Abstract
Systematic validation of pattern formation mechanisms revealed by molecular studies of development is essentially impossible without mathematical models. Models can provide a compact summary of a large number of experiments that led to mechanism formulation and guide future studies of pattern formation. Here, we realize this program by analyzing a mathematical model of epithelial patterning by the highly conserved EGFR and BMP signaling pathways in Drosophila oogenesis. The model accounts for the dynamic interaction of the feedforward and feedback network motifs that control the expression of Broad, a zinc finger transcription factor expressed in the cells that form the upper part of the respiratory eggshell appendages. Based on the combination of computational analysis and genetic experiments, we show that the model accounts for the key features of wild-type pattern formation, correctly predicts patterning defects in multiple mutants, and guides the identification of additional regulatory links in a complex pattern formation mechanism.
Keywords: computational modeling, Drosophila, signaling, systems biology
During Drosophila oogenesis, the 2-dimensional follicular epithelium that envelops the growing oocyte gives rise to an elaborate 3-dimensional eggshell (Fig. 1 A and B) (1). The formation of the respiratory eggshell appendages depends on the formation of the characteristic two-domain gene expression patterns in the follicular epithelium (Fig. 1D). One of the key regulators in this process is broad (br), a gene that encodes a transcription factor expressed in the cells forming the roof (upper part) of the future dorsal appendages (2–5). We have recently proposed a mechanism whereby the spatiotemporal pattern of br is governed by the sequential action of feedforward and feedback loops induced by the highly conserved epidermal growth factor receptor (EGFR) and bone morphogenetic protein (BMP) signaling pathways (Fig. 1C) (6).
Fig. 1.
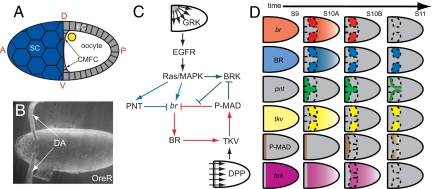
Regulatory network for Br expression in Drosophila oogenesis. (A) Schematic of a stage 10A egg chamber showing the follicular epithelium (FC), the oocyte, the stretch follicle cells (SC), and the centripetally migrating follicle cells (CMFC). Anterior (A), posterior (P), dorsal (D), and ventral (V) are shown. (B) Electron microscopy image of the wild-type Drosophila melanogaster eggshell showing the 2 dorsal respiratory appendages (DA). (Original magnification, 180×.) (C) The network diagram of br regulation by EGFR and DPP signaling pathways (see text for details). (D) Schematic representation of the dorsal views of the expression patterns of network components across 4 stages of oogenesis (stages 9–11) as observed in in situ hybridization and immunohistochemistry data (6). DPP signaling is monitored by following the spatial pattern of MAD phosphorylation (P-MAD) (6). B is reproduced from Yakoby et al. (6); C and D are modified from Yakoby et al. (6).
According to this mechanism, the EGFR ligand gurken (GRK), secreted from the dorsal anterior cortex of the oocyte, establishes the dorsoventral gradient of EGFR activation that induces br in the dorsal follicle cells (6–8). In the anterior dorsal midline, which corresponds to the highest level of EGFR activation, this gradient induces a localized repressor, most likely pointed (PNT), that counteracts the induction of br (8–10). In the anterior follicle cells, br is also repressed by signaling induced by DPP, a Drosophila BMP2/4-like ligand. DPP is secreted from the anteriorly located stretch and centripetally migrating follicle cells and acts through the uniformly expressed DPP receptors, establishing an anteroposterior gradient of DPP signaling (11–13). Thus, the EGFR and DPP pathways localize br expression to the 2 dorsolateral domains of the follicle cells (Fig. 1D). At later stages of oogenesis, BR controls the expression of thickveins (tkv), which encodes a type I DPP receptor essential for DPP signaling (6). Because DPP represses br, this initiates a negative feedback whereby BR controls its own transcriptional repression (6). Another layer of br regulation is provided by brinker (BRK), a transcriptional repressor of DPP signaling (14, 15) that is induced by EGFR and repressed by DPP in oogenesis (16, 17). BRK is likely to delay the repressive action of DPP in the roof cells until a sufficiently high level of BR is established.
Rigorous validation of patterning mechanisms at this level of complexity is essentially impossible without modeling approaches that can test the consistency of the proposed regulatory networks and suggest new experiments. One of the main goals for models is to predict the dynamic expression of multiple network components in multiple genetic backgrounds. With this in mind, we present here a mechanistic model of br regulation. We demonstrate that the model can successfully predict the dynamics of the network in the wild-type and mutant backgrounds. At the same time, we identify a number of inconsistencies between predicted and experimentally observed patterns and suggest changes in the mechanism that can explain them.
