Abstract
The second messenger cyclic dimeric GMP (c-di-GMP) regulates surface attachment and biofilm formation by many bacteria. For Pseudomonas fluorescens Pf0–1, c-di-GMP impacts the secretion and localization of the adhesin LapA, which is absolutely required for stable surface attachment and biofilm formation by this bacterium. In this study we characterize LapD, a unique c-di-GMP effector protein that controls biofilm formation by communicating intracellular c-di-GMP levels to the membrane-localized attachment machinery via its periplasmic domain. LapD contains degenerate and enzymatically inactive diguanylate cyclase and c-di-GMP phosphodiesterase (EAL) domains and binds to c-di-GMP through a degenerate EAL domain. We present evidence that LapD utilizes an inside-out signaling mechanism: binding c-di-GMP in the cytoplasm and communicating this signal to the periplasm via its periplasmic domain. Furthermore, we show that LapD serves as the c-di-GMP receptor connecting environmental modulation of intracellular c-di-GMP levels by inorganic phosphate to regulation of LapA localization and thus surface commitment by P. fluorescens.
Keywords: biofilm, c-di-GMP, LapA, HAMP domain
For many bacteria, commitment to surface attachment, and subsequent growth as a biofilm, is a highly regulated event (1, 2). Recent work has illuminated the central role of the intracellular second messenger bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP) in controlling surface attachment in many bacterial systems (3, 4). Although c-di-GMP appears to be a conserved signal, the outputs it regulates are diverse and vary among different systems. These outputs include regulation of extracellular polysaccharide production (5, 6), adhesin secretion and localization (7), flagellar function (6, 8), and transcription of genes that direct attachment (9). Although c-di-GMP signaling has been established as a conserved modality in biofilm regulation, the specific mechanisms by which the signal is received and acted upon are largely unknown.
At least 2 classes of c-di-GMP–binding proteins have been characterized. The first class, PilZ-domain proteins, has been linked to regulation of flagellar motility in Enterobacteria, Vibrio, and Caulobacter (10–12) and to the synthesis of alginate and type IV pili in Pseudomonas (13, 14). The second class is less defined but shares an amino acid motif, RxxD, first identified as the allosteric site of product feedback inhibition in the diguanylate cyclase (DGC) PleD (15). This motif is a common feature of DGCs but also has been identified in the c-di-GMP–binding protein PelD (16). Last, the transcription factor FleQ of Pseudomonas aeruginosa was shown recently to bind c-di-GMP (17). The mechanism by which FleQ binds c-di-GMP remains unknown, and it may represent a third class of effectors.
LapD is an inner-membrane protein required by Pseudomonas fluorescens for biofilm formation and for maintenance of the adhesin LapA on the cell surface. In this study we describe LapD as a c-di-GMP effector protein that binds c-di-GMP via a degenerate c-di-GMP phosphodiesterase (EAL) domain. Our analysis indicates that LapD is the c-di-GMP receptor in the signaling pathway by which inorganic phosphate (Pi) starvation controls biofilm formation. In contrast to c-di-GMP effectors identified to date, LapD is an inside-out signaling protein, communicating cytoplasmic c-di-GMP levels to the membrane-localized attachment machinery via a periplasmic output domain.
Results
lapD Is Required for Attachment via the LapA Adhesin.
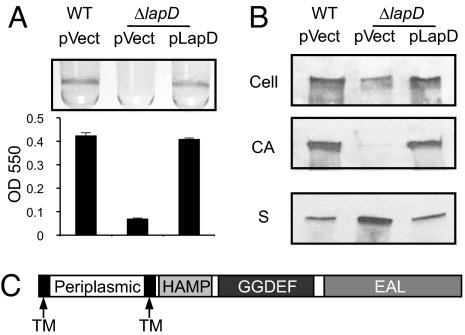
P. fluorescens requires the large adhesin LapA for stable attachment to surfaces (18). Prior work also identified the inner-membrane protein LapD as being required for biofilm formation and suggested that it played a role in the localization of LapA (Fig. 1A) (19). To define the effect of lapD on LapA localization, we assessed the distribution of LapA in the cellular fraction (Cell), cell-associated (CA) fraction, and supernatant fraction (S). The lapD mutant had decreased cellular levels of LapA compared with WT (Fig. 1B). Results from an earlier study suggest that this decrease is not caused by a difference in LapA transcription in the lapD mutant (19). We also have demonstrated that the abundance of LapA in the CA fraction is a strong indicator of a strain's propensity for biofilm formation (7, 19). Here we see that the biofilm-defective lapD mutant showed a nearly complete loss of LapA from the CA fraction (Fig. 1B). Interestingly, this difference in localization was not caused by a lack of secretion, because the culture supernatant of ΔlapD contained 2-fold more LapA than the WT. Thus it appears that the lapD mutant is unable to retain the LapA on the cell surface once it has been secreted (54% ± 2.2% of WT in the cell, 5.7% ± 3.2% in the CA, and 198% ± 25% for supernatant, n = 3) (Fig. 1B). Complementation of the lapD mutant with a plasmid carrying lapD restored WT LapA levels, localization, and biofilm (Fig. 1).
