Abstract
HIV-1 Tat enhances viral transcription elongation by forming a ribonucleoprotein complex with transactivating responsive (TAR) RNA and P-TEFb, an elongation factor composed of cyclin T1 (CycT1) and Cdk9 that phosphorylates the C-terminal domain of RNA polymerase II. Previous studies have shown that Lys-28 in the activation domain (AD) of Tat is essential for HIV-1 transcription and replication and is acetylated by p300/CBP-associated factor (PCAF), but the mechanistic basis of the Lys-28 requirement is unknown. Here, we show that Lys-28 acetylation modulates the affinity and stability of HIV-1 Tat–CycT1–TAR complexes by enhancing an interaction with the CycT1 Tat–TAR recognition motif. High-affinity assembly correlates strongly with stimulation of transcription elongation in vitro and Tat activation in vivo. In marked contrast, bovine lentiviral Tat proteins have evolved a high-affinity TAR interaction that does not require PCAF-mediated acetylation of the Tat AD or CycT1 for RNA binding, whereas HIV-2 Tat has evolved an intermediate mechanism that uses a duplicated TAR element and CycT1 to enhance RNA affinity and consequently transcription activation. The coevolution of Tat acetylation, CycT1 dependence, and TAR binding affinity is seen in viral replication assays using Tat proteins that rely on CycT1 for TAR binding but are acetylation deficient, where compensatory mutations rapidly accrue in TAR to generate high-affinity, CycT1-independent complexes reminiscent of the bovine viruses. Thus, lysine acetylation can be used to modulate and evolve the strength of a viral-host RNA–protein complex, thereby tuning the levels of transcription elongation.
Keywords: coevolution, P-TEFb, RNA polymerase II, lentivirus, histone acetyltransferase
The HIV-1 Tat protein activates transcription elongation by recruiting the P-TEFb elongation factor to the transactivating responsive (TAR) stem loop formed at the 5′ end of viral transcripts. Tat activity requires 2 domains (Fig. 1A), an activation domain (AD) and an arginine-rich motif (ARM), which functions as a RNA binding domain (RBD) and nuclear localization signal. Within the ARM, Arg-52 is essential for specific TAR recognition (1, 2), and this single Arg-mediated contact generates a complex of intrinsically low affinity (3). To achieve the high affinity needed for transcription activation, Tat forms a ternary complex with TAR and cyclin T1 (CycT1) (4). Tat uses its AD to interact with CycT1, positioning surfaces on Tat and CycT1, such as the Tat-TAR recognition motif (TRM), for proper RNA recognition (5). RNA binding is coupled to transcription activation through the CycT1-mediated recruitment of Cdk9, the catalytic subunit of P-TEFb (6), which phosphorylates the C-terminal domain (CTD) of RNA polymerase II (RNAP II) to facilitate the initiation to elongation transition (6, 7). Like HIV-1 Tat, HIV-2 and simian immunodeficiency virus (SIV) use CycT1-dependent RNA-binding modes for transcription activation (8), but, in marked contrast, bovine lentiviruses, such as bovine immunodeficiency virus (BIV) and Jembrana disease virus (JDV), have evolved CycT1-independent Tat–TAR binding modes (3), with P-TEFb required only for CTD phosphorylation (9).
Fig. 1.
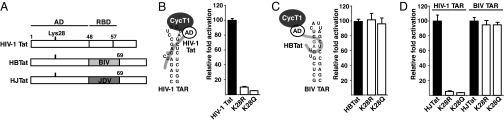
HIV-1 Tat Lys-28 is essential in a CycT1-dependent RNA-binding context. (A) Schematic of HIV-1 Tat and HBTat and HJTat chimeras composed of the HIV-1 Tat AD and BIV or JDV RBDs. (B) Schematic of HIV-1 ternary complexes, where HIV-1 TAR is bound by Tat interacting through its AD with CycT1 and transcription reporter activities are on an HIV-1 TAR reporter in HeLa cells. (C) Schematic of HBTat and the BIV Tat-TAR complex, where CycT1 is not required for TAR binding and transcription reporter activities are on a BIV TAR reporter in HeLa cells. (D) Activities of the Tat proteins indicated on HIV-1 and BIV TAR reporters.
Besides CTD phosphorylation, other posttranslational modifications can be important for transcription activation. For example, acetylation of coactivators can stabilize promoter complexes (10–12), in part by enhancing interactions with the bromo-homology domain (Brd) of histone acetyltransferases (HATs), such as p300/CBP-associated factor (PCAF) and CBP/p300 (13). These Brd/acetyl-Lys interactions help recruit or enzymatically activate HATs to modify their targets, for example by acetylating the N-terminal tails of histones to stimulate chromatin remodeling (13) or modifying specific lysines of transcription factors to modulate DNA-binding affinity (12, 14). HIV-1 Tat recruits PCAF and p300 to the integrated viral promoter (15, 16), where PCAF acetylates Lys-28 in Tat to slightly enhance (≈3-fold) the Tat–CycT1 interaction in vitro (17), whereas p300 acetylates Lys-50 in the RBD to trigger ternary complex disassembly (16, 17).