Results
Model Formulation.
The spatial arrangement of the midline, roof, and lateral cell fates in the follicular epithelium can be described using a 1-dimensional model, where the spatial coordinate measures the distance along a straight line that is drawn at an angle from the dorsal midline of the follicular epithelium (Fig. 2A). This line captures patterning along both the anterior–posterior as well as the dorsal–ventral axes. Although it misses subtle pattern variations in the midline domain, it allows us to accurately describe the expression patterns in the BR (roof) domain. In the model, local regulation modules that describe the cell-autonomous parts of the network, such as signaling and gene regulation, are coupled by the previously described reaction–diffusion modules that explain the gradients of EGFR and DPP activation (8, 11). We use switch-like models of gene regulation, where the dynamics of gene expression are governed by a piecewise-constant production function and a linear decay term (18–20) The use of such models is justified by the sharp expression boundaries for a large number of genes expressed in the follicular epithelium (21).
Fig. 2.
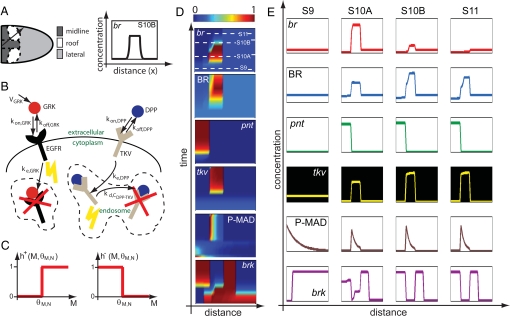
Model-based analysis of wild-type patterns. (A) Spatial arrangement of different cell fates (midline, roof, and lateral) in the 1-dimensional model (Left). The 1-dimensional system is shown by the black arrow. For the expression of br at stage 10B, the graph shows what the computational prediction of its concentration profile along our 1-dimensional system should look like (Right). (B) Processes included in the description of the GRK and DPP morphogens in the model. (C) Heaviside functions used to model switch-like gene regulation. (D) Simulation results for all of the network components in the wild-type background as shown by the color plots of the concentration of each component as a function of space and time. The concentrations are normalized so that their values range from 0 to 1. (E) Predicted concentration profiles of each network component in stages 9–11 of oogenesis. The colors of the graphs follow those of the expression pattern schematics shown in Fig. 1D. Numerical solution of model equations generates spatiotemporal distribution for each of the model components. The 4 chosen time points correspond to the cross-sections in the space–time plot for br expression. The 4 time points are chosen to both capture the lengths of individual stages of oogenesis and best represent the gene/protein localization data obtained from in situ hybridization and immunohistochemistry experiments.
Our model analyzes the regulatory interactions shown in Fig. 1C (experimental evidence for each of these interactions is presented in Table S1). Briefly, the model accounts for the spatial distribution of the GRK and DPP ligands and, consequently, EGFR and DPP signaling, and their effects on the expression of 4 genes: pnt, brk, tkv, and br. The GRK and DPP portions of the model are based on the previously published biophysical descriptions of these morphogens (8, 11). With the exception of br, we lump the transcript and the protein into one species characterized by a single time scale that is equal to the inverse of the degradation rate constant (kd in Eq. 3). Because it is known that the lifetime of BR protein is longer than that of the br transcript, br and BR are modeled separately, with the rate of BR production assumed to be linearly dependent in the level of br transcript.
We assume that the levels of both GRK and DPP signaling, [SEGFR] and [SDpp], are proportional to the occupancy of their receptors [for DPP signaling, it is proportional to the level of internalized ligand–receptor complexes (Fig. 2B)] (22):
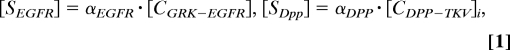 |
where [CGRK-EGFR] is the concentration of GRK–EGFR complexes and [CDPP-TKV]i is the concentration of internalized DPP–TKV complexes. The proportionality constants αEGFR and αDPP describe the combined effects of the EGFR and DPP pathway components downstream of activated receptors. Their values are equal to 1 in the wild-type background but are varied in mutants with defects in pathway activation. For example, αEGFR < 1 in the hypomorph mutant of Ras, a gene encoding an intracellular molecule required for EGFR signaling. Similarly, Mothers against DPP (MAD) and Medea (MED) are intracellular molecules required for BMP signaling. Thus, αDPP = 0 in clones of Mad− and Med− cells. Receptor occupancies are calculated based on the steady-state approximation for ligand–receptor kinetics (Fig. 2B; the derivations can be found in our previous work, and the description of parameters can be found in Table S2) (8, 11):
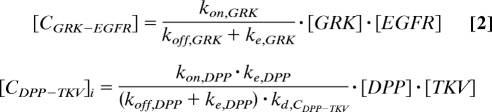 |
Thus, based on the distribution of cell surface receptors (EGFR and TKV) and sources of ligand production, we compute the distributions of extracellular GRK and DPP and the resulting patterns of EGFR and DPP signaling.