Fig. 1.
Biofilm formation and LapA localization phenotypes of the lapD mutant. (A) Quantitative analysis of biofilm formation by WT plus vector (WT pvect), ΔlapD plus vector (pvect), and ΔlapD plus pLapD (pLapD). (B) Western blots probed for LapA to analyze adhesin localization profiles for strains shown in (A). The fractions indicated are cellular (Cell), cell-associated (CA), and culture supernatant (S). (C) Predicted protein domains of LapD.
LapD Contains Degenerate and Inactive Diguanylate Cyclase and c-di-GMP Phosphodiesterase Domains.
The amino acid sequence of LapD contains 3 predicted domains: a HAMP domain, commonly present in transmembrane signaling proteins (20), as well as diguanylate cyclase (GGDEF) and EAL domains (Fig. 1C). GGDEF and EAL domain proteins regulate biofilm formation through c-di-GMP DGC and phosphodiesterase (PDE) activities, respectively (3). Alignment of the LapD protein sequence with those from empirically verified DGCs and PDEs reveals the absence of many residues known to be required for catalysis (supporting information (SI) Fig. S1),. For example, LapD's GGDEF domain harbors the amino acids RGGEF in the corresponding positions of this signature motif, replacing an acidic, catalytic residue with glycine. LapD lacks several critical EAL domain residues as well, including the glutamic acid of the EAL motif.
To test if LapD could synthesize or degrade c-di-GMP, a histidine-tagged form (LapD6H) was purified and tested alongside enzymatically active controls (PleD* and CC3396 of Caulobacter crescentus). We were unable to detect c-di-GMP synthesis or hydrolysis by LapD (Fig. S1), consistent with its GGDEF and EAL domains being enzymatically inactive. This result was not caused by interference of the His tag, because LapD6H can fully complement the ΔlapD mutation when provided in trans (data not shown).
LapD Binds c-di-GMP.
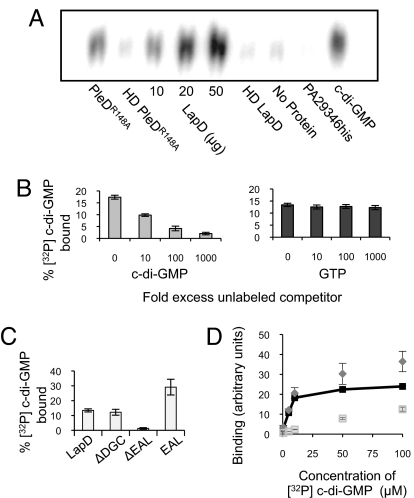
Degenerate GGDEF or EAL domains have been proposed to function as regulatory domains through the binding of c-di-GMP or other nucleotides (15, 25). To test if LapD could bind c-di-GMP, we evaluated the ability of LapD6H to interact specifically with this nucleotide. LapD6H and [32P]-c-di-GMP were mixed with Ni-silica resin, the resin was washed, and the nucleotide was eluted and resolved by TLC. c-di-GMP bound to LapD in a concentration-dependent manner, at levels comparable with those of PleDR148A, a known c-di-GMP–binding protein (Fig. 2A) (15). Little to no c-di-GMP was bound by heat-denatured protein, resin alone, or an unrelated, histidine-tagged protein (PA2934his) (21). The addition of unlabeled c-di-GMP blocked [32P]-c-di-GMP binding, whereas 1000-fold excess of GTP had no effect, demonstrating a specific interaction between LapD and c-di-GMP (Fig. 2B).
Fig. 2.