Here, we analyze the role of Tat acetylation in vivo and in the context of TAR binding by using Tat chimeras containing the HIV-1 Tat AD fused to different ARMs (3). The use of chimeras often uncovers new functions of transcription factors (3, 18, 19), and indeed, experiments with the Tat chimeras demonstrate another role for Tat acetylation in which the affinity of viral-host RNA–protein complexes is increased above a threshold needed for transcription activation. Strikingly, the related lentiviruses HIV-2, SIV, and BIV have evolved different mechanisms to achieve high-affinity RNA binding, such as duplication of TAR or generation of CycT1-independent TAR-binding modes, showing that acetylation is needed only when Tat–TAR complexes require CycT1. The results demonstrate how RNA-binding affinity can be modulated by a posttranslational modification that affects a key viral-host protein interaction and consequently the viral transcriptional program.
Results
HIV-1 Tat Lys-28 Is Required in the Context of CycT1-Dependent RNA Binding.
HIV-1 Tat can be acetylated at Lys residues in both its AD and RBD (16). We wanted to determine how these modifications affect transcription activation and particularly whether they might alter the assembly of RNA-binding complexes. To scan for candidate acetylated lysines in 2 different RNA-binding contexts, we measured the activities of Lys mutants in HIV-1 Tat and in a chimera containing the HIV-1 Tat AD fused to the RBD of BIV Tat (HBTat) (Fig. 1A) on transcriptional reporters containing HIV-1 or BIV TAR binding sites (Fig. 1 B and C, Fig. S1, and SI Text). Whereas HIV-1 Tat binds HIV-1 TAR with weak affinity and needs CycT1 for efficient binding, HBTat binds with high affinity to BIV TAR without the need to assemble a ternary complex with CycT1 (3, 9), allowing us to assess the importance of lysine residues in CycT1-dependent and -independent binding contexts. Using the same AD in the chimeric context allows us to directly assign any differences to RNA-binding modes rather than possible effects from sequence differences between HIV-1 and BIV Tat ADs (see Fig. 6A).
Fig. 6.
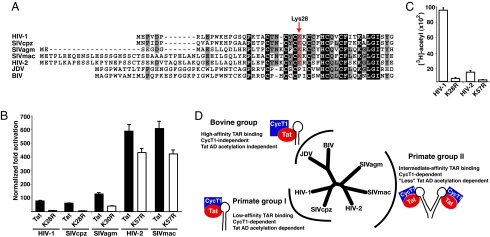
Comparison of lentiviruses based on differences in Tat–TAR affinities and CycT1 and Tat acetylation dependence. (A) ClustalW alignment of lentiviral Tat ADs. Lys-28 in HIV-1 Tat and the corresponding residues in the other Tats are indicated (red, and arrow). (B) HeLa cells were cotransfected with the indicated Tat or mutant, and transcription activation was assayed by using cognate promoter luciferase reporters. (C) HIV-2 Tat or K57R mutant were incubated with PCAF and [3H]-acetyl-CoA, and [3H]-acetyl incorporation was quantified by scintillation counting. (D) Unrooted phylogeny dendogram showing the genetic relationships among the 5 primate Tat proteins shown in A and the 2 bovine Tat proteins. The 3 phylogenetic branches of the dendogram correlate with evolutionary subgroups having similar requirements for transcription activation. Note that whereas BIV contains 2 TAR hairpins, full Tat activation is observed with just one (3), unlike HIV-2 or SIVmac (2).
Every Lys residue in the 2 contexts was mutated individually to Arg (maintaining the charge) or Gln (neutralizing the charge), which both eliminate possible acetylation. After cotransfection of each HIV-1 Tat mutant with the corresponding HIV-1 TAR reporter, we found that mutation of Lys-28 or Lys-41 in the Tat AD severely reduced activation (Fig. 1B and Fig. S1), as previously seen (4, 15, 17). In marked contrast, Lys-41, but not Lys-28, is important in the context of the BIV Tat–TAR interaction (Fig. 1C and Fig. S1), suggesting that Lys-28 is needed only for CycT1-dependent TAR binding. Interestingly, the position equivalent to Lys-28 in the BIV AD is Pro-44 (see Fig. 6A), consistent with its nonessential role in activation through BIV TAR.