The regulation of the transcriptional targets of GRK and DPP is described by using piecewise-linear models that greatly accelerate parametric studies of system dynamics (23). The production terms are equal to either zero or constant (γ in Eq. 3), depending on the levels of inductive signals (Fig. 2C). For example, to describe the fact that PNT is induced above a critical level of EGFR signaling θ[EGFR],[PNT], we model the transcriptional induction of PNT by a Heaviside step function of the local level of EGFR activation, which is provided by the GRK portion of the model. On the other hand, br is controlled by multiple signals: it is activated by EGFR signaling and repressed by both PNT and DPP signaling. Thus, the production term in the equation for br is equal to the product of the switch-like functions that describe both EGFR-dependent and constitutive activation of br, repression by PNT, and repression by DPP signaling. This model leads to the following equations (see also Table S2 and SI Text for the description of initial and boundary conditions):
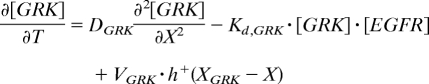 |
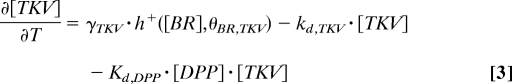 |
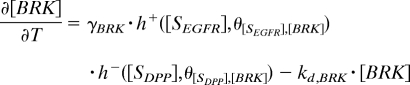 |
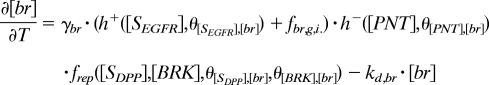 |
In these equations, h+(z) is the Heaviside step function, and h−(z) = 1 − h+(z) (Fig. 2C); frep models the BRK-dependent repressive effect of DPP signaling on br (6, 16). It is equal to 0 when DPP signaling is above the critical threshold θ[DPP],[br] and the level of BRK is below the critical threshold θ[BRK],[br], and it is 1 otherwise (see SI Text for details).
Parameter Selection and Wild-Type Patterning.
We nondimensionalized our model to reduce the number of free parameters and used a parameterization approach that emphasizes the qualitative analysis of observed patterns and their transitions (24). Details of nondimensionalization and parameter selection are given in the SI Text. Briefly, the spatial ranges of the inductive GRK and DPP signals have been estimated elsewhere (8, 11). For a number of parameters, such as the thresholds of the activation and repression functions, the numerical ranges that lead to wild-type transients are defined by inequalities that are suggested by the wild-type patterns themselves. For example, the threshold for the EGFR signaling required for the induction of the midline repressor (PNT) should be higher than that for br induction. This ensures that the width of the br domain is nonzero. As another example, the lifetime of BR protein has to be longer than the time scale of br transcript to be consistent with the fact that BR levels remain high in late stages of oogenesis, even when br mRNA is no longer detectable (3, 6).
Here, we focus on the dynamics of BR regulation in the roof cells. In agreement with the experimental observations, our model predicts that the GRK-dependent induction of br and BRK in the roof cells is followed by the change in the pattern of tkv, the change in the distribution of DPP ligand, the increase in DPP signaling in the roof cells, the DPP-dependent repression of BRK and, eventually, the DPP-dependent repression of br (Fig. 2D). The fact that the model prediction can be described as a sequence of well-defined transitions is an immediate consequence of the switch-like functions that are used to describe signal-dependent gene regulation. The dynamics predicted by the model are robust in a sense that changes in the choices of threshold values, without perturbing the constraints imposed by the wild-type patterns, lead to the same sequence of transitions that may be shifted in time and the same expression patterns that may be shifted in space. Thus, the proposed mechanism, based on the interaction of feedforward and feedback motifs, is sufficient to describe the dynamics of BR expression (Fig. 2 D and E).
Although patterning events in the roof cells are well-described by the model, predictions of dynamics in the midline area are less accurate. In contrast with experimental results, the expression of PNT is predicted to persist through stage 11 of oogenesis (Fig. 1D). This discrepancy can be attributed to the fact that GRK, assumed to be a static signal in the model, disappears at late stages of oogenesis because of the appearance of the vitelline membrane that provides a physical barrier between the oocyte and follicular epithelium (25).
Model-Based Analysis of Mutant Backgrounds.
Our model can be used as an efficient tool for predicting the dynamics of network components in mutant genetic backgrounds. The proposed mathematical model is able to recapitulate 30 experimentally observed pattering defects in 18 mutant backgrounds. A summary of model-based analysis of mutant backgrounds is shown in Tables S3–S5, and detailed model results for some mutant backgrounds can be found in the SI Text. Here, we present analysis of 2 mutants and experimental tests of model predictions.
