Abstract
The tumor suppressor p53 negatively regulates a number of genes, including the proto-oncogene c-Myc, in addition to activating many other genes. One mechanism of the p53-mediated c-Myc repression may involve transcriptional regulation. However, it is not clear whether microRNAs (miRNAs) play a role in the p53-mediated posttranscriptional regulation of c-Myc. In this study, we show that a putative tumor suppressor, miR-145, is expressed through the phosphoinositide-3 kinase (PI-3K)/Akt and p53 pathways. Importantly, p53 transcriptionally induces the expression of miR-145 by interacting with a potential p53 response element (p53RE) in the miR-145 promoter. We further show that c-Myc is a direct target for miR-145. Although miR-145 silences the expression of c-Myc, anti-miR-145 enhances its expression. This specific silencing of c-Myc by miR-145 accounts at least in part for the miR-145-mediated inhibition of tumor cell growth both in vitro and in vivo. Finally, the blockade of miR-145 by anti-miR-145 is able to reverse the p53-mediated c-Myc repression. Together, these results define the role of miR-145 in the posttranscriptional regulation of c-Myc by p53 and suggest that, as a new member of the p53 regulatory network, miR-145 provides a direct link between p53 and c-Myc in this gene regulatory network.
Keywords: miRNA, posttranscriptional regulation
The tumor suppressor p53 is a master gene regulator controlling diverse cellular pathways, by either activating or repressing downstream genes. Among such genes is the proto-oncogene c-Myc, which is negatively regulated by p53 (1). As a transcription factor, c-Myc regulates numerous genes directly or indirectly and thus plays an important role in cellular processes such as development, differentiation, cell proliferation and apoptosis. Deregulated expression of c-Myc has been detected in a wide variety of human cancers, including breast and colon, and is often associated with aggressive, poorly differentiated tumors (2). In breast cancer, c-Myc overexpression could account for 30–50% of patients (3). Therefore, there is a great interest in regulation of c-Myc. It have been reported that p53-mediated repression of c-Myc may involve transcriptional repression (4, 5). However, it is not clear whether other mechanisms may also account for this negative correlation between p53 and c-Myc. Specifically, no information is available as to whether microRNAs (miRNAs) play a role in the p53-mediated c-Myc regulation system.
miRNAs are small endogenous noncoding RNAs that have been shown to be crucial posttranscriptional regulators of gene expression (6, 7). Unlike short interfering RNAs (siRNAs), miRNAs typically silence multiple genes instead of a single gene because miRNAs target mRNAs at the 3′-untranslated region (3′-UTR) by partial sequence homology, leading to mRNA degradation or translation repression. It is predicted that an average miRNA can have >100 targets (8). Evidence suggests that miRNAs are often deregulated in human malignancies and can function as either oncogenes or tumor suppressors. In this regard, miR-145 belongs to a member of putative tumor suppressing miRNAs, because it is frequently underexpressed in many types of tumors (9, 10).
The goal of this study was to determine whether p53 negatively regulates c-Myc through the induction of miR-145, because we have evidence that miR-145 directly silences c-Myc expression. We show that miR-145 is expressed in response to stress through the phosphoinositide-3 kinase (PI-3K)/Akt and p53 pathways. Specific suppression of PI-3K/Akt causes up-regulation of miR-145 in a p53-dependent manner. We further show that p53 induces miR-145 expression by interacting with the miR-145 promoter, and that miR-145 directly targets the oncogene c-Myc. As a new member of the p53 regulatory network, miR-145 may play a critical role in the posttranscriptional regulation of c-Myc by p53. Therefore, our study provides a previously undescribed mechanism of p53-mediated repression of c-Myc.
Results
Expression of miR-145 Through the Akt and p53 Pathways.