To confirm that Lys-28 is needed only for CycT1-dependent TAR binding, we used a chimera between the HIV-1 Tat AD and the JDV Tat RBD, referred to as HJTat (Fig. 1A). The JDV RBD shares sequence features of both HIV-1 and BIV Tat RBDs and thus is able to recognize the 2 TAR sites, adopting different conformations in the 2 contexts (3, 20). In addition to the conformational differences, HJTat requires CycT1 for transcription activation through HIV-1 TAR, like HIV-1 Tat (Fig. 1B), but not through BIV TAR (Fig. 1C) (3). Moreover, the affinities and amino acid requirements of HJTat for the 2 TAR elements are very similar to the cognate HIV-1 and BIV Tat–TAR interactions (3) and thus the chimera is a good model to study transcription activation by the HIV-1 AD in CycT1-dependent and -independent RNA-binding contexts, simply by changing the RNA reporter. We infer that activation correlates with RNA-binding affinity, consistent with previous studies (3). Strikingly, K28R and K28Q mutants in HJTat sharply reduced activation through HIV-1 TAR but had no effect through BIV TAR (Fig. 1D), identical to the results observed with HIV-1 Tat and HBTat (Fig. 1 B and C). The interesting divergence in the evolution of these 2 types of viral complexes prompted us to further examine the acetylation properties of Lys-28 in HJTat and possible effects on RNA–protein complex formation.
PCAF Acetylates Tat Lys-28 to Enhance Formation of Ternary Complexes.
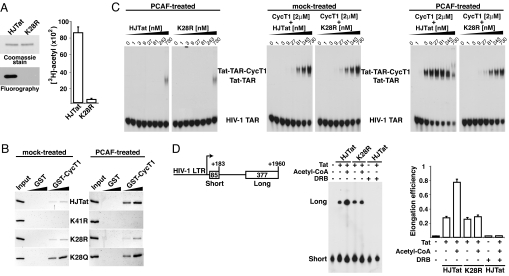
As shown for HIV-1 Tat (17), HJTat, which possesses the HIV-1 Tat AD (Fig. 1A), is efficiently acetylated by PCAF, whereas the K28R mutant is not (Fig. 2A), suggesting that Lys-28 is indeed a target for acetylation. To test the hypothesis that acetylation increases Tat affinity for CycT1 (17), we performed pull-down assays with GST-CycT1 and HJTat, the K28R mutant, or the transcriptionally inactive K41R mutant as a negative binding control (4). HJTat and the K28R mutant, but not K41R, interact with CycT1 (Fig. 2B). Furthermore, the interaction with wild-type HJTat, but not K28R, is stimulated ≈5-fold when preincubated with the catalytically-active PCAF HAT domain (Fig. 2B). To further confirm that Lys-28 acetylation enhances the CycT1 interaction, we created a K28Q mutant expected to be a constitutive mimic of acetylation because Gln, like N-acetyl-Lys, is a neutral amino acid with an amide group that can function as a hydrogen donor or acceptor (14, 21). Indeed, we observed an ≈5-fold enhanced interaction between CycT1 and the K28Q mutant, like for PCAF-treated HJTat, and no additional enhancement by PCAF (Fig. 2B), implying that Lys-28 acetylation enhances the Tat–CycT1 interaction.
Fig. 2.
Acetylation of Tat Lys-28 by PCAF enhances assembly of Tat–TAR–CycT1 complexes. (A) (Left) Recombinant HJTat and K28R proteins were incubated with PCAF and [3H]-acetyl-CoA, resolved by SDS/PAGE, and stained with Coomassie blue, and acetylated proteins were detected by [3H] fluorography. (Right) An identical acetylation reaction was performed, and [3H]-acetyl incorporation was quantified by scintillation counting. (B) GST-pull down assay with 1 or 5 μg of GST or GST-CycT1 bound to beads and HJTat or mutants, mock- or PCAF-treated before the pull-down. The input (20%) and bound proteins were separated by 15% SDS/PAGE and stained with Coomassie blue. (C) RNA-binding gel-shift assays with labeled HIV-1 TAR, without or with 2 μM His-tagged CycT1, and increasing amounts of HJTat or K28R mutant. Ternary complex formation was assessed with nonacetylated HJTat or K28R (mock-treated) or PCAF-treated proteins. (D) (Left and Center) Transcription reactions were performed with a template containing a double G-less cassette and HeLa nuclear extracts in the absence or presence of HJTat or K28R, with or without acetyl-CoA. Treatment with DRB was added as control to inhibit transcription elongation. (Right) Plot of calculated elongation efficiencies.