The first of these backgrounds is provided by flies with the hypomorphic allele of Ras, which leads to a lower level of signaling downstream of activated EGFR and results in ventralized eggshells with a single dorsal appendage (26). In our model, the level of EGFR signaling is a linear function of the local occupancy of cell surface receptors. All downstream components are viewed as a linear filter between activated receptors and gene regulation. In a Ras hypomorph mutant, the gain of this filter, denoted by αEGFR, is less than one, which is taken as the wild-type value. Lowering the value of αEGFR shifts the BR expression domain anteriorly. Below a critical level of αEGFR, the midline region, with no BR expression, disappears entirely (Fig. 3A). This corresponds to the single-appendage eggshell phenotype of the Ras hypomorph flies (26) and reflects a single domain of BR expression (Fig. 3A).
Fig. 3.
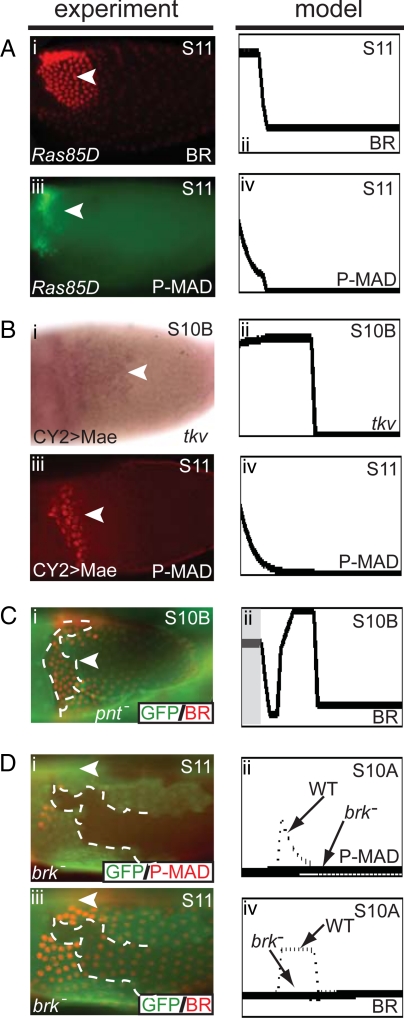
Analysis of pattern formation in mutant backgrounds. (A) BR expression in flies with hypomorphic allele of Ras: the dorsolateral BR patches are fused into a single domain and shifted anteriorly (i and ii). TKV expression is expected to follow the BR pattern, and thus P-MAD signaling also is shifted to the anterior (iii and iv). (B) (i) In situ hybridization image of tkv in egg chambers overexpressing Mae. (iii) Immunohistochemistry experiments show that in stage 11, P-MAD is expressed in an anterior domain instead of its wild-type eyebrow-shaped pattern. Overexpression of Mae results in tkv expression that is expanded anteriorly and late P-MAD signaling that is shifted to the anterior (i–iv). (C) In an anterior clone of pnt−, ectopic BR expression is observed (i and ii). The clone is marked by the absence of GFP, and BR is shown in red (i). Gray box marks the location of the computationally generated pnt− clone (ii). (D) Clonal analysis shows that a brk− clone results in the reduction of both BR and P-MAD (i and iii). The brk− clone is marked by the absence of GFP, whereas P-MAD (i) and BR (iii) are both shown in red. The model predicts that a large brk− clone results in loss of P-MAD and significant reduction of BR expression (ii and iv). Arrowheads mark the dorsal midline, and dotted lines mark the perimeter of the clones; anterior is to the left. (Original magnification, 20×.)
Based on the analysis of eggshell phenotypes, the disappearance of the midline fate upon reduction of EGFR signaling has been attributed to the loss of PNT, which is activated by high levels of EGFR signaling and was proposed to repress BR (3, 10). To test this prediction, we examined the pattern of BR in the egg chambers with uniform expression of mae, a gene encoding a repressor of PNT (9). This should influence the effect of EGFR signaling (by negating the repressive effect of PNT), but not the spatial pattern of signaling itself. In our model, this amounts to setting PNT = 0 at all times throughout the system. As expected based on the eggshell phenotype with a single broad appendage, we observed a single patch of BR expression (Fig. 3B and Table S3), with the “filled” midline and a ventral boundary at the wild-type position. In addition, the model correctly predicts lateral shifts of the BR patches upon increase of the GRK dose, as well as ectopic expression of BR in the midline clones of Ras− and pnt− cells (Fig. 3C and Table S3) (6, 7). Thus, the model serves as a compact representation of a large number of experiments and naturally explains the fact that the size of the midline is set by the level of EGFR signaling (27).
