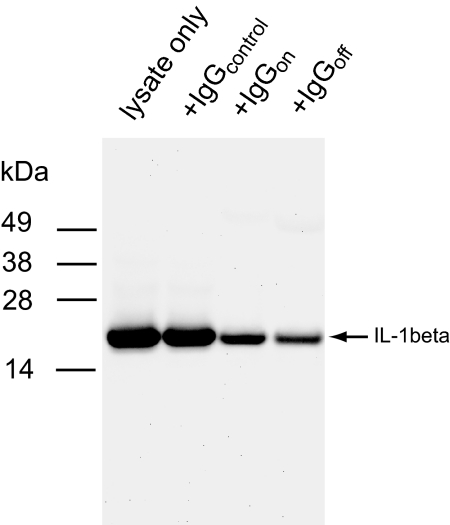
To investigate whether the antibodies have similar effects on caspase-1-mediated cleavage in cells, we examined the processing of pro-IL-1ß, a well characterized natural caspase-1 substrate, in the presence of each of the 2 IgGs. THP-1 cells were stimulated by LPS for 4 h to increase pro-Il-1ß expression, and cell extract was prepared as described (7). Western blots probed for mature Il-1ß showed that IgGoff significantly blocked caspase-1-mediated cleavage of pro-Il-1ß (Fig. 6). IgGon also inhibited processing of pro-IL-1 ß but to a lesser extent possibly by sterically blocking the larger protein substrate binding. These results suggest that IgGoff can effectively inhibit caspase-1 activity in cells by sequestering the enzyme in the off-state.
Fig. 6.
Inhibition of caspase-1 by IgGoff in THP-1 extracts. LPS-stimulated THP-1 cell extracts were incubated with IgGs at 4 °C overnight and probed by Western blot analysis using an antibody specific for mature IL-1ß (Cell Signaling).
Localization of Active Caspase-1 Conformer in Cells.
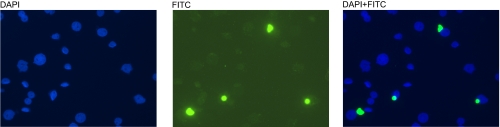
The ability of IgGon to bind selectively to the on-form of caspase-1 and not to procaspase-1 makes it potentially useful for tracking caspase-1 activation in cells. THP-1 cells can be differentiated into macrophages with PMA treatment, which then become adherent to glass coverslips. After fixation and mild detergent permealization, IgGon was added as the probe for the on-state of caspase-1. Immunofluorescent microscopy revealed that most of cells stimulated with LPS/ATP showed a general increase in diffuse fluorescent signal (Fig. S3). Intriguingly, ≈10% of these cells contained a single bright, near-spherical structure located in the cytosol (Fig. 7). That we observed this structure only when staining with IgGon and not with IgGoff suggests it is highly enriched for the on-state of caspase-1.
Fig. 7.
Probing active conformers of caspase-1 in THP-1 cells. LPS-stimulated THP-1 cells were stained with DAPI (A) and IgGon (B). The merged image is shown in C. Note the spherical body found in ≈10% of cells.
Discussion
We present a general strategy for directly obtaining conformation-selective antibodies that was applied to study distributions of on- and off-conformers of caspase-1. The differences in conformation for the on- and off-states as viewed by X-ray crystallography are relatively modest. Thus, that high-affinity antibodies specific for the 2 states could be isolated suggests the wide applicability of this strategy.
The key elements of this approach are the availability of ligands to trap specific conformations of the protein of interest coupled with an in vitro selection method to isolate specific binding proteins. Although we used covalent ligands to lock specific conformations, one could also employ noncovalent ligands, so long as one worked at a high enough concentration to maintain saturation. It should also be possible to trap these alternate target conformations with natural protein-binding partners or using engineered disulfide or chemical cross-linkers.
State-selective binding partners have been identified before. It has been reported that using phage display to isolate peptides specific for estradiol- or tamoxifen-activated estrogen receptor (20). Fabs have also been selected for GTP-bound Ras (21). However, neither study selected for more than 1 state. Recent elegant work by Grutter and coworkers (22) has shown that it is possible to isolate conformation-specific binding proteins to caspase-2 without using conformation-trapped antigens. However, this approach required the assaying of >100 individual clones following selections from a combinatorial library of designed ankyrin repeat proteins (DARPin) to identify the allosteric inhibitors.