Because acetylation of Lys-28 increases the Tat–CycT1 interaction, and because Lys-28 is required for HJTat-mediated activation through HIV-1 TAR but not BIV TAR, we reasoned that the modification assists in forming ternary complexes with HIV-1 TAR, rather than modulating the Tat–TAR (Fig. 2C) or CycT1–TAR interaction directly. Indeed, gel-shift assays show that the amount of ternary complex formed with acetylated HJTat is ≈110-fold more than with unacetylated HJTat or the K28R mutant (Fig. 2C), with similar results observed with HIV-1 Tat instead of HJTat (see Fig. S2). Interestingly, the constitutive K28Q HJTat mutant also shows an ≈65-fold increase in ternary complex formation, which correlates with the increased Tat-CycT1 protein–protein interaction (Fig. 2B). Although the K28Q mutant enhances the assembly of the complex in vitro, it does not activate transcription through HIV-1 TAR (Fig. 1D), suggesting that a deacetylation step may be important for cycles of Tat activity. Although other possibilities could explain this mutant phenotype, mutation of Lys-28 does not affect subcellular localization (see Fig. S3) or steady-state levels (Fig. S1).
To directly test whether the acetylation-induced ternary complex assembly correlates with an increase in transcription efficiency, we performed in vitro transcription elongation assays with an HIV-1 LTR double G-less cassette template (Fig. 2D). This template synthesizes transcripts that contain 2 RNaseT1-resistant regions (G-less cassettes) of different sizes (22). One is located proximal to the promoter and produces a 183-nt (short) fragment upon RNase T1 digestion that approximates the extent of transcription initiation, and the second produces a 1,960-nt (long) fragment approximating the extent of elongation. Only short transcripts are generated without Tat, whereas long transcripts are observed upon HJTat addition (Fig. 2D). Elongation efficiency increases substantially (0.85 vs. 0.22) with acetylated HJTat, generated by adding acetyl-CoA to the extracts, and requires Lys-28 (Fig. 2D). To corroborate that enhancement is specific to elongation, we preincubated reactions with 5,6-dichloro-1-β (DRB) like in reporter assays, suggesting that a step of Lys-28 deacetylation may be part of the transcriptional cycle.
PCAF-Mediated Tat Lys-28 Acetylation Is Required in Vivo.
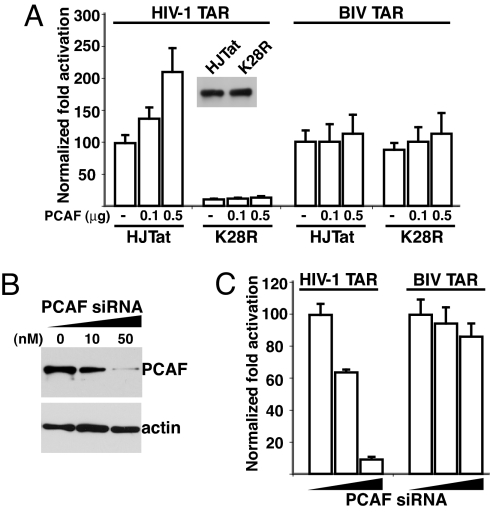
To examine Lys-28 acetylation in vivo and the role of PCAF, we overexpressed PCAF in Tat activation assays by cotransfecting PCAF with HJTat or the K28R mutant along with HIV-1 or BIV TAR reporters (Fig. 3). PCAF stimulated HJTat-mediated activation of an HIV-1 TAR reporter by ≈2.5-fold, but not when using the equally well-expressed K28R mutant (Fig. 3A). PCAF-mediated stimulation was not observed on a BIV TAR reporter (Fig. 3A) or when using a catalytically inactive PCAF HAT domain. Similar levels of PCAF-mediated stimulation were observed with HIV-1 Tat, but not the K28R mutant, on an HIV-1 TAR reporter (Fig. S4), further confirming that stimulation is observed only in the HIV-1 Tat–TAR context. RNAi knockdown experiments support the role of PCAF in Lys-28 acetylation and enhancement of ternary complex formation. Transfecting HeLa cells with a PCAF siRNA (Fig. 3B), but not a scrambled siRNA, efficiently reduced PCAF expression (Fig. 3B) and strongly decreased HJTat activation from an HIV-1 TAR but not a BIV TAR reporter (Fig. 3C). Our results support a model in which Lys-28 acetylation stimulates Tat activation by enhancing the affinity of Tat–CycT1–HIV-1 TAR ternary complexes.
Fig. 3.
PCAF-mediated Tat Lys-28 acetylation is required in vivo when TAR binding is CycT1-dependent. (A) Transcription activation on HIV-1 or BIV TAR reporters and stimulation by increasing amounts of PCAF in HeLa cells. A Western blot with the Flag antibody shows expression levels of HJTat and K28R. (B) Western blot of PCAF expression after RNAi knockdown and β-actin control. (C) Transcription activation of HJTat on HIV-1 or BIV TAR reporters in HeLa cells pretreated as in B with increasing amounts of PCAF siRNA.
