Abstract
The T cell receptors from the regulatory IL-10-secreting T cell line induced by the random amino acid copolymers poly(F,Y,A,K,)n in SJL mice (H-2s) have been characterized, cloned, sequenced and expressed both in 293T cells and in 2 different TCR α−/β− T cell hybridomas. The usage of TCR α and β V regions in the cell line was oligoclonal. Four TCR α/β pairs cloned from single cells of the T cell line were inserted into a retrovirus vector linked by an oligonucleotide encoding the 2A peptide that spontaneously cleaves in vivo. After cotransfection of this vector with a CD3 vector into the 293T cells, the TCR were surface expressed. Moreover, after transduction into the 2 T cell hybridomas, all 4 were functional as evidenced by their response to stimulation by poly(F,Y,A,K)n. All 4 pairs were Vα3.2(3.5)/Vβ14, a prominent clonotype found in the poly(F,Y,A,K)n-specific T cell line. These V regions are identical to those recently found in a regulatory T cell line that secretes both IL-4 and IL-10 induced in B10.PL mice with a different MHC hapotype (H-2u) by a small peptide obtained from an autoimmune TCR of that strain. These data lead to a hypothesis regarding the origin of the epigenetic modifications that lead to selective cytokine secretion in T cells.
Keywords: epigenetics, Foxp3, FYAK, glatiramer acetate, retrovirus vector
Regulation of immune responses is currently a central focus of immunology. Several types of cells are involved in this regulation, including macrophages (1), dendritic cells (2), and T cells. Two principle types of regulatory T cells have been described, cytokine-secreting antigen-nonspecific regulatory T cells that mediate bystander immunosuppression (3–5) and forkhead box P3 (Foxp3+) antigen-specific T cells (natural Treg) that require cell–cell contact for their activity (6, 7).
The random amino acid copolymer Copaxone [poly(Y,E,A,K)n (YEAK), glatiramer acetate (GA), Copolymer 1 (Cop1)] has been widely used for the therapy of multiple sclerosis in humans and for the amelioration of experimental autoimmune encephalomyelitis (EAE) in mice for several decades (8, 9). More recently, a random copolymer, poly (F,Y,A,K)n, called FYAK, that appears to be more effective than GA in several mouse models of EAE has been extensively studied (10). Both copolymers ameliorate EAE by induction of regulatory T cells that secrete the immunomodulatory cytokine IL-10 principally, and other cytokines in smaller amounts (10). These IL-10 secreting T cells belong to the family of cytokine-secreting T cells, including Tr1 and Th3 cells (3, 11), which mediate bystander immunosuppression. The amino acid copolymers are presented by dendritic cells and/or macrophages that also appear to have a role in the phenomenon (12, 13).
At the same time in a second line of investigation immunization with peptide derived from the Vβ TCR segment or DNA encoding a Vβ peptide was found to lead to amelioration of autoimmune disease (14–18). In parallel work, TCR blocking antibodies were also shown to be useful in therapy (19, 20). Moreover, immunization with peptides derived from TCR Vβ segments may induce T cells that secrete a large amount of IL-10 in addition to IL-4 (16, 21, 22). Thus, the mechanisms by which immunization with TCR peptide and that in which blocking antibody ameliorate disease are distinct.
The purpose of the present research was to characterize structurally and functionally the T cell receptors of an IL-10 secreting FYAK-specific T cell line. Only a few reports of the T cell receptors of cytokine-secreting or of Foxp3+ nTreg have appeared (18, 23), and we will discuss these reports in the context of the present work.
Results
TCR Vβ Repertoire of an FYAK-Specific T Cell Line.
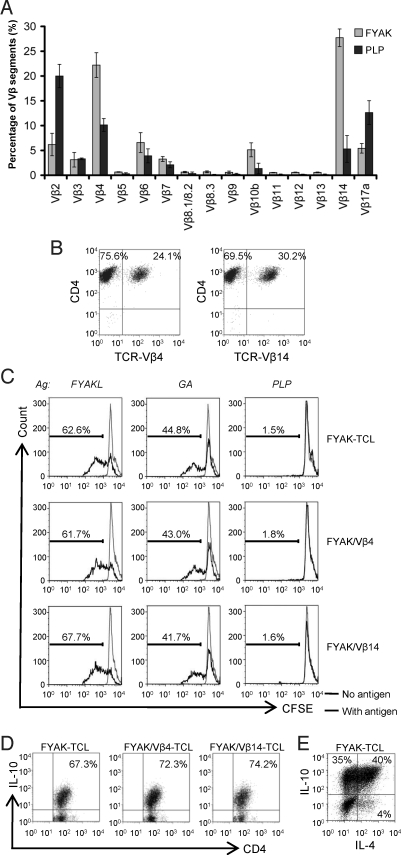
T cell lines were established from spleens of SJL mice as described, using either the random amino acid copolymer FYAK or the autoantigen PLP139–151 as the immunogen (10, 24–26). The primary lines were restimulated 3 times at 2-week intervals in IL-2. T cell lines were analyzed after the last restimulation using a panel of murine TCR Vβ-specific mAb. At least 3 different batches of the cell lines were analyzed. For the FYAK-specific cell lines, Vβ4 and Vβ14 were preferentially used, whereas, in the PLP139–151-specific cell lines, Vβ2 and Vβ17a were preferentially used. Some PLP-specific cell lines also used Vβ4 and Vβ14 (Fig. 1A). The frequency of TCR Vβ4 and Vβ14 in the FYAK-specific cell lines averaged 22% and 28%, respectively, and the highest frequency observed was 24% for Vβ4 and 30% for Vβ14, a total of 54% (Fig. 1B).