Analysis of c-di-GMP binding by LapD. (A) TLC resolution of [32P] c-di-GMP co-purified with histidine-tagged proteins. Each reaction contained 50 μg of protein unless otherwise indicated. HD = heat denatured. (B) Increasing amounts of unlabeled c-di-GMP decrease binding of LapD to [32P] c-di-GMP (Left), whereas unlabeled GTP does not compete with [32P] c-di-GMP binding at concentrations up to 1 mM (1000-fold excess) (Right). (C) c-di-GMP binding by LapD lacking the GGDEF or EAL domain or by the EAL domain alone. (D) Binding of c-di-GMP to E. coli membranes containing LapD (diamonds) or the ΔEAL protein (open squares) at increasing concentrations of ligand. Specific binding (dark squares) is binding to LapD minus binding to ΔEAL.
To determine which domain(s) of the LapD protein participates in binding c-di-GMP, LapD6H variants lacking the GGDEF (R247-A387) or EAL (H412-G649) domain were tested. The protein lacking the GGDEF domain was able to bind c-di-GMP at levels comparable to WT, whereas LapD6H lacking the EAL domain (ΔEAL) did not show significant binding above background levels (student's t test P = 0.074) (Fig. 2C). Purification of the EAL domain alone (G391-H655) yielded an ≈30 kD protein capable of binding c-di-GMP as well as, or better than, the full-length protein (Fig. 2C and data not shown). Taken together, these data suggest that LapD's binding activity resides in the EAL domain.
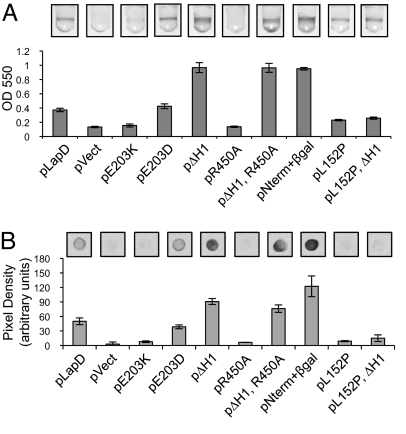
We further defined the c-di-GMP binding site of LapD by constructing alanine replacements in candidate residues chosen for their involvement in substrate binding and their proximity to the active site of the well-characterized PDE RocR (22). LapD variants with the mutations K446A, R450A, K581A, and E617A were stably expressed in the membrane in vivo at WT levels (Fig. 3A). Each mutant protein was purified and tested for c-di-GMP binding in vitro, and all showed a reduction in binding compared with the WT protein (Fig. 3B). These alleles also were defective for complementation of the lapD mutant, indicating that c-di-GMP binding by LapD is required for biofilm formation (Fig. 3C).
Fig. 3.
Biochemical and phenotypic analysis of EAL-domain point mutants. (A) Membrane preparations were assayed for in vivo levels of LapD by Western blot. (B) c-di-GMP binding by LapD variants and WT LapD. (C) Biofilm assay performed on ΔlapD complemented with pLapD or EAL-domain mutants. (D) Strains with the ΔlapD, WT, or K446A allele of lapD are compared for biofilm formation when the DGC PA1107 is expressed.
We next tested whether stimulating biofilm formation by increasing cellular c-di-GMP levels would require binding by LapD. The DGC(s) that controls attachment via LapD is not known, so we used a heterologous DGC, PA1107. Expression of PA1107 boosted biofilm formation by the WT by 82% but had no effect on a strain with the K446A mutation in LapD (Fig. 3D). These data are consistent with LapD sensing changes in c-di-GMP and show that c-di-GMP binding is required for LapD function in vivo.
Estimation of LapD's Dissociation Constant (Kd) for c-di-GMP.
To measure the affinity of membrane-bound LapD for [32P]-c-di-GMP, we used a filter-binding assay. The c-di-GMP was mixed with membranes purified from Escherichia coli overexpressing either LapD6H or the ΔEAL protein, and unbound nucleotide was removed by washing and vacuum filtration. Membranes containing LapD6H bound c-di-GMP in a concentration-dependent and saturable manner, at levels well above those with the ΔEAL protein (Fig. 2D). The specific binding data (binding of LapD minus binding of ΔEAL) were analyzed by non-linear regression, and the Kd of LapD for c-di-GMP was estimated to be 5.5 ± 2.8 μM (n = 3) (Fig. S2).
LapD's HAMP Domain Modulates Biofilm Formation.