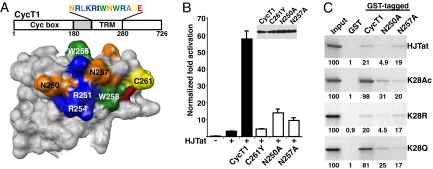
A Binding Site for Lys-28 Acetylated Tat in the Tat–TAR Recognition Motif of CycT1.
The importance of the acetylated Tat–CycT1 interaction prompted us to search for regions of CycT1 potentially involved in recognition. HATs use a conserved Asn in their Brd to hydrogen bond to the acetyl-Lys side-chain oxygen (21, 24). Whereas CycT1 does not possess a Brd fold, we observed 2 Asn residues (at positions 250 and 257) in the TRM (Fig. 4A), where Asn-250, but not Asn-257, is important for unacetylated Tat binding (4). To test the requirement of Asn-257 in HJTat-mediated transcription activation, we performed reporter assays with CycT1 and mutants in murine NIH 3T3 cells, which encode a CycT1 nonfunctional for Tat activation caused by a C261Y mutation, but can be complemented by expressing human CycT1 (4, 25). In addition to the known requirements of Cys-261 and Asn-250 (4), Asn-257 is important for activity, with all CycT1 variants expressed to similar degrees (Fig. 4B). Because Asn-257 has not been implicated in Tat or TAR binding (4), we asked whether it might be important for binding acetylated HJTat (K28Ac) by using pull-down experiments with GST-tagged CycT1 and point mutants (Fig. 4C). As observed (4), N250A reduced binding ≈3- to 5-fold whether or not Lys-28 was acetylated, whereas, strikingly, N257A reduced binding only of K28Ac by ≈5- to 6-fold (Fig. 4C). Furthermore, binding of K28Ac to CycT1 increased ≈5-fold compared with unacetylated Tat, and this increase was abolished by the N257A mutation. K28R behaves similarly to unacetylated Tat, whereas the K28Q behaves like K28Ac (Fig. 4C). These data provide evidence that acetylation of Lys-28 in Tat may stabilize the Tat–CycT1 complex in part by interacting with Asn-257 in the CycT1 TRM, and that this interaction is critical in the context of ternary complex formation.
Fig. 4.
A possible binding site for Tat acetylated Lys-28 in the CycT1 TRM. (A) Schematic of CycT1 with its N-terminal cyclin box and TRM sequence indicated and surface representation of CycT1 (residues 140–280) displayed by using UCSF chimera (37), with the TRM highlighted. (B) NIH 3T3 cells were cotransfected with the HIV-1 TAR reporter alone (−), in the presence of HJTat (+), or with wild-type human CycT1 or point mutants, and transcription activation levels were determined. (Inset) Western blot with an anti-Flag antibody shows transfected CycT1 expression levels. (C) GST-pull down assays using GST or GST-fusions to wild-type CycT1, or mutants bound to beads, and either HJTat, PCAF-acetylated HJTat (K28Ac), or mutants. The input (20% of total) and bound proteins were separated by 15% SDS/PAGE, stained with Coomassie blue, and quantified.
Evolution of CycT1-Independent Tat–TAR Interactions in the Absence of Lys-28.
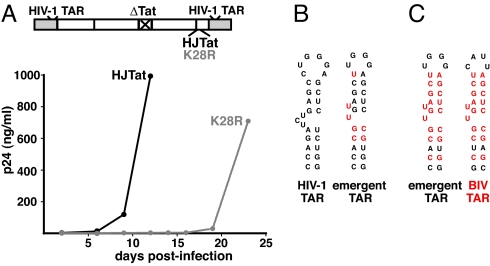
If acetylation of Lys-28 is needed to increase the affinity of Tat–CycT1–TAR ternary complexes and consequently for HIV-1 replication (15), we reasoned that viruses containing a Tat K28R mutation must evolve alternative ways to assemble high-affinity Tat–TAR complexes. To test this hypothesis, we constructed an HIV-1 proviral clone in which Tat was deleted and HJTat containing the K28R mutation was inserted (Fig. 5A). Similar viruses constructed with HJTat replicate as well as HIV-1, and are useful for our experiments because they allow the possibility to readily evolve BIV-like binding modes (26). As expected, the K28R mutant virus showed delayed replication kinetics but ultimately reached high levels of p24 expression 22 days postinfection (Fig. 5A). We cloned and sequenced integrated proviral DNA fragments corresponding to the Tat coding region and HIV-1 promoter from the emergent viruses and, of 10 clones analyzed, the only changes observed were in TAR (Fig. 5B). The RNA structure generated resembles BIV TAR (Fig. 5C), with all of the characteristics needed to bind HJTat in a high-affinity binding mode (3, 26). Interestingly, TAR sequences derived from viruses collected 8–15 days postinfection showed intermediate stepwise changes that suggest pathways in which the RNA structure may be “naturally selected” for binding (Fig. S5). As expected, an HJTat K28R virus preengineered with BIV TAR replicated efficiently, without the need to accrue mutations in TAR for transcription activation (Fig. S6).