Fig. 1.
Predominance of the TCR Vβ4 and Vβ14 usage in FYAK-specific CD4+ T cell lines. (A) FACS analysis of Vβ segments. Frequencies of each segment are shown as means of at least 3 experiments. Error bars show SD values. (B) Staining of FYAK-specific T cell line with antibodies against CD4 and Vβ4 or Vβ14. (C) Sorted CD4+Vβ4+ and CD4+Vβ14+ populations were restimulated with FYAK, GA or PLP139–151 for 72 h. Proliferation of unsorted and sorted populations to the different antigens was evaluated by CFSE-labeling. (D) In the last 4 h of restimulation, unsorted or Vβ sorted FYAK-specific T cell populations were incubated with PMA, ionomycin, and BFA; then, IL-10 expression was analyzed by intracellular staining. (E) IL-4 and IL-10 expression on one FYAK-specific T cell line was analyzed by intracellular staining.
Next, the 2 populations of Vβ4 and Vβ14 containing TCR from a FYAK-specific cell line were sorted by FACS. Proliferation assays with CFSE-labeling showed that the 2 sorted populations each proliferated significantly to FYAK and less to GA but not to PLP139–151, as was also the case with the unsorted population (Fig. 1C). All 3 populations produced IL-10 with 67–78% of the cells IL-10 positive by intracellular staining (Fig. 1D). For one of the FYAK-specific T cell lines, 35% of the cells secreted IL-10, 40% secreted both IL-10 and IL-4 and 4% secreted IL-4 only using intracellular staining for detection (Fig. 1E).
TCR Vα Repertoire.
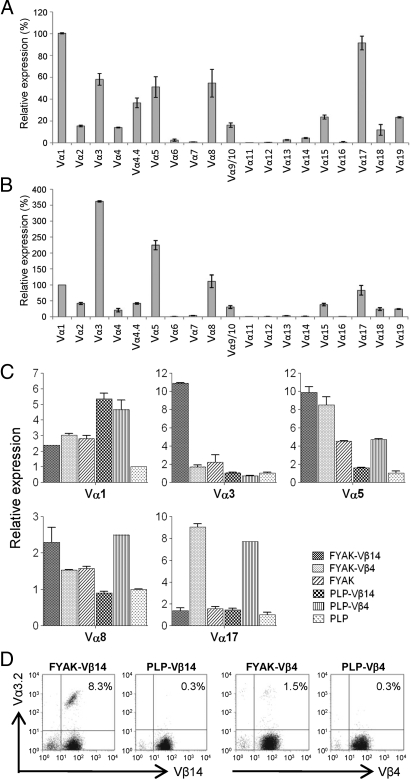
Only a limited number of TCR Vα-specific mAb are available (Vα2, Vα3.2, Vα8.3, and Vα11.1/11.2). Analysis of the Vα repertoire in FYAK-specific T cell line was therefore carried out by quantitative PCR (qPCR). Among 19 Vα segments analyzed, 12 were used in the unsorted population, 2 were used at relatively high levels, 4 were used at intermediate level, and 6 were used at low levels (Fig. 2A). In the sorted Vβ14-expressing FYAK-specific T cell population, Vα3 predominated, Vα5 was present at an intermediate level, and the other 8 were present at lower levels (Fig. 2B). The expression of the 5 most frequently used Vα segments represented in both unsorted and Vβ14+ sorted populations was further examined in the 3 FYAK populations (unsorted, Vβ14-expressing and Vβ4-expressing) and in corresponding PLP139–151-specific populations. Strikingly, the Vα3 segment was expressed only by the Vβ14-expressing FYAK-specific T cells (Fig. 2C). By contrast, the usage of Vβ14 and Vα3 in the PLP139–151 cell line was very low (Figs. 1A and 2C). The selective expression of Vα3.2 in these cells was confirmed by flow cytometry analysis, using Vα3.2-specific mAb (Fig. 2D).
Fig. 2.
TCR Vα repertoire analysis. RNA was isolated from T cells and reverse-transcribed into cDNA for qPCR with specific 5′ Vα primers and a 3′ Cα primer. (A and B) Expression of Vα1 was designated as 100%, and relative expression of each Vα segment on established unsorted FYAK-specific T cell line (FYAK-TCL) (A) or Vβ14-expressing sorted FYAK-TCL (B) are shown. (C) The top 5 Vα segments with the highest expression in both A and B were compared by qPCR among different T cell populations. They are (from left to right) Vβ14+ FYAK-TCL, Vβ4+ FYAK-TCL, unsorted FYAK-TCL, Vβ14+ PLP-TCL, Vβ4+ PLP-TCL, and unsorted PLP-TCL. The expression of each Vα segment on unsorted PLP-TCL was designated as 1. All of the qPCR data are shown as means of triplicates. (D) Flow cytometry showing the expression of Vα3.2 on different Vβ sorted T cell populations.