According to the network in Fig. 1C, loss of the midline fate upon either reduction of EGFR signaling or removal of PNT should be accompanied by the midline expression of TKV, a DPP receptor that is transcriptionally activated by BR (Fig. 1C). Our previous genetic mosaic experiments have shown that TKV is required both for transducing the DPP signaling and limiting the diffusion of the anteriorly secreted DPP ligand (6). Thus, in both Ras hypomorph and mae-overexpressing flies, the wild-type gradient of DPP signaling should be shifted to the anterior boundary of the single-patched domain of BR expression. Fig. 3 A and B shows that this is indeed predicted by the computational model. We tested and confirmed this prediction experimentally by visualizing the spatial patterns of tkv and MAD phosphorylation in both of these backgrounds. This demonstrates the ability of our model to predict the expression of multiple network components in multiple mutants.
Computational Analysis of the Patterning Function of BRK.
BRK, a transcriptional repressor of DPP signaling, was placed in the patterning network based on the analysis of its effects on both BR expression and eggshell structures (Fig. 1C). Specifically, BR expression is cell-autonomously abolished in clones of brk− cells, and eggshells derived from these mosaics can be characterized by complete loss of dorsal appendages (16). The distribution of brk mRNA and its changes in mutants revealed that BRK is activated by EGFR and repressed by DPP. In combination with the repressive effect of DPP signaling on BR (6), this suggests that BRK antagonizes the repressive action of DPP during the early stage of eggshell patterning, when the DPP pathway is activated in an anteroposterior gradient. At this stage, the pattern of DPP signaling reflects a combination of anteriorly secreted ligand and uniformly expressed receptors (6, 11, 17)
These effects are readily reproduced in our computational model: BR expression is lost in the brk− region because of the fact that the inductive action of EGFR is overridden by the repressive action of the early anteroposterior gradient of DPP activation. At the same time, our computational results revealed that brk− clones lead to a loss of DPP signaling at later stages of oogenesis. This reflects the action of the negative feedback loop in the model: loss of BR leads to a loss of TKV in the roof cell domain at late stages of oogenesis. Because TKV is essential for DPP signaling, this should lead to a loss of DPP signaling. To test this prediction, we examined the pattern of phosphorylated MAD at late stages of oogenesis (Fig. 1D). The wild-type pattern of P-MAD looks like the 2 laterally placed “eyebrows” and can be interpreted in terms of a model where an anteriorly secreted ligand diffuses across the follicular epithelium with a spatially nonuniform (2-domain) pattern of TKV (6, 28). We found that P-MAD is cell-autonomously lost in the GFP-marked brk− clones, in agreement with our computational prediction (Fig. 3D). Thus, according to our computational results, BRK has a dual role in oogenesis. First, it counteracts the repressive effect of DPP signaling, just as it does in other stages of development (14, 29, 30). This earlier effect has an “echo” at later stages of oogenesis, when BR, protected from DPP signaling by BRK, begins to control TKV expression and, as a consequence, the spatial distribution of DPP ligand and the spatial pattern of DPP signaling (Fig. 1C).
Positive Feedback Loop Suggested by the Model.
In analyzing the computational predictions of our model in multiple mutant backgrounds, we discovered that it fails to predict the absence of tkv expression in egg chambers with uniformly inhibited DPP signaling. Experimentally, this was implemented by uniform expression of Daughters against DPP (DAD), an intracellular inhibitor of DPP signaling; the efficiency of this inhibition is confirmed by the fact that the P-MAD is undetectable in egg chambers with uniformly expressed DAD (data not shown and ref. 31). In our model, DAD overexpression is described by setting αDPP = 0 for all locations at all times (see Eq. 1).
By in situ hybridization, we found that tkv expression is greatly reduced in this background (Fig. 4A). In contrast to this observation, our model predicts that the pattern of TKV expression would be unaffected by uniform inhibition of DPP signaling (Fig. 4B Upper). Indeed, inhibition of DPP signaling should eliminate its repressive effect on BR which, according to the model, induces TKV at later stages of oogenesis. The necessity of this regulatory connection was based on the fact that tkv expression and P-MAD were eliminated in clones of br− cells (6). At the same time, these previously published experiments have not explored whether BR is sufficient to induce tkv, and thus whether it is the only activator of tkv during late stages of oogenesis.
Fig. 4.