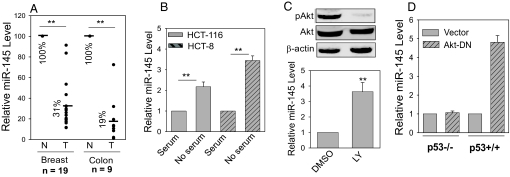
It has been reported that miRNAs are frequently deregulated in malignant tissues (11). Among them is miR-145, which is underexpressed in several types of tumors (9, 10, 12). Consistent with these reports, we found that miR-145 was underexpressed in breast and colon cancer specimens, compared with matched normal tissue samples (Fig. 1A), suggesting that miR-145 is a putative tumor suppressor. In particular, down-regulation of miR-145 was more prominent in colon cancer than in breast cancer. These findings prompted us to investigate the regulation of miR-145 in both colon cancer and breast cancer cells. Because activation or overexpression of Akt is often observed in colon cancer (13, 14), we examined levels of the phosphorylated Akt (pAkt) in colon cancer specimens by immunohistochemistry and confirmed that most of the tumor specimens expressed higher levels of pAkt than normal tissue specimens (Fig. S1A, and Table S1). Because Akt is a key regulatory factor in the mitogen-mediated signaling pathway, we then tested the effect of serum starvation on miR-145. We showed that serum starvation caused a significant induction of miR-145 in both colon cancer HCT-116 and HCT-8 cells (Fig. 1B). In agreement with this result, the PI3-K inhibitor LY294002 strongly suppressed pAkt and at the same time, it significantly enhanced miR-145 expression (Fig. 1C), highlighting the importance of the Akt pathway in miR-145 expression. A number of regulatory genes are controlled by the Akt pathway; 2 important downstream transcription factors involved in tumor suppression are forkhead (Foxo) and p53 (15). Because transient transfection experiments indicated that Foxo has no significant effect on miR-145, we then tested whether p53 is involved in miR-145 expression. It is well known that, whereas the active Akt phosphorylates MDM2, which in turn stimulates p53 degradation, inactivation (dephosphorylation) of Akt increases the stability of p53 (16). Consistent with this, we confirmed that both serum starvation and the PI3-K inhibitor LY294002 induced p53 in HCT-116 cells (Fig. S1B). A known p53 inducer, doxorubicin (Doxo), also induced miR-145 expression (Fig. S1C). Therefore, we determined whether Akt-mediated miR-145 expression is through activated p53 using a dominant negative Akt (Akt-DN) (17). As shown in Fig. 1D, Akt-DN induced miR-145 in a p53-dependent manner. In p53+/+ HCT-116, the level of miR-145 in the Akt-DN-transfected cells was >4-fold higher than in vector control (Fig. 1D). However, in p53−/− HCT-116 cells, Akt-DN revealed no effect on miR-145 expression, demonstrating the important role of p53 in Akt-mediated miR-145 expression. Therefore, it would be of interest to further define the role of p53 in the induction of miR-145.
Fig. 1.
Expression of miR-145 through the Akt and p53 pathways. (A) Expression of miR-145 in the matched breast and colon tumor specimens, respectively. In breast tumor tissue, the average level of miR-145 was 31% of that in the matched normal tissue; in colon tumor tissue, the corresponding number was 19%. (B) Induction of miR-145 by serum starvation. Cells (HCT-116 and HCT-8) were grown in the medium containing 10% or no FBS for 24 h. (C) Induction of miR-145 by PI-3K inhibitor LY294002. HCT-116 cells were treated with 50 μM LY294002 for 16 h. (D) Dominant negative Akt (Akt-DN) induces miR-145 expression. HCT-116 cells were transfected with vector alone (pCMV) or Akt-DN for 24 h. Values in B–D are means ± SE of 3 separate experiments and normalized to their respective controls as 1. **, P < 0.01.
p53 Induces miR-145.
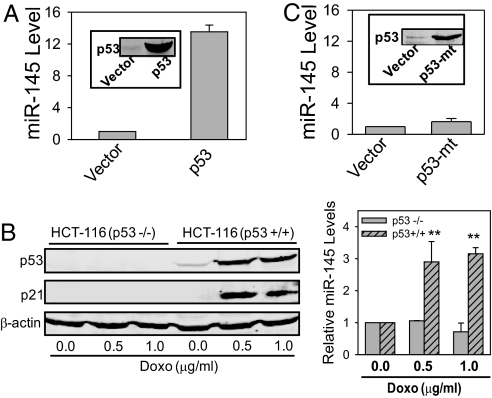
Bioinformatics analysis of the putative miR-145 promoter using the MatInspector module of the Genomatix database further suggested that p53 is a potential miR-145 regulator, because there are 2 putative p53 response elements in a 1.5-kb DNA fragment upstream to the pre-miR-145. Therefore, we tested whether p53 can induce miR-145 in breast cancer MCF-7 cells (Fig. 2A Inset), in addition to colon cancer HCT-116 cells in which p53 induced miR-145 expression. As expected, the miR-145 level was significantly up-regulated by p53 (Fig. 2A). To determine whether the endogenous p53 can also induce miR-145, we treated MCF-7 with Doxo, which caused up-regulation of the endogenous p53 (Fig. S2A). From the same treated cells, we detected an ≈4-fold increase in the miR-145 level as compared with no drug treatment (Fig. S2B). To better define the role of p53 in miR-145 induction, we compared p53+/+ and p53−/− HCT-116 cell lines. As shown in Fig. 2B, Doxo induced p53 expression, as did the p53 target gene p21. It is of importance that this occurred in p53+/+ cells but not in p53−/− cells (Fig. 2B). Consistent with the p53 induction, we detected an ≈3-fold increase in miR-145 levels in Doxo-treated p53+/+ cells compared to no Doxo control. In contrast, there was no such induction of miR-145 in p53−/− cells (Fig. 2B). Because p53 (R175H) is the most frequent mutant in colon cancer (18), we tested the effect of this mutant p53 on miR-145. The mutant p53 revealed no significant effect on miR-145 (Fig. 2C), demonstrating an important role of wild type p53 in the induction of miR-145.