Diversity of the Complementarity-Determining Region 3 (CDR3).
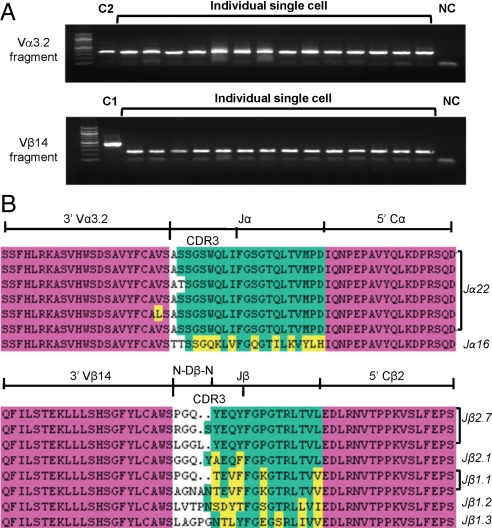
To examine the CDR3 sequences, the Vα3.2 and Vβ14 double positive cells (Fig. 2D) were sorted by single-cell sorting into 96-well plates. Then the primers specific to Vα3.2 and Cα were used to amplify the CDR3 regions of Vα3.2 from 14 different cells; similarly, the primer specific to Vβ14 and Cβ universal primer were used to amplify the CDR3 regions of Vβ14 from another 14 different cells (Fig. 3A). Seven PCR products of the Vα3.2 fragment and 8 of the Vβ14 fragments were purified and subjected to DNA sequencing. Translated protein sequences were aligned, showing that the diversity of the CDR3 region was more pronounced in the Vβ14 segments than in the Vα3.2 segments (Fig. 3B). These TCR sequences were also analyzed by IMGT-V-QUEST (IMGT; available at http://imgt.cines.fr, Montpellier, France) for the usage of J regions. The predominant J regions used were Jα22 and to a slightly lesser extent, Jβ2.7 (Fig. 3B).
Fig. 3.
CDR3 analysis of TCR Vα3.2 and Vβ14 fragments. (A) The CD4+Vα3.2+Vβ14+ T cells from FYAK-TCL were sorted at 1 cell per well into 96-well PCR plate for specific amplification of TCR fragments by single-cell RT-PCR. The PCR products from 29 single cells are shown: 14 cells for the Vα3.2 fragment, 14 cells for the Vβ14 fragment, and 1 cell for the β-actin gene (C1). RNA from a pool of FYAK-specific T cells was used as a positive control for the PCR amplification (C2). NC, negative control with no cells. (B) PCR products from single cells (7 for Vα3.2; 8 for Vβ14) were subjected to sequencing. Alignments of the translated protein sequences are shown. Red, V and C regions; green, J region; yellow, those amino acids that differ from the most frequent residues appearing at the same position. The N-Jα or N-Dβ-N region is not colored.
Cloning of Full-Length TCR Pairs Used by the Vα3.2+/Vβ14+ FYAK-Specific T Cells.
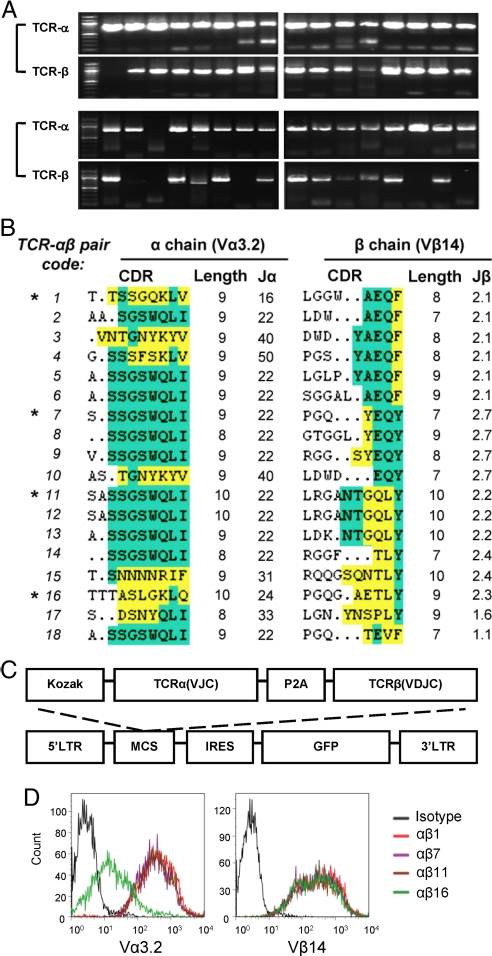
Cloning of full-length TCRα and TCRβ cDNAs from single cells of a FYAK-specific T cell line was initiated for further work. Single cells were again sorted into a 96-well plate for RT-PCR. Pairs of full-length TCRα and β segments with suitable quality were obtained from 18 cells (Fig. 4A). In these experiments, cell lysis for total RNA, reverse transcription and PCR amplification were done in 1 step, using primers that were designed based on the published sequences for these segments (see Methods). Sequencing of these 18 clones confirmed that they all contained Vβ14; 17 of them contained Vα3.2 while 1 of them contained Vα3.5 (sequences as shown in reference (27)). The alignment of protein sequences again indicated substantial diversity in both CDR3 regions, especially in the Vβ14 CDR3 region, represented by different amino acids and length ranges (8–10 aa for Vα3.2 and 7–10 aa for Vβ14) (Fig. 4B). Four pairs of full-length α and β cDNAs (αβ1, -7, -11, and -16) were selected with reasonably diverse sequences. The α and β segments of each of these pairs were linked through overlapping PCR, with an oligonucleotide encoding the 2A peptide derived from Porcine teschovirus-1 (P2A; see ref. 28 for the sequence). After transcription and translation this peptide mediates spontaneous cleavage between the 2 linked proteins. The TCRα(VJC)-2A-TCRβ(VDJC) was inserted into the multicloning sites of the retrovirus vector pMIG (M, MSCV; I, IRES; G, GFP) (Fig. 4C). To evaluate the expression of these cloned TCR pairs, the plasmid pMIG-TCRα-2A-TCRβ was cotransfected with pMIG-CD3 (the plasmid encoding all 4 CD3 subunits) into 293T cells. After a 48-h culture, the 293 T cells were analyzed by flow cytometry, using Vα3.2- and Vβ14-specific mAbs. All 4 cultures were positive with both antibodies, although the group “αβ16” that used Vα3.5 showed a weaker staining of Vα3.2 mAb than the other 3 (Fig. 4D). These experiments established that single-cell RT-PCR cloning had yielded at least 4 pairs of TCRα and TCRβ segments that would properly combine in vivo to express a TCR on the surface of 293T cells.