The HAMP domain plays an essential role in many signaling proteins. These domains typically relay signals across the cytoplasmic membrane, altering their conformation in response to activation of an extracellular input domain and propagating that conformational change to a cytoplasmic output domain (20). Structure/function studies of HAMP-domain proteins have shown that mutations in this domain can have profound effects, constitutively activating or inactivating signaling output (23). To investigate a role for this domain in regulating LapD function, we mutated E203, one of the few amino acids that are well conserved among all HAMP domains (24). A non-conservative substitution of this residue, E203K, led to a loss of biofilm formation as well as a defect in c-di-GMP binding in vitro (7.5% ± 1% of WT) (Fig. 4A). This finding shows that the HAMP domain can regulate LapD function, and it suggests that LapD E203K could be locked in an inactive conformation that eliminates binding to its ligand and stimulation of biofilm formation. A conservative substitution at the same position, E203D, resulted in WT LapD function in these assays (150% ± 12% of WT c-di-GMP binding) (Fig. 4A). Both alleles are stably expressed in vivo (Table S1).
Fig. 4.
Effects of lapD mutations on biofilm formation and cell-surface LapA levels. (A) Mutant forms of lapD are compared with pLapD in their ability to complement ΔlapD for biofilm formation. (B) Quantification of cell-surface levels of LapA in the strains in (A) by densitometry on α-LapA dot blots (n = 3, ± SD; representative blots are shown).
To identify more mutations in the HAMP domain that affect LapD function, we constructed 3 deletions: a short deletion in each predicted α-helix and a deletion of the entire HAMP domain. Interestingly, deletions in each individual helix, M180-A186 (ΔH1) or V207-Q213 (ΔH2), or in the entire HAMP domain all led to a hyperbiofilm phenotype (Figs. 4A and S3A). These data demonstrate that the HAMP domain controls LapD's effect on biofilm formation and suggest that gross perturbations to the HAMP structure lead to misregulated, constitutive output from LapD, resulting in increased biofilm formation.
A possible explanation for the phenotype of the HAMP deletion alleles was that these mutations increase LapD's affinity for c-di-GMP, and increased c-di-GMP binding could promote biofilm formation. In our in vitro assay, however, the ΔH1 protein binds c-di-GMP at levels comparable to WT (94% ± 5% of WT). If the hyperbiofilm resulting from these HAMP mutations was caused by increased c-di-GMP binding in vivo, we reasoned that mutations that block binding would suppress these effects. Intriguingly, the ΔH1 R450A double-mutant allele yielded a hyperbiofilm phenotype but produced a protein that was unable to bind c-di-GMP in vitro (4.9% ± 5% of WT) (Fig. 4A). Thus the affect of the ΔH1 mutation on biofilm is not realized through changes in LapD's c-di-GMP binding function. Instead, this mutation must uncouple the necessity of c-di-GMP binding from LapD's function in biofilm formation by locking it in an active state.
Evidence for an Inside-Out Signaling Mechanism.
We considered two models for how LapD may act as a signaling protein. In the first, LapD receives signals from the periplasm that effect binding of c-di-GMP in the cytoplasm. This model seemed unlikely, because the hyperbiofilm phenotype of the ΔH1 mutation in the HAMP domain is not dependent on c-di-GMP binding. In the second model, conformational change caused by c-di-GMP binding/dissociation in the cytoplasm is communicated to the periplasm through the HAMP domain. If this inside-out mechanism were correct, we hypothesized, then the periplasmic portion of LapD should be responsible for LapD's output: promoting biofilm formation.
To test this hypothesis we expressed the N-terminal portion of LapD consisting only of the periplasmic domain and both transmembrane domains (TMD). This protein (pNterm), although unstable, was capable of restoring biofilm formation to ΔlapD (Fig. S3). We stabilized this LapD variant by fusing it to β-galactosidase (β-gal) after the second TMD. Expression of the stabilized periplasmic domain (pNterm + β-gal) led to a hyperbiofilm phenotype comparable to that of the ΔH1 mutation (Fig. 4A). This gain of function relative to WT is consistent with the output of the periplasmic domain being uncoupled from regulatory input from the cytoplasmic domains. Neither expression of the entire cytoplasmic portion of LapD nor the EAL domain alone complemented the lapD mutant (Fig. S3), providing further evidence that the periplasmic domain is responsible for LapD's output.
We identified a mutation in the periplasmic domain, L152P, that reduces biofilm formation (Fig. 4A). This mutant provided an avenue for further testing the necessity of the periplasmic domain for LapD output and the directionality of LapD signaling. We hypothesized that if this mutation causes impaired output function, it should suppress the constitutive output that results from deletions in the HAMP domain. When the L152P and ΔH1 mutations were combined, the periplasmic L152P mutation fully suppressed the hyperbiofilm phenotype of the HAMP domain mutation (Fig. 4A). These data underscore the role of the periplasmic domain as the output of LapD signaling and provide compelling evidence that LapD regulates biofilm formation via an inside-out signaling mechanism.