Fig. 2.
Induction of miR-145 by p53. (A) MCF-7 cells were transduced with adenoviral pAd-GFP (Vector) or pAd-GFP-p53 (p53) for 24 h and then harvested for extraction of protein or RNA. Insert, detection of p53 expression. (B) HCT-116 (p53−/−) or HCT-116 (+/+) were treated with Doxo at 0.5 or 1.0 μg/ml for 16 h. The cells were separately harvested for protein or RNA. p21 was used as a positive control. (C) MCF-7 cells were transiently transfected with vector (pCMV) or mutant p53 (R175H) and incubated for 24 h. Insert, detection of mutant p53. Values in A, B, and C are means ± SE of 3 separate experiments and normalized to their respective controls as 1. **, P < 0.01.
p53 Directly Binds the Promoter of miR-145 at the p53 Response Element.
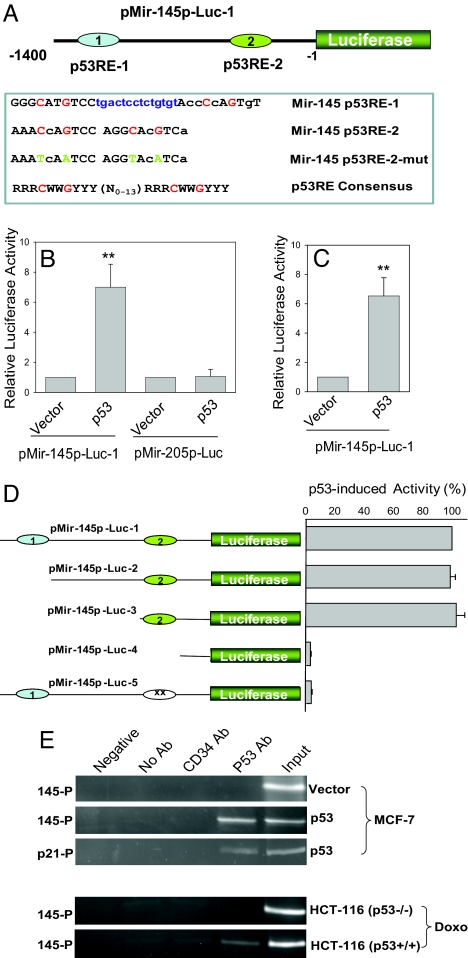
To determine whether p53 activates the miR-145 promoter, we cloned a 1.4-kb putative miR-145 promoter carrying the 2 p53 response elements (p53REs) into a pGL3 basic vector to generate pMir-145p-Luc-1 (Fig. 3A). We first tested pMir-145p-Luc-1 in NIH/3T3 cells. As shown in Fig. 3B, p53 increased miR-145 promoter activity by 7-fold. This p53 induction of luciferase activity was specific to the miR-145 promoter, because it had no effect on a different promoter from miR-205 with the same length of insert (pMir-205p-Luc) (Fig. 3B). We also tested pMir-145p-Luc-1 in MCF-7 cells. Similarly, p53 also increased miR-145 promoter activity in MCF-7 cells (Fig. 3C). These results support the idea that p53 induces not only the endogenous miR-145 level, but also the miR-145 promoter activity.