In vitro selection allowed antiselection in solution (23) with the noncognate form of the protein, which enabled the dramatic enrichment of selective binders. Although we found phage display to be particularly effective in vitro selection, there are a variety of other methods one could apply, including ribosome display, yeast display, and others (for review, see refs. 24 and 25). Fab display is particularly effective, because it is well established for naïve selections and has been made even more effective recently by using codon-restricted CDR libraries enriched for Ser and Tyr residues (12, 14). However, there is a variety of other scaffolds that could be potentially used, such as DARPins (26), Affibodies (27), fibronectin type III domains (28), zinc finger DNA-binding domains (29), and many others. Moreover, one could imagine using this approach to identify conformation-selective antibodies to other biopolymers such as alternate forms of DNA or RNAs such as riboswitches, etc.
Our data suggest that the Fabs we identified are specific for the conformers of caspase-1 they were selected for. First, at the selection stage, we found strong enrichments for the specific antigen used in the selection, and binders survived the antiselection steps with noncognate antigens. The consensus sequences for the Fabon and Faboff were very different from each other, and they bound to their cognate antigen with 20- to 500-fold higher affinity than to the noncognate forms. Strong evidence for conformational specificity was provided by the SPR data, which showed that the increased affinity for the cognate antigen was due to a faster on-rate rather than a slower off-rate. Interestingly, the fact that the antibodies can bind to the noncognate form at all suggests each of the inhibitor-locked antigens is dynamic, albeit the inhibitor-locked cognate conformer is much more populated than the noncognate conformer. In addition, both Fabs did not cross-react with caspase-4 and -5, which have the highest sequence similarity to caspase-1 (Fig. S4). We do not know the exact epitopes where these Fabs bind on caspase-1. However, it is very unlikely their epitopes include the inhibitor itself. In fact, the structure of the inhibitor was alternated during positive selection to avoid antibodies that interacted directly with the inhibitor.
The binding kinetics of these Fabs to the apo form of caspase-1 suggests strongly that the enzyme in solution is dynamically interchanging between conformations because the affinity for Fabs is intermediate between the locked forms of caspase-1 (Fig. 3B). However, the apo enzyme sits overwhelmingly in the off-form. For example, the enzyme binds with an affinity that is much closer to the locked off-form of caspase-1 than to the on-form. The ratios of KD values for the Faboff to Fabon vary >4 logs; the unlabeled caspase-1 is within 1 log unit of the locked-off form yet 3 log units away from the locked-on form of caspase-1. Thus, the average ensemble of conformations for apo caspase-1 is intermediate between the on- and off-states but populates the off- over the on-state by a factor of ≈1,000. It is noteworthy that X-ray structures show the conformation of the apo-form of caspase-1 (9) is virtually identical to the off-form generated by the allosteric tethered ligand (8).
The Fab fragments have marked functional effects on the apo caspase-1 that are consistent with previous reports that the enzyme exhibits positive cooperativity (8, 11). The titration of apo-caspase-1 with Fabon causes >3-fold activation of activity (Fig. 4A) probably by binding and stabilizing the active conformer. Moreover, the EC50 for activation by Fabon (11 nM) is significantly lower than the KD for binding (300 nM; Fig. 4B). These data suggest that the presence of substrate can stabilize the active conformer and thus improve apparent affinity. Likewise, Faboff inhibits the enzyme. The IC50 for inhibition (920 nM) is significantly above the KD for binding of Faboff (5 nM) and reflects the fact that substrate stabilizes the on-form and thus competes for binding of Faboff. We previously showed that labeling caspase-1 with Ac-YVAD-cmk and the allosteric inhibitor are mutually exclusive: binding of the active site inhibitor promotes a conformation that is incompatible with binding of the allosteric inhibitor and vice versa (8). Similarly the Fabs appear to bind to mutually exclusive conformations and away from these inhibitor binding sites. Thus, the enzyme can be allosterically activated or inhibited through binding of conformation-specific antibodies on different surfaces.
One of the advantages of using Fabs is that they are readily converted into IgGs that allow one to study possible avidity effects. Interestingly, the IgGoff bound 40 times tighter than Faboff to apo-caspase-1, whereas IgGon bound with nearly identical affinity to Fabon. This may reflect the different position of the binding epitope on caspase-1 and resultant stoichiometry such that 1 IgGoff molecule can bind to both subunits of caspase-1 simultaneously, whereas 1 IgGon molecule cannot. The enhancement in affinity seen for IgGoff for Faboff is almost entirely due to enhanced on-rate, which may reflect that the dimer has a greater probability of productive binding per collision.