Fig. 4.
TCR Cloning. (A) Every TCR pair including the full-length α chain and the full-length of β chain was PCR amplified from the same single CD4+Vα3.2+Vβ14+ FYAK-specific T cell. PCR products from 32 single cells are shown. Only those pairs with good amplification of both α and β chain were used for further subcloning. (B) CDR3 amino acid sequences for Vα3.2 and Vβ14 TCR from 18 individual cells. The conserved residues CAXS or CAWS flanking the 5′ CDR3 of Vα3.2 and Vβ14 respectively, or FGXG flanking the 3′ CDR3 is not shown. Residues found in the J region are indicated in green. The amino acids different from the most frequent residues of the J region are indicated in yellow. The asterisk denotes the 4 TCR αβ pairs used for subcloning. (C) Structure of retroviral TCR construct. Amplified TCR α and β chain cDNA sequences were linked with the 2A peptide sequence of porcine teschovirus-1 (P2A) by recombinant PCR and then subcloned into a cassette vector, MSCV-IRES-GFP (pMIG), for retrovirus-mediated expression. (D) 293T cells cotransfected with TCR construct and CD3 vector (pMIG-CD3δγεζ) were stained with mAbs against TCR Vα3.2 and Vβ14. Flow cytometry showing the expression of cloned TCR on the surface of transfected 293T cells.
Functionality of the FYAK-Specific Vα3.2/Vβ14 TCR αβ Pairs.
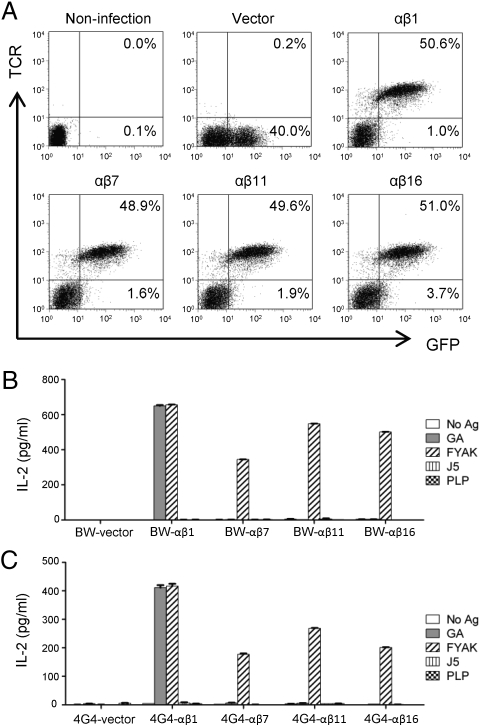
To examine the functionality of the cloned TCRαβ pairs, the 4 pairs described in the preceding section were expressed in 2 different TCRα−/β− T-hybridoma cells, BW-1 and 4G4.CD4. A full complement of CD3 genes (γ, δ, ε, and ζ) had been inserted into both recipient cell lines and CD4 had also been inserted into the second cell line. These 2 recipient cell lines are programmed for IL-2 secretion that results from insertion of any functional TCR αβ pair into them. For this experiment, pMIG-TCRα-2A-TCRβ together with pEQ-Pam3(-E) and pVSVg that encode retrovirus packaging proteins were cotransfected at a ratio of 2:2:1 into 293T cells. The supernatants of 293T cells were collected starting from 36 h after transfection and used for generating the retrovirus-producing GP+E86 cells. Recipient BW-1 and 4G4.CD4 cells were then infected by retroviral supernatant produced by GP+E86 cells. GFP expression on recipient cells showed the infection efficiency, and the majority of GFP-positive T-hybridoma cells, except for the group in which vector alone was used, were expressing TCR (Fig. 5A). Sorted GFP+ or GFP+/TCR+ recipient T-hybridoma cells were stimulated with FYAK, GA, J5 [an immunomodulatory peptide whose sequence was based on a binding motif for GA (26)], and PLP139–151. All 4 BW-1 or 4G4.CD4 cell lines that received TCRαβ-containing vectors were functional as evidence by secretion of IL-2 on stimulation with FYAK. They differed, however, in the extent of stimulation that was paralleled in the 2 different recipients. In addition, only 1 of these 4 TCR-transduced cell lines was stimulated by GA, and none was stimulated by the immunomodulatory peptide J5 or by PLP139–151 (Fig. 5 B and C).