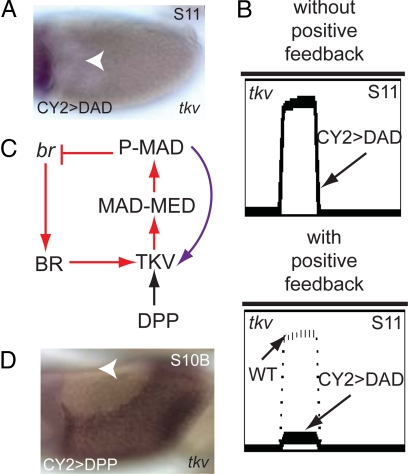
Positive feedback loop suggested by the model. (A) In situ hybridization of tkv in CY2-DAD background shows that tkv expression is greatly reduced. (B) Simulations without the positive feedback loop predict that tkv expression in this background is indistinguishable from its wild-type expression pattern (Upper). When the positive feedback loop is included, however, simulations predict that tkv expression is greatly reduced in CY2-DAD egg chambers (Lower). (C) A positive feedback involving DPP signaling regulating the expression of its own receptor TKV is added (purple arrow) into the network presented in Fig. 1C. Here, only part of the network is shown. (D) In situ hybridization image of tkv in a CY2-DPP egg chamber. A shows dorsal view and D shows lateral view with dorsal on top; anterior to the left. Arrowheads mark the dorsal midline. (Original magnification, 20×.)
The discrepancy between the observed and predicted patterns of tkv led us to hypothesize that in addition to BR, the expression of TKV requires DPP signaling. Thus, we proposed that during the late stages of eggshell patterning, TKV controls its own expression via a positive feedback loop (Fig. 4C). To test the feasibility of this feedback, we used the CY2-Gal4 driver to express DPP uniformly throughout the follicular epithelium. This induced a dramatic change in the pattern of tkv expression. The low levels of tkv in the lateral and posterior areas of the wild- type follicular epithelium have been replaced with a strong domain of tkv expression (Fig. 4D). Overexpression of DPP in the posterior region by using the E4-Gal4 driver (active in the posterior of the egg chamber) also results in local up-regulation of tkv (data not shown).
These experiments support the proposed positive feedback loop. Because the mechanism of this feedback is currently unknown, it can be modeled in a number of different ways. DPP signaling can either positively regulate tkv transcription or stabilize the tkv transcript. Our computational results show that either of these mechanisms can repair the discrepancy between the predicted and observed patterns of tkv in the egg chambers with uniformly inhibited DPP signaling (Fig. 4B Lower; see SI Text for details). Importantly, the addition of this regulatory connection does not compromise the ability of the model to account for patterning defects in other mutants.
Discussion
We have developed a mechanistic model for BR regulation by the EGFR and DPP pathways in Drosophila oogenesis. Our model is relatively simple: it is quasi 1-dimensional, describes only a part of the genes acting in eggshell patterning, and employs switch-like nonlinearities to describe gene regulation. At the same time, the analysis of this model clearly demonstrates its ability to summarize a large number of experimental observations, predict dynamics of network components in genetic backgrounds of essentially arbitrary complexity, and guide further experimental studies of eggshell patterning. The model provides further support for the previously proposed feedforward-feedback mechanism and makes a number of testable predictions (6). For example, the model correctly predicted that BRK, which in other stages of fruit fly development antagonizes the effects of DPP signaling, indirectly controls the level of DPP signaling in late oogenesis by regulating the action of the negative feedback loop in the BR-patterning network. By systematically exploring network dynamics in mutant backgrounds, we identified a number of inconsistencies between the experimentally observed and model-predicted patterns. For instance, contrary to the model prediction, uniform inhibition of DPP signaling in the follicular epithelium leads to disappearance of the tkv transcript. This observation suggested the presence of an additional positive feedback loop between DPP signaling and DPP receptor expression. The possibility of this feedback is further supported by gain-of-function experiments.
In the future, our model can be extended to account for the 2-dimensional character of expression patterns in the follicular epithelium and other components of the eggshell-patterning network (21). For instance, a static description of the EGFR activation pattern in our model neglects the fact that at later stages of oogenesis, the oocyte-derived GRK input disappears and EGFR activation is mediated by Spitz, secreted by the rhomboid-expressing cells (25, 33–35). Because the model involves a feedback loop, the continuous description of GRK production in this model may lead to oscillatory behavior of the system in even later stages of oogenesis; i.e., the repression of br by DPP signaling results in an eventual reduction of P-MAD, which is then unable to repress br, allowing its expression. This is contrary to experimental results that show that br expression remains undetectable throughout later stages of oogenesis. Replacing EGFR ligand from GRK to Spitz at late stages of oogenesis will eliminate EGFR signaling in the wide dorsal domain, preventing the system from entering the oscillatory regime.