Fig. 5.
Forced viral evolution with a Tat K28R mutant. (A) SupT1 cells were infected with the 2 hybrid viruses indicated and p24 levels were monitored. (B) Sequence comparison of wild-type HIV-1 and emergent TARs (changes in red) obtained from K28R emergent viruses 20 days postinfection. (C) Sequence comparison of the BIV and emergent TARs, with shared residues in red.
Lentiviruses Evolved Different Dependencies on Tat Acetylation.
Given the different requirements for Lys-28 acetylation between HIV-1 and BIV, we wanted to assess the Tat binding strategies used by other lentiviruses. We aligned the Tat ADs of consensus HIV-1, simian chimpanzee (SIVcpz), African green monkey (SIVagm), macaque (SIVmac), and HIV-2, and in contrast to the bovine viruses, all primate viruses strictly conserved the Lys-28 equivalent (Fig. 6A). We mutated the appropriate Lys to Arg in each Tat and measured transcription activation on its cognate promoter. For both HIV-1 and SIVcpz, K28R disrupts activation by ≈90% irrespective of the isolate (Fig. 6B and Fig. S7). In SIVagm, K30R shows a somewhat more moderate ≈75% decrease in activity, whereas, unexpectedly, K57R in SIVmac or HIV-2 Tat shows only an ≈20% decrease (Fig. 6B). Interestingly, these latter viruses use a duplicated TAR element (Fig. 6D) that appears to increase RNA-binding affinity (8) and consequently may depend less on acetylation to assemble a high-affinity ternary complex. Given that the HIV-2 Tat K57R mutation only reduced activation slightly we tested whether the HIV-2 Tat AD is a PCAF substrate. We found that HIV-2 Tat is acetylated by PCAF in vitro but only to ≈15%, the level observed with HIV-1 Tat, whereas the K57R control is not acetylated (Fig. 6C). A comparative analysis of lentiviral Tat sequences (Fig. 6A) shows 3 different evolutionary groups (Fig. 6D) where, interestingly, members of each group have similar requirements for Tat activation. Thus, it appears that Tat activity can be tuned by altering the balance of Tat–TAR affinity, and CycT1 and Tat AD acetylation dependence for ternary complex formation (Fig. 6D).
Discussion
We have shown that acetylation of Lys-28 in HIV-1 Tat by PCAF strengthens the assembly of Tat–TAR–CycT1 ternary complexes and thereby enhances HIV-1 transcription and replication. This acetylation requirement can be bypassed by recruiting Tat to the promoter with an inherently high-affinity protein–RNA interaction, like BIV Tat–TAR. A comparison of transcriptional programs in related lentiviruses, like HIV-1 and BIV, indicates that tight Tat complexes are assembled by a balance of protein–RNA and protein–protein interactions where each relies on individual interactions to different extents, much like bacteriophage N protein–NusA antitermination complexes (27). Interestingly, the requirement for HIV-1 Tat Lys-28 acetylation in ternary complex assembly may allow for additional steps of transcription regulation, perhaps involving cycles of acetylation and deacetylation. Indeed, the SIRT1 deacetylase is part of a positive feedback HIV-1 transcriptional circuit that operates via the Tat RBD (28, 29). Our data suggest that such fine-tuning control mechanisms may arise during the evolution of lentiviral transcriptional programs. Other posttranslational modifications (13), yet to be discovered, may further modulate the affinity of HIV-1 or other lentiviral Tat–TAR complexes.
Lys acetylation often occurs at protein–protein interfaces and generates a neutral and hydrophobic side chain that retains intrinsic hydrogen-bonding capacity (21). In HIV-1 Tat, acetylation of Lys-28 slightly enhances the interaction with CycT1 and may remove an unfavorable charge at the CycT1 interface and/or change its conformation to simultaneously recognize CycT1 and TAR more efficiently. Asn-257 in the TRM of CycT1 is essential for recognition of acetylated Tat and Tat-dependent activation and may directly contact N-acetyl-Lys-28. The TRM of CycT1 is rather flexible in solution (25), but also adopts a helical structure and may become ordered or change its conformation upon interaction (30). An Asn residue in Brd-containing proteins recognizes N-acetyl-Lys through an amide nitrogen-acetyl carbonyl group interaction (21), and even though CycT1 does not contain a Brd fold, it may use a related recognition principle.