The mechanism we propose for LapD signaling depends on the topology of LapD in the inner membrane. The predicted topology of LapD was confirmed experimentally by translationally fusing lacZ or phoA to each predicted TMD. Fusing PhoA in the place of the predicted periplasmic domain yielded an active phosphatase, whereas a similar fusion in place of the cytoplasmic domain did not (Fig. S4). The converse was true of analogous fusions of LapD and β-gal, confirming that the portion of LapD between the 2 TMDs does localize to the periplasm (Fig. S4).
LapD Regulates Localization of LapA to the Cell Surface.
Aberrant localization of LapA to the culture supernatant from the cell surface probably is the basis for the lapD mutant's inability to form a biofilm (Fig. 1B). To determine if retention of LapA on the cell is the basis of LapD's regulation of biofilm formation, the relative levels of cell-surface LapA on the strains discussed previously were quantified. In every case, the amount of LapA on the cells was reflective of a strain's biofilm phenotype (Figs. 4B and S3B), indicating that LapA localization is regulated by LapD signaling. Additionally, lapA is required for biofilm formation by strains with lapD hyperbiofilm alleles, consistent with LapA regulation being sufficient to explain their phenotypes (data not shown).
LapD's Role in the Phosphate-Dependent c-di-GMP Signaling Pathway.
We have shown previously that environmental Pi can impact LapA localization through changes in cellular c-di-GMP levels. Specifically, the c-di-GMP PDE RapA is expressed in low-Pi conditions as a member of the Pho regulon. RapA-mediated reduction in the cellular level of c-di-GMP inhibits retention of LapA at the cell surface (7). Noting the phenotypic similarities between Pi starvation and the lapD mutant, and LapD's regulation of cell-surface LapA levels, we examined LapD's role in this environmentally relevant c-di-GMP signaling pathway.
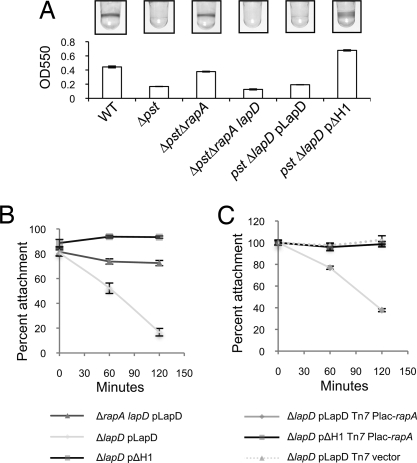
Mutants in the pst system constitutively express the Pho regulon, including rapA, and are defective for biofilm formation even in Pi-replete conditions (Fig. 5A) (7). Deletion of rapA restores biofilm formation to pst mutants (Fig. 5A) (7) and leads to an increase in cellular c-di-GMP levels in vivo (7). Biofilm formation by a ΔpstΔrapA mutant requires lapD, because a ΔpstΔrapA lapD mutant could not form a biofilm, consistent with LapD's role as a c-di-GMP receptor in this pathway (Fig. 5A).
Fig. 5.
The role of LapD in the phosphate-regulated c-di-GMP signaling pathway. (A) Quantitative assay of biofilm formation by the strains indicated. (B) Surface attachment by the indicated strains was monitored in high- and low-Pi media. The percentage of attachment in low-Pi medium relative to high-Pi medium is given at 0, 60, and 120 min after Pi starvation. (C) Surface attachment by the indicated strains in high-Pi medium at 0, 60, and 120 min after ectopic induction of rapA expression (given as a percentage of attachment by the isogenic strain without inducer at each time point).
We next tested the prediction that constitutively active mutants of LapD should be insensitive to decreases in cellular c-di-GMP associated with Pho regulon expression. In support of this hypothesis, the ΔH1 allele of LapD was epistatic to the pst mutation, restoring biofilm formation to the pst mutant (Fig. 5A). These data show that a change in LapD structure and function is sufficient to suppress the effect of constitutive Pho regulon expression on biofilm formation.