Fig. 5.
Expression and function of retroviral TCR in T-hybridoma cells. Supernatant from the retrovirus-producing GP+E86 cells were collected for infection of the TCR-deficient BW-1 and 4G4.CD4 T-hybridoma cells. (A) BW-1 cells were stained with TCRβ antibody. Flow cytometry shows the expression of all 4 TCR pairs (αβ1, αβ7, αβ11, and αβ16) on infected (GFP-positive) cells. No TCR expression was detected on BW-1 cells infected by vector only. (B and C) The TCR-bearing and vector-only infected T-hybridoma cells were sorted for GFP+TCRβ+ (groups αβ1, -7, -11, and -16) or GFP+ (vector) populations and stimulated with different antigens presented by APC. Supernatant from stimulated BW-1 (B) or 4G4.CD4 (C) cells were harvested at 48 h, and IL-2 production was measured by ELISA.
Discussion
An important question which these experiments seeks to approach is whether the rearrangement of the T cell receptor genes has some role in the phenotype of regulatory T cells, i.e., whether it has a role in the epigenetic changes that lead to different functions or to selective cytokine secretion by different T cells. In the case of natural regulatory T cells that are identified by the expression of the transcription factor Foxp3 (6, 7) and by the surface expression of CD39 and CD73 (nucleotide hydrolases that lead to generation of immunomodulatory adenosine from ATP) (29) epigenetic modifications clearly occur. These Treg are generated in the thymus, are antigen-specific, and require cell–cell contact to mediate immunosuppression. The mechanism of their central generation is largely unknown. However, the TCR sequences from Foxp3+CD4+CD25+ T cells are polyclonal and in one study appear to mirror the sequences found in the self-reactive nonregulatory T cell repertoire, at least in part (23), supporting the idea that the former are derived by epigenetic conversion of the latter. They can also be generated in the periphery. For example, administration of anti-DEC-205-HA to mice and its binding to tolerogenic immature dendritic cells leads to immune tolerance to HA that appears to result from the conversion of CD4+ T cells recognizing HA to Foxp3+CD4+CD25+ Treg (30). [Anti-DEC-205-HA is a fusion monoclonal antibody comprised of a dendritic cell-specific monoclonal antibody, anti-DEC-205, to which the immunogenic peptide hemagglutinin 107–119 (HA) has been linked to the C terminus of both of the heavy chains in the antibody.] In addition introduction of Foxp3 by retrovirus vectors into cytotoxic T cells converts these cells from cytotoxic T cells to regulatory T cells (31).
A second type of Treg, cytokine-secreting autoantigen non-specific Treg that mediate bystander immunosuppression, is generated in the periphery. Two main types have been described, Tr1 cells that secrete IL-10 (although some IL-10-secreting T cells also secrete IL-4) (3–5) and Th3 cells that secrete TGFβ (although some Th3 cells secrete both TGFβ and IL-10) (11). A failure of either of these regulatory mechanisms in vivo and of the generation of natural regulatory T cells as occurs in Foxp3 deficiency (6) could be involved in the genesis of autoimmune diseases.