Interestingly, Tat provides an example in which acetylation modulates assembly of an RNA–protein complex through increased protein–protein association. Although the effect of acetylation on RNA-binding affinity is indirect, it can be compared with cases where acetylation modulates DNA-binding affinity. One well-studied example is the transcription factor p53, where its low-affinity interaction with some promoters can be enhanced by site-specific Lys acetylation (13, 14). In other cases, acetylation of transcription factors also has been shown to trigger transcription activation by recruiting/displacing factors (10, 31), stabilizing complexes assembled at promoters (32), or providing a catalytic switch to activate transcription (12).
Our data show that Tat acetylation is coupled to RNA binding and has coevolved with its dependence on the CycT1 host protein interaction. It is especially interesting that the 3 branches of the lentiviral evolutionary tree show a segregation of their RNA-binding modes according to the intrinsic affinities of their Tat–TAR interactions, their use of duplicated TAR elements to enhance affinity (8, 33), their use of Tat AD acetylation to enhance affinity, and their CycT1 dependence for RNA binding (Fig. 6D). Primate group I, including HIV-1, has a low Tat–TAR affinity that is enhanced by CycT1 and strongly depends on Tat acetylation; primate group II, including HIV-2, has an intermediate Tat–TAR affinity with a duplicated TAR that is still CycT1-dependent but modestly enhanced by Tat acetylation; and the bovine group has a high Tat–TAR affinity that does not require CycT1 or Tat acetylation. Our virus evolution experiments are consistent with this coevolution model, showing that HIV-1 containing a Tat K28R mutant deficient for PCAF-mediated acetylation sequentially accrues changes in TAR to evolve a CycT1-independent Tat–TAR interaction reminiscent of the bovine group (3, 26). Such an evolutionary mechanism underscores the intimate dependence of HIV-1 transcription on the host machinery, extending to posttranslational modifications.
Materials and Methods
Cell Culture, Transcription Reporter Assays, and RNAi.
HeLa, 293T, and NIH 3T3 cells were cultured in DMEM with 10% FBS and penicillin-streptomycin. SupT1 CD4+ cells were cultured in RPMI medium 1640 supplemented with 10% FBS. Cells were transfected as reported (34). Firefly luciferase reporter activities were normalized to a constitutive CMV Renilla luciferase expressor. For RNAi knockdown, HeLa cells were plated to a density of 5 × 105 cells per well in 6-well plates and analyzed by Western blot 72 h after transfecting PCAF siRNA (sc-36198) or scrambled siRNA (sc-37007) (Santa Cruz).
Plasmids and Mutagenesis.
Tat proteins were expressed in pSV2 vectors and reporters were as described (34). CMV-PCAF and CMV-PCAFΔHAT were kindly provided by K.-T. Jeang (National Institutes of Health, Bethesda). CycT1 and mutants were cloned into pcDNA4/TO with a C-terminal Flag tag. Mutagenesis was carried out by using Pfu Ultra (Stratagene). See SI Text for lentiviral Tat accession numbers.
Recombinant Protein Expression and RNA Binding Assays.
An N-terminal fragment of human CycT1 (residues 1–280) was cloned into pET21d, expressed as a C-terminal His-tagged fusion at 30 °C, and purified by using a Ni-NTA column. GST-CycT1 was cloned into pGEX2T, expression was induced at 30 °C, and protein was purified on glutathione agarose and digested with thrombin as needed. Tat, mutants, and chimeras were cloned into pGEX2T and expressed at 30 °C. GST-PCAF HAT domain (residues 493–658) was induced at 20 °C overnight. Gel-shift assays were performed as described (3).
Nuclear Extract Preparation and in Vitro Transcription.
Nuclear extracts were prepared as described (35). Transcription reactions were performed by using a template with 2 G-less cassettes (22) and recombinant Tat proteins. Elongation efficiency was calculated as the molar ratio of long to short transcripts. Radioactivity incorporated into each product was quantified by densitometry and normalized for uridine content.
HAT Assays.
The PCAF HAT domain was used for in vitro acetylation of GST-tagged Tat, HJTat, and mutant substrates. Reactions were performed in 20 mM Hepes (pH 7), 1 mM DTT, 2 mM sodium butyrate, 5% glycerol, and 0.5 μL of [3H]-acetyl-CoA (65 mCi/mmoL; ICN) for 1 h at 30 °C and either run on SDS/PAGE gels and fluorographed with NAMP100 or spotted on P81 filters and quantified by scintillation counting.
Viral Replication Assays.
SupT1 T cells were infected with viral stocks R7/HTAR-HJTat, R7/BTAR-HJTat, or K28R mutants as described (26). Viral replication was analyzed by p24 ELISA, and emergent viruses were sequenced (34).