Previous studies indicate that procaspase-1 is monomeric (4, 30), yet very little is known about the conformation of this form in vitro or in cells. We find the IgGoff binds modestly to procaspase-1 but neither IgGon nor Fabon nor Faboff bind detectably (Fig. 5A). This suggests that the pro-form is much closer to the off-form of caspase-1 in conformation and that dimerizing procaspase-1 by binding to IgGoff promotes a conformation more like that of apo caspase-1. Nonetheless, the pro-form has a conformation that is not identical to the off-form because IgGoff bound to procaspase with a KD of 130 nM compared with binding to caspase-1 with a KD of 0.38 nM. These results were mirrored in the immunoprecipitation experiments (Fig. 5B). The IgGoff can pull down both procaspase-1 and caspase-1 in THP-1 cells, whereas IgGon can pull down only the mature form.
The additional advantage for converting the Fabs to IgGs was that these can be used with standard immunostaining reagents to localize specific conformations of caspase-1 in cells. In THP-1 cells probed with IgGoff, we saw only diffuse staining of the cells whether stimulated with LPS or not (data not shown). In contrast, when THP-1 cells were treated with LPS and stained with IgGon, we found intense staining of a single supermolecular cytosolic structure (1∼2 μm in diameter) in ≈10% of the cells. Alnemri and coworkers have reported the existence of a virtually identical supermolecular structure, which they termed the pyroptosome, in 15–30% of THP-1 cells stimulated with LPS using GFP-tagged ASC (apoptosis-associated speck-like protein containing a CARD domain) (30). That we can stain the supermolecular structure only with IgGon not IgGoff suggests strongly that virtually all of the caspase-1 is processed to the mature form and rests in the active conformation in the pyroptosome-like structure. This would indicate that caspase-1 is either actively catalyzing proteolysis, or perhaps more likely that binding to the ASC scaffold stabilizes the on-form of caspase-1. In this regard, ASC may function like the on-state antibody; however, its binding site does not overlap with that of IgGon.
Overall, these studies demonstrate a general approach for selecting binding proteins to specific protein conformations using small molecule ligands to lock conformations of interest and phage display to identify conformation-selective antibodies. The Fabs and IgG derivatives were useful for defining and localizing the specific forms of caspase-1 in vitro and in cells. Moreover, this approach could be useful for generating specific inhibitors or even activators of proteins in cells extracts and possibly in cells by using appropriate intracellular antibody delivery technology (31, 32).
Materials and Methods
Caspase-1 Expression and Purification.
The p20 subunit (residues 120–279) and p10 subunit (residues 317–404) of wild-type human caspase-1 were separately expressed in E. coli as inclusion bodies from a pRSET expression vector (Invitrogen). The purification and refolding of protein from inclusion bodies was performed as described (8). The Cys285Ala mutant of caspase-1 was made by refolding Cys285Ala mutated p20 with wild-type p10 inclusion bodies. A form of procaspase-1 lacking the CARD domain (CARDless procaspase, residues 120–404) was cloned into a pET23b expression vector (Novagen) with a C-terminal His6 tag and transformed into E. coli BL21(DE3) strain. The expression was induced with 0.2 mM IPTG induction for 20 min at OD600 ≈0.6. Cell pellets were lysed by 5 passes through a microfluidizer in ice-cold lysis buffer (100 mM Tris, pH 8.0, 100 mM NaCl). The lysate was cleared by centrifugation at 48,500 × g for 15 min at 4 °C. The supernatant was first loaded on a 5-mL HisTrap HP column (GE Healthcare), and bound protein was eluted with a 0- to 200-mM imidazole gradient after washing. The eluate were diluted into 20 mM Tris, pH 8.0, 5% glycerol, and loaded on a 5-mL HiTrap Q HP column. The p32 was eluted with a 0- to 0.5-M NaCl gradient and aliquots were frozen immediately in an ethanol-dry ice bath.
Caspase-1 Labeling.