Because of the modular structure of our mechanism and its mathematical representation, new components can be incorporated relatively straightforwardly into our computational model. As an example, recent experiments have shown that BR represses Cad74A, an atypical cadherin that is down-regulated in the roof cells and important for robust dorsal appendage morphogenesis (36). In combination with the simple repressive connection between BR and Cad74A, our model can predict how Cad74A responds to various perturbations of eggshell-patterning signals. Finally, we note that all of the regulatory connections in our model are yet to be verified by the detailed analysis of the cis-regulatory modules in the eggshell-patterning network.
Materials and Methods
Genetics.
The following stocks were used in this study: wild-type OreR, X7;28.20 (which contains 4 copies of P[w+ grk+] (2P{GRK}), ref. 37) and UAS-Dad, CY2-UAS-Mae(edl) (gifts from J. Duffy, Worcester Polytechnic Institute, Worcester, MA), UAS-Dpp (gift from T. Schüpbach, Princeton University), CY2-Gal4 (38, 39), and E4-Gal4 (38). To generate the Ras hypomorph mutant flies, Ras85DE62K and Ras85D05703ry (506)cv-c flies were crossed (both were gifts from C. Berg, University of Washington, Seattle). The FLP/FRT mitotic recombination system (40, 41) was used to generate clones of mutant follicle cells marked by the absence of GFP. The following flies were used to generate Ras− clones: a null allele of Ras: e22c-Gal4 UAS-FLP; FRT82B rasΔC40b/FRT82B ubi-GFP (32, 42). To generate pnt− clones, w;pntLacZ/ e22c-Gal4 UAS-FLP;pntΔ88 FRT82B (gift from J. Duffy) were used, whereas brkCA54 FRT19A; e22c-Gal4 UAS-FLP (gift from T. Schüpbach) were used to generate brk− clones. Flies were grown on agar cornmeal medium; baker yeast was added to the fly medium 24 h before ovary harvesting; all crosses were done at 23 °C.
In Situ Hybridization, Immunofluorescence, and Microscopy.
In situ hybridization was carried out as described previously (6). The primary antibody of rabbit anti-P-Smad1/5/8 (1:3000) was a generous gift from D. Vasiliauskas (New York University, New York) and S. Morton, T. Jessell, and E. Laufer (Columbia University Medical Center, New York). Secondary antibodies AlexaFluor 568 anti-mouse, Oregon Green 488 anti-rabbit, and AlexaFluor 488 anti-sheep were used (1:2000; Molecular Probes). Images of immunofluorescence experiments were taken with a PerkinElmer RS3 Spinning Disk Confocal microscope and a Nikon Eclipse E800 compound microscope. Images were processed with ImageJ (Rasband, 1997–2006) and Photoshop (Adobe Systems).
Supplementary Material
Acknowledgments.
We thank Trudi Schüpbach for advice and encouragement during the course of this work and all members of the Shvartsman lab for helpful discussions. We are grateful to Dan Vasiliauskas, Susan Morton, Tom Jessell, and Ed Laufer for providing the P-MAD antibody. We also thank Joe Duffy, Celeste Berg, and Trudi Schüpbach for fly stocks and reagents. This work was supported by National Institutes of Health Grants P50 GM071508 and R01 GM078079.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810728106/DCSupplemental.
References
- 1.Berg CA. The Drosophila shell game: Patterning genes and morphological change. Trends Genet. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Tzolovsky G, Deng WM, Schlitt T, Bownes M. The function of the broad-complex during Drosophila melanogaster oogenesis. Genetics. 1999;153:1371–1383. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- 4.James KE, Berg CA. Temporal comparison of Broad-Complex expression during eggshell-appendage patterning and morphogenesis in two Drosophila species with different eggshell-appendage numbers. Gene Expr Patterns. 2003;3:629–634. doi: 10.1016/s1567-133x(03)00136-4. [DOI] [PubMed] [Google Scholar]
- 5.Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol. 2004;267:320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Yakoby N, Lembong J, Schupbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135:343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- 7.Atkey MR, Lachance JF, Walczak M, Rebello T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133:2115–2123. doi: 10.1242/dev.02369. [DOI] [PubMed] [Google Scholar]
- 8.Goentoro LA, et al. Quantifying the gurken morphogen gradient in Drosophila oogenesis. Dev Cell. 2006;11:263–272. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada T, Okabe M, Hiromi Y. EDL/MAE regulates EGF-mediated induction by antagonizing Ets transcription factor Pointed. Development. 2003;130:4085–4096. doi: 10.1242/dev.00624. [DOI] [PubMed] [Google Scholar]
- 10.Morimoto AM, et al. Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development. 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- 11.Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A. 2008;14:1469–1477. doi: 10.1089/ten.tea.2008.0098. [DOI] [PubMed] [Google Scholar]