To test how this pathway might function more dynamically, we examined the effect of physiological Pho regulon induction on surface attachment by ΔrapA lapD pLapD, ΔlapD pLapD, and ΔlapD pΔH1, when these strains were starved for Pi. The Pho regulon is induced after 4 hours of growth in low-Pi medium (data not shown), and a mutation in rapA partially restores biofilm formation in low-Pi conditions (7). Before Pho activation, all strains showed a comparable percentage of attachment in Pi-depleted vs. Pi-replete conditions (Fig. 5B, 0 min). As Pi starvation ensued, attachment of ΔlapD pLapD dropped to 17% of that in the replete condition. Deletion of the PDE rapA partially restored attachment to 72%, consistent with previous data (7). The strain carrying the ΔH1 allele of LapD shows complete insensitivity to Pi starvation, maintaining the same level of attachment throughout the course of the assay (Fig. 5B).
To specifically assess the role of c-di-GMP in controlling attachment, independent of Pi starvation, we ectopically expressed an inducible copy of rapA in the ΔlapD pLapD and ΔlapD pΔH1 backgrounds and monitored attachment in Pi-replete media. Induction of rapA was sufficient to decrease attachment by ΔlapD pLapD significantly over a 120-min period to 37% of that by the uninduced control (Fig. 5C). As predicted, rapA induction had little effect on attachment by ΔlapD pΔH1, demonstrating that the ΔH1 allele is resistant to the activity of RapA in vivo. These data, in conjunction with our previous work (7), describe the pathway by which expression of the Pho regulon controls surface attachment via LapA: RapA reduces cellular c-di-GMP levels, and concomitant loss of c-di-GMP binding by LapD results in loss of LapA from the cell surface.
Discussion
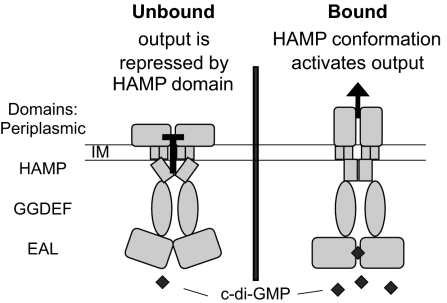
Here we present LapD, a protein with predicted GGDEF and EAL domains that binds but cannot synthesize or degrade c-di-GMP. We propose that LapD acts as an effector protein linking this intracellular signaling molecule to the function of an extracellular adhesin, LapA, and does so through an inside-out signaling mechanism (Fig. 6). According to this model, in the absence of c-di-GMP binding, the conformation of LapD is such that the HAMP domain represses the periplasmic output. When LapD binds c-di-GMP, the HAMP domain assumes a conformation that activates the periplasmic output. In vivo and in vitro analyses of HAMP domain mutants suggest that the ΔH1 and E203K mutations lock LapD in the bound and unbound conformations, respectively. Many HAMP-domain proteins have been shown to transmit extracellular signals into the cell; LapD, however, has an intracellular input and an extracellular output.
Fig. 6.
A model for inside-out signaling through LapD. (Left) In the absence of c-di-GMP binding, the periplasmic output is repressed via the HAMP domain. (Right) When c-di-GMP is bound by LapD, the HAMP domain assumes a conformation that activates output. LapD is depicted as a dimer, because other HAMP domain proteins are known to be dimers.
Since the identification of GGDEF and EAL domains and their enzymatic activities, many proteins containing these domains have been shown to have broad phenotypic effects through the synthesis and degradation of c-di-GMP (3). In contrast, relatively few c-di-GMP–binding proteins have been characterized, leaving open the question of how this signal is received and translated into phenotypic outputs. The occurrence of degenerate GGDEF and EAL domains has led many to speculate that some are inactive and serve as effectors (15, 25). LapD provides a key example of an effector protein with degenerate GGDEF and EAL domains that specifically binds c-di-GMP. Interestingly, it is the EAL domain, and not the GGDEF domain, of LapD that is necessary for binding. Functional DGCs are known to bind c-di-GMP as an allosteric inhibitor through the RxxD motif, but these residues are absent in LapD (Fig. S1A) (15).
LapD is not the first protein with predicted GGDEF or EAL domains for which an alternative function has been demonstrated. CsrD of E. coli, GpdS of Staphylococcus epidermidis, and CdgG of Vibrio cholerae also have functions distinct from their predicted activities (26–28). Romeo and colleagues have shown that CsrD does not synthesize or degrade c-di-GMP but instead binds the regulatory RNAs, CsrB and CsrC (26). The Staphylococcal protein GpdS stimulates biofilm formation but does not show DGC activity in vitro, nor does it require its GGDEF domain for its function in vivo (27). Last, CdgG of V. cholerae is a degenerate and inactive GGDEF-domain protein that requires the RxxD motif but not the GGDEF motif for its function in regulating rugosity (28). As Beyhan et al. speculate, CdgG may be a c-di-GMP–binding protein. These studies, as well as our work on LapD, highlight the diverse functional potential of proteins containing these ubiquitous domains.