To prepare the on-form caspase-1, wild-type caspase-1 was incubated with 4-fold excess of active-site inhibitor (Ac-YVAD-cmk or z-WEHD-fmk) at 4°C overnight in the labeling buffer (50 mM Hepes, pH 8.0, 200 mM NaCl, 50 mM KCl, 200 μM ß-ME). Protein precipitate was removed by centrifugation, and the labeling was confirmed by the mass shift observed by LC-MS (Waters). To prepare the off-form of caspase-1, a catalytic-inactive caspase-1 Cys285Ala was incubated with 150 μM of the allosteric inhibitor [compound 34 or compound 11 (8)] at 4 °C overnight in the same labeling buffer containing 1 mM ß-ME. For random biotinylation, the off-form of caspase-1 was incubated with 15-fold excess sulfo-NHS-LC-biotin (Pierce) for 45 min at ambient temperature, and the reaction was stopped by buffer exchange using a NAP-25 column (GE Healthcare).
Library Construction and Sorting.
We modified the Fab-template phagemid (pV-0116c) (12) to have TAA stop codons in all 3 heavy chain CDRs and the light chain CDR-L3 to reduce wild-type Fab background. For the construction of naïve libraries, the resulting phagemid was used as the “stop template” in a mutagenesis reaction with oligonucleotides designed to repair simultaneously the stop codons and introduce designed mutations at the desired sites, as described (16).
In sorting for on-form specific Fabs, the phage pool was cycled through rounds of binding selection with the active conformer of caspase-1 that was directly immobilized on 96-well Maxisorp plate (Thermo Fisher). Bound phage were eluted with 100 mM HCl and neutralized with 1 M Tris, pH 8.0. Phage were amplified in E. coli XL1-blue (Stratagene) with the addition of M13-KO7 helper phage (New England Biolabs). In sorting for the off-form specific Fabs, a solution-phase binding strategy was adapted for better control over the selection and anti-selection process. The phage pool was incubated for 2 h at room temperature with biotinylated allosteric conformer before being captured on neutravidin or streptavidin (Pierce) coated Maxisorp plates. The bound phage were then eluted and propagated as described above. After selection, individual clones were picked and grown in a 96-well deep well plate with 2YT broth supplemented with carbenicillin and M13-KO7. The culture supernatants were used in phage ELISAs to identify binding clones (33).
Antibody Purification and Kinetic Analysis by SPR.
The phage display phagemid was converted into the Fab expression vector by deleting the sequence encoding for the cP3 minor phage coat protein and inserting a λ terminator sequence (GCTCGGTTGCCGCCGGGCGTTTTTTAT) downstream of the stop codon at the end of CH1 domain. Fab protein was secreted from E. coli 34B8 strain transformed with individual plasmids in low-phosphate medium at 30 °C for 26 h, as described (18). To generate IgG proteins, the variable domains were subcloned into vectors designed for transient IgG expression in CHO cells (18). Fab proteins were purified with protein A affinity chromatography and IgG proteins were purified with protein G affinity chromatography.
Kinetic binding analyses were performed by surface plasmon resonance (SPR) using a BIAcore 3000 (GE Healthcare). Ligand-bound or free caspase-1 dimers were immobilized on CM5 chips and serial dilutions of Fabs or IgGs were injected. Binding responses on flow cells with immobilized caspase-1 variants were corrected by subtraction of responses on a blank reference flow cell. A 1:1 Languir model in BIAevaluation software (GE Healthcare) was used for fitting the sensograms and the KD values were calculated from the ratios of koff/kon.
Immunoprecipitation from THP-1 Cell Extracts.
THP-1 cells were grown to a density of 1 × 106 cell/ml and harvested by centrifugation at 150 × g for 5 min. Cells were lysed by Dounce homogenizer in ice-cold buffer (20 mM Hepes-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM Na-EDTA, 1 mM Na-EGTA, 0.1 mM PMSF, and the Roche protease inhibitor mixture). Cell lysates were cleared by centrifugation at 500 × g, 3,000 × g, and 22,000 × g for 10 min each. Aliquots were incubated with or without IgGs overnight at 4 °C and immunocomplexes were recovered by protein-G agarose beads. Presence of procaspase-1 and caspase-1 were visualized by Western blot analysis.
Immunofluoresence Microscopy.
THP-1 cells were grown to the density of 5 × 105 cell/ml and differentiated with 0.5 μM PMA for 3 h and allowed to attach to no. 1½ glass coverslips overnight. Cells were treated with 1 mM LPS for 2 h followed by 5 mM ATP for 30 min before fixation and mild detergent permeabilization. After blocking with 10% BSA for 1 h, IgGon was added at 100 μM concentration for 1 h. After 3 washes with PBS + 0.1% Triton X-100, the cells were stained for 1 h with Alexa Fluor 488 conjugated goat anti-human IgG antibody (Invitrogen). Cells were washed 3 times and mounted with ProLong Gold containing DAPI (Invitrogen). Images were recorded on a Nikon 6D High-Throughput Microscope equipped with a Photometrics Coolsnap HQ2 Camera.