Administration of amino acid copolymers results in generation (presumably expansion) of IL-10-secreting regulatory T cells, similar to Tr1 cells. The expansion of these cells likely results from proliferation of precursors generated in response to regulatory phenomena associated with the control of bacterial invasion (32). The gastrointestinal tract is the site at which many different bacteria are kept in homeostatic equilibrium with host tissues and consequently the gut-associated lymphoid tissue is the presumable site of their generation (33). The generation of these induced Treg as the result of co polymer treatment is an important part of the mechanism that leads to their amelioration of some autoimmune diseases in mice and in humans. In particular, the Vα3/Vβ14 rearrangement, found in a reasonably large number of the inducible Treg studied here, occurred only at very low frequency in the other T cell lines examined. No sequences have been reported for TCR derived from Tr1 or Th3 Treg. However, regulatory T cells derived by vaccination with DNA encoding a Vβ peptide from an autoimmune TCR or by immunization with Vβ peptides secrete IL-10 and IL-4 and are likely to belong to this category (16, 21, 22). In the 1 case studied in detail, B10.PL mice (H-2u) were immunized with a peptide (B5) from the framework region of a Vβ8.2 TCR found in mice with induced EAE (18), the polyclonal Treg obtained were predominantly Vβ14. Moreover, of the 11 Vα sequences examined, 4 were Vα4.3 and 2 were Vα3.2. The Vα3.2/Vβ14 clones are strikingly similar to those studied here that were derived from SJL mice (H-2s), using a different Treg-inducing immunogen, FYAK. These data suggest that the question whether specific TCRα/β pairs may characterize IL-10-secreting Treg requires careful study and that the epigenetic change(s) that leads to selective cytokine secretion in T cells may be dependent, at least in part, on the structure of the TCR. This view is supported by a study of immune tolerance induced by myelin basic protein (MBP)-specific CD8+ T cells. Two TCR-transgenic mouse lines (C3H background, H-2K+) expressing different MBP79-87-specific TCR (Vα8/Vβ6 or Vα8/Vβ8) were generated. T cells expressing Vα8/Vβ6 TCR were subjected to both central and peripheral tolerance, while T cells expressing the other TCR showed no evidence of tolerance induction, implying that the TCR itself may confer epigenetic regulation (34).
Further in vivo experiments using these TCR to generate retrogenic mice (28, 35, 36) may provide an answer to this question. This approach may have therapeutic implications because the techniques used are adaptable to human studies.
FYAK and other amino acid copolymers of 50–70 aa are synthesized randomly and contain a multiplicity of 15 mers that are presented by Class II MHC proteins to produce the immune response that leads to generation of the inducible Treg. It is clear that more than 1 peptide in FYAK is able to stimulate FYAK-specific cell lines and may account for some of the diversity of CDR3 sequences in the TCR studied. The parallel extent of secretion of IL-2 in the 4 transfectants in each of the 2 recipient cell lines shown in Fig. 5 B and C suggest that these 4 TCR may recognize distinct peptides within these copolymers. Moreover, only 1 of them is stimulated by both FYAK and GA. This is not surprising because these 2 random copolymers, FYAK and GA (YEAK), share 3 amino acids. In addition, the “αβ1” TCR transfectant that recognizes both FYAK and GA must recognize an epitope distinct from the epitope in GA that cross-reacts with the J5 immunomodulatory peptide (26) (that can also induce regulatory T cells) because J5 stimulates neither the TCR transductant nor the cell line from which the “αβ1” TCR was obtained (10). In further experiments, it will be important to use J5 and other immunomodulatory peptides with defined sequences to examine these issues.
Materials and Methods
Mice.
SJL/J female mice (8–10 weeks of age) were purchased from The Jackson Laboratory and maintained according to the Guidelines of the Committee on Animal Care of Harvard University and the Committee on Care and Use of Laboratory Animal Resources, National Research Council (Department of Health and Human Services Publication 85-23, revised 1987).
Copolymers and Peptide.
Copolymer FYAK and peptide PLP139–151 (HSLGKWLGHPDKF) were synthesized as described in ref. 10. FYAK (PI-2301) was a gift from Peptimmune, Inc.. Copaxone was the commercial product of Teva.
Generation of T Cell Lines.
FYAK or PLP139–151-specific T cell lines were generated as described in refs. 10 and 24–26. Briefly, SJL/J mice were immunized with copolymer or peptide emulsified in CFA. Ten days later, splenocytes and lymph nodes cells were isolated and stimulated with corresponding antigens in the presence of antigen-presenting cells (APC, irradiated SJL splenocytes). A total of 3 rounds of restimulation were performed with 2-week intervals of culture in IL-2.
FACS Analysis and Cell Sorting.
T cells were stained using a standard protocol with specific antibodies for mouse CD4, TCRβ, TCR-Vβ4, Vβ14, and Vα3.2 (BD Biosciences). TCR-Vβ analysis was performed by using the mouse Vβ TCR screening panel from BD Biosciences, which contains 15 FITC-conjugated mAbs. Flow cytometry analysis was performed on a FACSCalibur (BD Biosciences), and cell sorting was performed on a MoFlo (DakoCytomation) or FACSAria (BD Biosciences).
Proliferation Assay Through CFSE-Labeling.
T cells (5 × 105) were labeled with CFSE (Molecular Probes) following the manufacturer's instruction. Antigen stimulation was set up right after the labeling process by mixing the T cells with unlabeled APC and antigens. Cultures were maintained at 37 °C for 72 h then analyzed by flow cytometry.
Cytokine Measurement by Intracellular Staining and ELISA.
For intracellular staining, PMA (20 ng/mL), ionomycin (250 ng/mL) and BFA (3 μg/mL) were added during the last 4 h of the restimulation. T cells were then collected and stained for surface markers followed by fixation and permeabilization for intracellular staining. Monoclonal antibody against IL-10 was purchased from (eBioscience) for staining of T cells. Supernatant for ELISA was harvested from hybridoma-T cells stimulated with antigens and APC after 48 h as described in ref. 10. The IL-2 ELISA kit was purchased from eBioscience.
Quantitative PCR Analysis of TCR-Vα Gene Expression.