In P. fluorescens we have shown that environmental Pi is an important signal governing surface attachment. Among other effects, this signal impacts cellular c-di-GMP pools through the PDE RapA (7). Although our previous work indicated that loss of the LapA adhesin from the cell surface was a key phenotypic consequence of RapA induction, the mechanism by which intracellular c-di-GMP levels could affect adhesin localization remained in question. This study identifies LapD as an important player in this signaling pathway, binding c-di-GMP in the cytoplasm and communicating this signal to the extracellular machinery of attachment, LapA.
How does LapD control LapA localization? It is possible that LapD's periplasmic domain physically stabilizes LapA on the cell surface by direct interaction or through a protein complex. Alternatively LapD could regulate LapA function indirectly through the activity of another protein or proteins in the periplasm. Further study investigating how LapD functions as a signaling protein and the precise nature of its output is currently underway. Uncovering these details will bring us closer to a complete understanding of how this c-di-GMP signaling pathway links an environmental signal (Pi) to a complex biological output.
Materials and Methods
Strains and Growth Conditions.
Bacteria strains listed in Table S2 were cultured routinely on lysogeny broth. K10T media were prepared as described (29). E. coli S17–1 λ-pir was used for maintenance and conjugal transfer of plasmids. Yeast strain InvSc1 (Invitrogen) was cultured as described (30). Gentamycin (10 μg ml−1 for E. coli, 30 μg ml−1 for Pseudomonas) and kanamycin (30 μg ml−1) were used where appropriate.
Static Biofilm Assay.
Static biofilm assays were performed and quantified as described (7) using K10T-1 medium and an incubation time of 6 h. The quantitative biofilm assay also was used to monitor attachment dynamically at 4–6 h after inoculation using K10T-1 medium as the Pi-replete condition and K10T-π as the low-Pi condition.
LapA Localization Assay.
Localization of LapA protein was performed using a functional HA-tagged variant of LapA as described (7) with minor modifications. Overnight cultures were subcultured in 45 ml of K10T-1 broth for 6 h. Samples from all fractions were normalized to total protein using the bicinchoninic acid assay (Pierce).
Quantitative LapA Blotting Assay.
Bacteria were grown as described for the LapA localization assay. Aliquots of cultures were pelleted, washed once in K10T-1 broth, and then resuspended in K10T-1. Cell suspensions were normalized to the OD of the parent culture, serially diluted in K10T-1, and 5-μl aliquots of each dilution were spotted onto a nitrocellulose membrane. After drying, membranes were probed for LapA as was done for Western blot analyses (7). Additional controls can be found in the SI Text.
Deletion and Complementation of lapD.
A chromosomal deletion of the lapD reading frame (Pfl01_0131) was constructed with pMQ-LapDKO, a derivative of allelic exchange vector pMQ83, using established techniques (7). This mutant lacks bases 142526–144476 of the annotated P. fluorescens Pf0–1 genome. The plasmid pLapD, and variants, were used to transform the ΔlapD strain by electroporation. Construction of pLapD and variants is described in the SI Text; primers are listed in Table S3.
Protein Purification.
E. coli expressing PleDR148A was obtained from Urs Jenal (University of Basel, Switzerland), and the protein was purified as described (15, 31). Purification of LapD6H and variants from E. coli was performed as previously described (32), with the addition of 1% Triton to all buffers. Protein purifications ranged from 79% to 96% purity, as determined by SDS PAGE gel staining and densitometry, and molar concentrations of each protein were corrected according to relative purity before all assays.
c-di-GMP–Binding Assay.
[32P]-c-di-GMP was synthesized as previously described but without HPLC fractionation (31). Binding of [32P]-c-di-GMP to LapD was assessed by co-purification of the nucleotide with protein bound to a nickel resin. Each reaction contained 1% Triton X-100, 75 mM Tris pH 8, 150 mM NaCl, 10 mM MgCl2, 1 μM c-di-GMP, and 300 pmol of protein in a total volume of 100 μl. After 15-min incubation, 50 μl of His-link Ni-silica resin (Promega) was added and gently agitated for 15 min. Next, resin was pelleted and washed twice in 150 μl of buffer. Nucleotide was eluted by boiling resin in 20 μl of TLC loading buffer and then was fractionated by TLC and quantified by exposure to a phosphor storage screen as described (7). Binding assays with cold competitors contained 1 μM labeled c-di-GMP for GTP competition and 10 μM for c-di-GMP competition. Unlabeled, chemically synthesized c-di-GMP was obtained from GLSynthesis Inc.