Supplementary Material
Acknowledgments.
We thank S. Birtalan, Y. Zhang, B. Li, G. Fuh, J. Scheer, V. Chiang, and Y. Chen for advice and assistance. We also thank the Protein Engineering Department and the sequencing, oligonucleotide synthesis, and fermentation teams at Genentech for generous support, as well as K. Thorn and the Nikon Imaging Center at University of California, San Francisco, for help with immunofluorescence microscopy. This work was supported by National Institutes of Health Grant 5R01AI070292–02 and the Sandler Family Foundation gift (to J.A.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812952106/DCSupplemental.
References
- 1.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 2.Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: A danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Dinarello CA. Mutations in cryopyrin: Bypassing roadblocks in the caspase 1 inflammasome for interleukin-1beta secretion and disease activity. Arthritis Rheum. 2007;56:2817–2822. doi: 10.1002/art.22841. [DOI] [PubMed] [Google Scholar]
- 4.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 5.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 6.Mariathasan S, Monack DM. Inflammasome adaptors and sensors: Intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 8.Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanowski MJ, Scheer JM, O'Brien T, McDowell RS. Crystal structures of a ligand-free and malonate-bound human caspase-1: Implications for the mechanism of substrate binding. Structure. 2004;12:1361–1371. doi: 10.1016/j.str.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Rano TA, et al. An allosteric circuit in caspase-1. J Mol Biol. 2008;381:1157–1167. doi: 10.1016/j.jmb.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta D, Scheer JM, Romanowski MJ, Wells JA. An allosteric circuit in caspase-1. J Mol Biol. 2008;381:1157–1167. doi: 10.1016/j.jmb.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a 4-amino-acid code: A dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birtalan S, et al. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 14.Fellouse FA, et al. Molecular recognition by a binary code. J Mol Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 15.Deshayes K, et al. Rapid identification of small binding motifs with high-throughput phage display: Discovery of peptidic antagonists of IGF-1 function. Chem Biol. 2002;9:495–505. doi: 10.1016/s1074-5521(02)00129-1. [DOI] [PubMed] [Google Scholar]
- 16.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 17.Jackson JR, Sathe G, Rosenberg M, Sweet R. In vitro antibody maturation. Improvement of a high affinity, neutralizing antibody against IL-1 beta. J Immunol. 1995;154:3310–3319. [PubMed] [Google Scholar]
- 18.Lee CV, et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 19.Johnson G, Wu TT. Kabat database and its applications: 30 years after the first variability plot. Nucleic Acids Res. 2000;28:214–218. doi: 10.1093/nar/28.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norris JD, et al. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 21.Horn IR, Wittinghofer A, de Bruine AP, Hoogenboom HR. Selection of phage-displayed fab antibodies on the active conformation of ras yields a high affinity conformation-specific antibody preventing the binding of c-Raf kinase to Ras. FEBS Lett. 1999;463:115–120. doi: 10.1016/s0014-5793(99)01617-8. [DOI] [PubMed] [Google Scholar]
- 22.Schweizer A, et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: Specificity, structure, and inhibition mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Li B, et al. Minimization of a polypeptide hormone. Science. 1995;270:1657–1660. doi: 10.1126/science.270.5242.1657. [DOI] [PubMed] [Google Scholar]
- 24.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipovsek D, Pluckthun A. In-vitro protein evolution by ribosome display and mRNA display. J Immunol Methods. 2004;290:51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Binz HK, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 27.Nord K, et al. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 28.Koide A, Koide S. Monobodies: Antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- 29.Cho GS, Szostak JW. Directed evolution of ATP binding proteins from a zinc finger domain by using mRNA display. Chem Biol. 2006;13:139–147. doi: 10.1016/j.chembiol.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes-Alnemri T, et al. The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visintin M, Tse E, Axelson H, Rabbitts TH, Cattaneo A. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc Natl Acad Sci USA. 1999;96:11723–11728. doi: 10.1073/pnas.96.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 33.Sidhu SS, et al. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J Mol Biol. 2004;338:299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 34.Kabat EA, et al. Sequences of Proteins of Immunological Interest. 4th Ed. Bethesda: National Institutes of Health; 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.