- 12.Jekely G, Rorth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 2003;4:1163–1168. doi: 10.1038/sj.embor.7400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peri F, Roth S. Combined activities of Gurken and Decapentaplegic specify dorsal chorion structures of the Drosophila egg. Development. 2000;127:841–850. doi: 10.1242/dev.127.4.841. [DOI] [PubMed] [Google Scholar]
- 14.Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell. 1999;96:563–573. doi: 10.1016/s0092-8674(00)80660-1. [DOI] [PubMed] [Google Scholar]
- 15.Pyrowolakis G, Hartmann B, Muller B, Basler K, Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev Cell. 2004;7:229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Schupbach T. The role of brinker in eggshell patterning. Mech Dev. 2006;123:395–406. doi: 10.1016/j.mod.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007;134:2261–2271. doi: 10.1242/dev.02856. [DOI] [PubMed] [Google Scholar]
- 18.Casey R, de Jong H, Gouze JL. Piecewise-linear models of genetic regulatory networks: Equilibria and their stability. J Math Biol. 2006;52:27–56. doi: 10.1007/s00285-005-0338-2. [DOI] [PubMed] [Google Scholar]
- 19.de Jong H, Geiselmann J, Batt G, Hernandez C, Page M. Qualitative simulation of the initiation of sporulation in Bacillus subtilis. Bull Math Biol. 2004;66:261–299. doi: 10.1016/j.bulm.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Plahte E, Mestl T, Omholt SW. A methodological basis for description and analysis of systems with complex switch-like interactions. J Math Biol. 1998;36:321–348. doi: 10.1007/s002850050103. [DOI] [PubMed] [Google Scholar]
- 21.Yakoby N, et al. A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell. 2008;15:725–737. doi: 10.1016/j.devcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi YG, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 23.Perkins TJ, Jaeger J, Reinitz J, Glass L. Reverse engineering the gap gene network of Drosophila melanogaster. PLoS Comput Biol. 2006;2:e51. doi: 10.1371/journal.pcbi.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J Theor Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 26.Schnorr JD, Berg CA. Differential activity of Ras1 during patterning of the Drosophila dorsoventral axis. Genetics. 1996;144:1545–1557. doi: 10.1093/genetics/144.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuman-Silberberg FS, Schupbach T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development. 1994;120:2457–2463. doi: 10.1242/dev.120.9.2457. [DOI] [PubMed] [Google Scholar]
- 28.Mantrova EY, Schulz RA, Hsu T. Oogenic function of the myogenic factor D-MEF2: Negative regulation of the Decapentaplegic receptor gene thick veins. Proc Natl Acad Sci USA. 1999;96:11889–11894. doi: 10.1073/pnas.96.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: Regulation of Dpp targets by brinker. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- 30.Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T. brinker is a target of Dpp in Drosophila that negatively regulates Dpp dependent genes. Nature. 1999;398:242–246. doi: 10.1038/18451. [DOI] [PubMed] [Google Scholar]
- 31.Tsuneizumi K, et al. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- 32.James KE, Dorman JB, Berg CA. Mosaic analyses reveal the function of Drosophila Ras in embryonic dorsoventral patterning and dorsal follicle cell morphogenesis. Development. 2002;129:2209–2222. doi: 10.1242/dev.129.9.2209. [DOI] [PubMed] [Google Scholar]
- 33.Sapir A, Schweitzer R, Shilo BZ. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development. 1998;125:191–200. doi: 10.1242/dev.125.2.191. [DOI] [PubMed] [Google Scholar]
- 34.Peri F, Bokel C, Roth S. Local Gurken signaling and dynamic MAPK activation during Drosophila oogenesis. Mech Dev. 1999;81:75–88. doi: 10.1016/s0925-4773(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 35.Ruohola-Baker H, et al. Spatially localized rhomboid is required for establishing the dorsal-ventral axis in Drosophila oogenesis. Cell. 1993;73:953–965. doi: 10.1016/0092-8674(93)90273-s. [DOI] [PubMed] [Google Scholar]
- 36.Zartman JJ, et al. Cad74A is regulated by BR and is required for robust dorsal appendage formation in Drosophila oogenesis. Dev Biol. 2008;322:289–301. doi: 10.1016/j.ydbio.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- 38.Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 39.Goentoro LA, Yakoby N, Goodhouse J, Schupbach T, Shvartsman SY. Quantitative analysis of the GAL4/UAS system in Drosophila oogenesis. Genesis. 2006;44:66–74. doi: 10.1002/gene.20184. [DOI] [PubMed] [Google Scholar]
- 40.Duffy J, Harrison D, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- 41.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 42.Hou XS, Chou TB, Melnick MB, Perrimon N. The torso receptor tyrosine kinase can activate Raf in a Ras-independent pathway. Cell. 1995;81:63–71. doi: 10.1016/0092-8674(95)90371-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.