Sequence Alignment and Phylogenetic Analyses.
Alignments were done with ClustalW (36), and phylogenetic trees were built by using the neighbor-joining method. Support for the trees was assessed by using 1,000 nonparametric bootstrap replicates.
Supplementary Material
Acknowledgments.
We thank R. Andino, J. Gross, T. Aragón, M. Daugherty, and V. Calabro for comments; H. Luecke (National Institutes of Health, Bethesda, MD), B. Hahn (University of Alabama, Birmingham, AL), K.-T. Jeang, B. Xie (University of California, San Francisco), C. Sune (Instituto de Parasitología y Biomedicina Lopéz Neyra, Granada, Spain), and M. Garcia-Blanco (Duke University Medical Center, Durham, NC) for reagents; and J. Cox for use of the BSL3 laboratory. This work was supported by a postdoctoral fellowship from the Human Frontier Science Program and American Foundation for AIDS Research Mathilde Krim Fellowship in Basic Biomedical Research 106988-43-RFNT (to I.D.) and National Institutes of Health Grants AI29135 and P50 GM082250 (HIV Accessory and Regulatory Complexes Center) (to A.D.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900012106/DCSupplemental.
References
- 1.Puglisi JD, Chen L, Frankel AD, Williamson JR. Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tao J, Frankel AD. Electrostatic interactions modulate the RNA binding and transactivation specificities of the HIV and SIV Tat proteins. Proc Natl Acad Sci USA. 1993;90:1571–1575. doi: 10.1073/pnas.90.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith CA, Calabro V, Frankel AD. An RNA-binding chameleon. Mol Cell. 2000;6:1067–1076. doi: 10.1016/s1097-2765(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 4.Garber ME, et al. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter S, Ping YH, Rana TM. TAR RNA loop: A scaffold for the assembly of a regulatory switch in HIV replication. Proc Natl Acad Sci USA. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Bres V, Yoh SM, Jones KA. The multitasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Analysis of the effect of natural sequence variation in Tat and in cyclin T on the formation and RNA binding properties of Tat–cyclin T complexes. J Virol. 1999;73:5777–5786. doi: 10.1128/jvi.73.7.5777-5786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barboric M, Taube R, Nekrep N, Fujinaga K, Peterlin BM. Binding of Tat to TAR and recruitment of P-TEFb occur independently in BIV. J Virol. 2000;74:6039–6044. doi: 10.1128/jvi.74.13.6039-6044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 11.Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 12.Li AG, et al. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28:408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Yang XJ, Seto E. Lysine acetylation: Codified cross-talk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J, et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bres V, Kiernan R, Emiliani S, Benkirane M. Tat acetyl-acceptor lysines are important for HIV-1 replication. J Biol Chem. 2002;277:22215–22221. doi: 10.1074/jbc.M201895200. [DOI] [PubMed] [Google Scholar]
- 16.Kaehlcke K, et al. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell. 2003;12:167–176. doi: 10.1016/s1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- 17.Kiernan RE, et al. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 19.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: The functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calabro V, Daugherty MD, Frankel AD. A single intermolecular contact mediates intramolecular stabilization of both RNA and protein. Proc Natl Acad Sci USA. 2005;102:6849–6854. doi: 10.1073/pnas.0409282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taverna SD, Li H, Ruthenburg A, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sune C, Goldstrohm AC, Peng J, Price DH, Garcia-Blanco MA. An in vitro transcription system that recapitulates EIAV Tat-mediated inhibition of HIV-1 Tat activity demonstrates a role for P-TEFb and associated proteins in the mechanism of Tat activation. Virology. 2000;274:356–366. doi: 10.1006/viro.2000.0480. [DOI] [PubMed] [Google Scholar]
- 23.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 24.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 25.Das C, Edgcomb SP, Peteranderl R, Chen L, Frankel AD. Evidence for conformational flexibility in the Tat-TAR recognition motif of cyclin T1. Virology. 2004;318:306–317. doi: 10.1016/j.virol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Xie B, Wainberg MA, Frankel AD. Replication of HIV engineered with heterologous Tat-transactivation response element interactions. J Virol. 2003;77:1984–1991. doi: 10.1128/JVI.77.3.1984-1991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 28.Pagans S, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:210–220. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 30.Baumli S, et al. The structure of P-TEFb, its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Munshi N, et al. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 33.Berkhout B. Structural features in TAR RNA of HIV and SIV: A phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Orso I, Grunwell JR, Nakamura RL, Das C, Frankel AD. Targeting Tat inhibitors in the assembly of HIV-1 transcription complexes. J Virol. 2008;82:9492–9504. doi: 10.1128/JVI.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera, a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.