Total RNA was extracted from specific T cell populations by using the Absolute RNA kit according to the manufacturer's protocol, and reverse transcribed to cDNA by using The AffinityScript QPCR cDNA Synthesis Kit (Stratagene). Quantitative PCR for TCR-Vα analysis was carried out with the Stratagene MX3000p system using Brilliant SYBR Green QPCR Master Mix (Stratagene) and the specific primers described in ref. 37.
Single-Cell RT-PCR.
Single cells from a FYAK-specific T cell line were sorted based on their expression of CD4, TCR-Vα3.2, and TCR-Vβ14 into a 96-well PCR plate. Reverse transcription and PCR were carried out sequentially in the same well by using a OneStep RT-PCR kit (QIAGEN).
Generation of TCR Retroviral Vectors.
TCRα and TCRβ chains were cloned from single cells from FYAK-specific T cell line. TCR encoding 2A peptide-linked constructs were generated by recombinant PCR and confirmed by DNA sequencing, then were inserted into pMIG [a gift from D. Vignali (St. Jude Children's Research Hospital, Memphis, TN)], an MSCV-based retroviral vector containing an IRES-GFP cassette (28, 35).
Retroviral transduction of TCR.
Retrovirus containing TCR genes was produced as described in refs. 28, 35, and 38. Briefly, 4 μg of TCR vector together with 4 μg of pEQ-Pam3(-E) and 2 μg of pVSVg (gifts from D. Vignali) were cotransfected into 293T cells using FuGNEN HD Transfection Reagent (Roche). Supernatant was then collected after 36 h twice daily and used to infect GP+E86 cells (gift from D. Vignali). Transduced GP+E86 cells were sorted for GFP expression and expanded as retroviral producers. Their supernatant was used to infect TCR-deficient BW-1 (39) [gift from J. Kappler and P. Marrack (University of Colorado, Denver, CO)] and 4G4.CD4 (38) [gift from T. L. Geiger (St. Jude Children's Research Hospital)] T hybridoma cells.
Acknowledgments.
We thank Dario Vignali and Kate Vignali for essential advice in addition to reagents. This work was supported by National Institutes of Health Grant AI49524 and National Multiple Sclerosis Society Grant RG 3796A3/1).
Footnotes
Conflict of interest statement: J.L.S. is a member of the Scientific Advisory Board of Peptimmune, Inc., who are developing FYAK for clinical trial.
References
- 1.Mantovani A, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu Rev Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 3.Battaglia M, Gregori S, Bacchetta R, Roncarolo MG. Tr1 cells: From discovery to their clinical application. Semin Immunol. 2006;18:120–127. doi: 10.1016/j.smim.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Hawrylowicz CM, O'Garra A. Potential role of interleukin-10-secreting regulatory T cells in allergy and asthma. Nat Rev Immunol. 2005;5:271–283. doi: 10.1038/nri1589. [DOI] [PubMed] [Google Scholar]
- 5.Izcue A, Coombes JL, Powrie F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol Rev. 2006;212:256–271. doi: 10.1111/j.0105-2896.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- 6.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 7.Kim JM, Rudensky A. The role of the transcription factor Foxp3 in the development of regulatory T cells. Immunol Rev. 2006;212:86–98. doi: 10.1111/j.0105-2896.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- 8.Arnon R, Sela M. Immunomodulation by the copolymer glatiramer acetate. J Mol Recognit. 2003;16:412–421. doi: 10.1002/jmr.628. [DOI] [PubMed] [Google Scholar]
- 9.Farina C, Weber MS, Meinl E, Wekerle H, Hohlfeld R. Glatiramer acetate in multiple sclerosis: Update on potential mechanisms of action. Lancet Neurol. 2005;4:567–575. doi: 10.1016/S1474-4422(05)70167-8. [DOI] [PubMed] [Google Scholar]
- 10.Stern JN, et al. Amino acid copolymer-specific IL-10-secreting regulatory T cells that ameliorate autoimmune diseases in mice. Proc Natl Acad Sci USA. 2008;105:5172–5176. doi: 10.1073/pnas.0712131105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner HL. Induction and mechanism of action of transforming growth factor-beta-secreting Th3 regulatory cells. Immunol Rev. 2001;182:207–214. doi: 10.1034/j.1600-065x.2001.1820117.x. [DOI] [PubMed] [Google Scholar]
- 12.Hussien Y, Sanna A, Soderstrom M, Link H, Huang YM. Glatiramer acetate and IFN-beta act on dendritic cells in multiple sclerosis. J Neuroimmunol. 2001;121:102–110. doi: 10.1016/s0165-5728(01)00432-5. [DOI] [PubMed] [Google Scholar]
- 13.Weber MS, et al. Type II monocytes modulate T cell-mediated central nervous system autoimmune disease. Nat Med. 2007;13:935–943. doi: 10.1038/nm1620. [DOI] [PubMed] [Google Scholar]
- 14.Vandenbark AA, Hashim G, Offner H. Immunization with a synthetic T-cell receptor V-region peptide protects against experimental autoimmune encephalomyelitis. Nature. 1989;341:541–544. doi: 10.1038/341541a0. [DOI] [PubMed] [Google Scholar]
- 15.Howell MD, et al. Vaccination against experimental allergic encephalomyelitis with T cell receptor peptides. Science. 1989;246:668–670. doi: 10.1126/science.2814489. [DOI] [PubMed] [Google Scholar]