Filter-Binding Assay.
E. coli cells were prepared as for protein purification (described in previous sections). Membranes were purified from the clarified lysate by ultracentrifugation (1 h at 100,000 × g, 4 °C) and were resuspended at a concentration of 1 mg protein ml−1 in buffer: 75 μM Tris pH 8, 150 mM NaCl, 10 mM MgCl2, and 1 X complete EDTA-free protease inhibitors (Roche). Western blot analysis on membrane preparations was performed to confirm the presence of equivalent concentrations of LapD6H and ΔEAL-6H. Binding reactions contained 50 μl of membranes and [32P]-c-di-GMP at the concentrations indicated. Filtration, washing, and radiolabel quantification by phosphor storage screen exposure was done essentially as described (18).
Construction and Analysis of Strains with Inducible DGC or PDE.
The DGC PA1107 was expressed from pMQ72 by the addition of 0.018% arabinose in ΔlapD strains with lapD or K446A reintroduced into the native locus (SI Text). For PDE expression, rapA (Pfl01_1678) was cloned downstream of Plac to place it under the control of LacI; this cassette, consisting of lacI-Plac-rapA, was integrated into the chromosome as described in the SI Text. In attachment assays, rapA expression was induced 4 h after inoculation by the addition of 50 μM isopropyl β-D-1-thiogalactopyranoside.
Supplementary Material
Acknowledgments.
We thank Urs Jenal for supplying plasmids, P. Cushing and D. Madden for help with Kd calculations, J. Merritt for cloning PA1107, and C. Boyd for thoughtful reading of the manuscript. This work was supported by a National Institutes of Health T32 GM08704 predoctoral fellowship to P.D.N. and by National Science Foundation grant 9984521 to G.A.O.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808933106/DCSupplemental.
References
- 1.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: A genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 2.Pruss BM, Besemann C, Denton A, Wolfe AJ. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J Bacteriol. 2006;188(11):3731–3739. doi: 10.1128/JB.01780-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Römling U, Gomelsky M, Galperin MY. c-di-GMP: The dawning of a novel bacterial signaling system. Mol Microbiol. 2005;57(3):629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe AJ, Visick KL. Get the message out: Cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190(2):463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325(15):279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 6.Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007;189(22):8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monds RD, Newell PD, Gross RH, O'Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0–1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol. 2007;63(3):659–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 8.Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genetics. 2007;3(9):1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol Microbiol. 2006;62(4):1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 10.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: The PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281(41):30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 11.Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282(17):12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christen M, et al. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA. 2007;104(10):4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178(1):46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65(4):876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 15.Christen B, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281(42):32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 16.Lee VT, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65(6):1474–1488. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69(2):376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 19.Hinsa SM, O'Toole GA. Biofilm formation by Pseudomonas fluorescens WCS365: A role for LapD. Microbiology. 2006;152(Pt 5):1375–1383. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- 20.Taylor BL. Aer on the inside looking out: Paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol Microbiol. 2007;65(6):1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacEachran DP, et al. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect Immun. 2007;75(8):3902–3912. doi: 10.1128/IAI.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: A study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol. 2008;190(10):3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Appleman JA, Chen LL, Stewart V. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J Bacterial. 2003;185(16):4872–4882. doi: 10.1128/JB.185.16.4872-4882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126(5):929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 25.Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: Recent advances and new puzzles. J Bacteriol. 2006;188(24):8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20(18):2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland LM, et al. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J Bacteriol. 2008;190(15):5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008;190(22):7392–7405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monds RD, Newell PD, Schwartzman JA, O'Toole GA. Conservation of the Pho regulon in Pseudomonas fluorescens Pf0–1. Appl Environ Microbiol. 2006;72(3):1910–1924. doi: 10.1128/AEM.72.3.1910-1924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. A yeast-based molecular tool kit for manipulation of gram-negative bacterial genes. Appl Environ Microbiol. 2006;72(7):5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280(35):30,829–30,837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 32.MacEachran DP, Stanton BA, O'Toole GA. Cif is negatively regulated by the TetR family repressor CifR. Infect Immun. 2008;76(7):3197–3206. doi: 10.1128/IAI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.