- 16.Waisman A, et al. Suppressive vaccination with DNA encoding a variable region gene of the T-cell receptor prevents autoimmune encephalomyelitis and activates Th2 immunity. Nat Med. 1996;2:899–905. doi: 10.1038/nm0896-899. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, et al. Induction of a type 1 regulatory CD4 T cell response following V beta 8.2 DNA vaccination results in immune deviation and protection from experimental autoimmune encephalomyelitis. Int Immunol. 2001;13:835–841. doi: 10.1093/intimm/13.6.835. [DOI] [PubMed] [Google Scholar]
- 18.Madakamutil LT, Maricic I, Sercarz EE, Kumar V. Immunodominance in the TCR repertoire of a [corrected] TCR peptide-specific CD4+ Treg population that controls experimental autoimmune encephalomyelitis. J Immunol. 2008;180:4577–4585. doi: 10.4049/jimmunol.180.7.4577. [DOI] [PubMed] [Google Scholar]
- 19.Acha-Orbea H, et al. Limited heterogeneity of T cell receptors from lymphocytes mediating autoimmune encephalomyelitis allows specific immune intervention. Cell. 1988;54:263–273. doi: 10.1016/0092-8674(88)90558-2. [DOI] [PubMed] [Google Scholar]
- 20.Sakai K, et al. Involvement of distinct murine T-cell receptors in the autoimmune encephalitogenic response to nested epitopes of myelin basic protein. Proc Natl Acad Sci USA. 1988;85:8608–8612. doi: 10.1073/pnas.85.22.8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar V, Sercarz E. Induction or protection from experimental autoimmune encephalomyelitis depends on the cytokine secretion profile of TCR peptide-specific regulatory CD4 T cells. J Immunol. 1998;161:6585–6591. [PubMed] [Google Scholar]
- 22.Vandenbark AA, et al. Therapeutic vaccination with a trivalent T-cell receptor (TCR) peptide vaccine restores deficient FoxP3 expression and TCR recognition in subjects with multiple sclerosis. Immunology. 2008;123:66–78. doi: 10.1111/j.1365-2567.2007.02703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 24.Stern JN, et al. Amelioration of proteolipid protein 139–151-induced encephalomyelitis in SJL mice by modified amino acid copolymers and their mechanisms. Proc Natl Acad Sci USA. 2004;101:11743–11748. doi: 10.1073/pnas.0403832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Illes Z, et al. Modified amino acid copolymers suppress myelin basic protein 85–99-induced encephalomyelitis in humanized mice through different effects on T cells. Proc Natl Acad Sci USA. 2004;101:11749–11754. doi: 10.1073/pnas.0403833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern JN, et al. Peptide 15-mers of defined sequence that substitute for random amino acid copolymers in amelioration of experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2005;102:1620–1625. doi: 10.1073/pnas.0409022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 28.Holst J, Vignali KM, Burton AR, Vignali DA. Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat Methods. 2006;3:191–197. doi: 10.1038/nmeth858. [DOI] [PubMed] [Google Scholar]
- 29.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kretschmer K, et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 31.Allan SE, et al. Generation of potent and stable human CD4+ T regulatory cells by activation-independent expression of FOXP3. Mol Ther. 2008;16:194–202. doi: 10.1038/sj.mt.6300341. [DOI] [PubMed] [Google Scholar]
- 32.Wohlfert E, Belkaid Y. Role of endogenous and induced regulatory T cells during infections. J Clin Immunol. 2008;28:707–715. doi: 10.1007/s10875-008-9248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamanaka M, et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–952. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Perchellet A, Stromnes I, Pang JM, Goverman J. CD8+ T cells maintain tolerance to myelin basic protein by “epitope theft”. Nat Immunol. 2004;5:606–614. doi: 10.1038/ni1073. [DOI] [PubMed] [Google Scholar]
- 35.Holst J, et al. Generation of T-cell receptor retrogenic mice. Nat Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- 36.Arnold PY, Burton AR, Vignali DA. Diabetes incidence is unaltered in glutamate decarboxylase 65-specific TCR retrogenic nonobese diabetic mice: Generation by retroviral-mediated stem cell gene transfer. J Immunol. 2004;173:3103–3111. doi: 10.4049/jimmunol.173.5.3103. [DOI] [PubMed] [Google Scholar]
- 37.Blish CA, et al. Chronic modulation of the TCR repertoire in the lymphoid periphery. J Immunol. 1999;162:3131–3140. [PubMed] [Google Scholar]
- 38.Alli R, Nguyen P, Geiger TL. Retrogenic modeling of experimental allergic encephalomyelitis associates T cell frequency but not TCR functional affinity with pathogenicity. J Immunol. 2008;181:136–145. doi: 10.4049/jimmunol.181.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White J, et al. Two better cell lines for making hybridomas expressing specific T cell receptors. J Immunol. 1989;143:1822–1825. [PubMed] [Google Scholar]