Fig. 3.
p53 induces the miR-145 promoter activity through binding to p53RE-2. (A) A schematic description of the putative miR-145 promoter with 2 potential p53 response elements, p53RE-1, and p53RE-2 and the mutated p53RE-2 (p53RE-2-mut), as compared with the p53RE consensus, where R = A+G; W = A+T and Y = C+T. Small letters denote deviations from the consensus. The conserved nucleotides C and G are highlighted in red. The sequences between the 2 half sites are indicated with blue color. In p53RE-2-mut, the conserved C and G were replaced by T and A, respectively. (B) and C, Luciferase assays with pMIR-145p-Luc-1 in NIH/3T3 and MCF-7 cells, respectively. pMIR-205p-Luc was used as a negative control. (v) vector. (D) Deletion and site-directed mutagenesis analysis identifies the importance of miR-145 p53RE-2 in p53-mediated induction of the luciferase activity. (E) ChIP assay reveals that p53 specifically interacts with p53RE-2. p21 serves as a positive control. Sequences from the upstream region of the human calcium-activated chloride channel protein 2 gene without p53 binding sites serves as a negative control. 145-p, mir-145 promoter; p21-p, p21 promoter. Values in B–D are means ± SE of 3 separate experiments and normalized to respective controls as 1 (B and C) or 100% (D). **, P < 0.01.
The sequences of p53RE-1 and p53RE-2 are shown in Fig. 3A as compared with the p53RE consensus (19). To determine whether any of the 2 elements directly interacts with p53 to mediate the induction of the miR-145 promoter by p53, we generated a series of deletion constructs (Fig. 3D Left). Based on luciferase assays, it was evident that only p53RE-2 was responsible for p53-mediated miR-145 induction, because the deletion or mutation of p53RE-2 abolished the induction activity whereas the deletion of p53RE-1 had no effect on p53-mediated induction of the miR-145 promoter activity (Fig. 3D Right). To further define the role of p53RE-2 in miR-145 induction, we performed chromatin immunoprecipitation (ChIP) assays. Consistent with the luciferase results, we detected 1 specific PCR product derived from p53RE-2 (Fig. 3E) but not from p53RE-1 after transfection with p53. Moreover, we treated p53+/+ and p53−/− HCT-116 cells with Doxo. Again, we detected a specific band in p53+/+ HCT-116 cells but not in the p53−/− cells (Fig. 3E). Therefore, p53RE-2 was responsible for the p53-mediated induction of miR-145 promoter activity, suggesting that p53 induces miR-145 by directly interacting with its promoter.
c-Myc Is a Direct Target of miR-145.
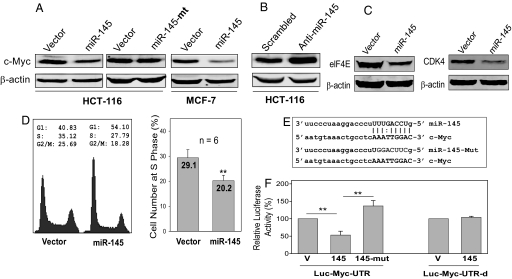
Bioinformatics searches (see SI Text) combined with assays of luciferase reporters carrying the 3′-untranslated region (3′-UTR) identified several putative miR-145 target genes, including c-Myc and MMP-11. In particular, c-Myc is a well known oncogene that plays an important role in neoplastic transformation and apoptosis (20) and can be negatively regulated by p53 (1). However, the underlying mechanism of c-Myc regulation by p53 is not well understood. To determine whether miR-145-mediated silencing of c-Myc is another mechanism responsible for c-Myc repression by p53, we ectopically expressed miR-145 in both HCT-116 and MCF-7 cells. As expected, c-Myc expression was significantly lower in the miR-145 transfected cells than in vector control cells (Fig. 4A). In both cell lines, the level of suppression was ≈60% (not shown). In contrast, the mutant miR-145, in which the seed sequence was mutated (Fig. 4E), had no effect on c-Myc. To further determine the specificity of the miR-145 targeting of c-Myc, we used the antisense locked nucleic acid (LNA) oligo of miR-145 (anti-miR-145) to knockdown the miR-145 level, which was confirmed by TaqMan real-time PCR (Fig. S3). As shown in Fig. 4B, anti-miR-145 was able to enhance c-Myc expression. Immunostaining further confirmed that the ectopic expression of miR-145 substantially suppressed c-Myc expression (Fig. S4). At the same time, real-time RT-PCR analysis of the same transfected cells indicated that c-Myc mRNA was also suppressed by >50% in the miR-145 transfected cells as compared with vector control (Fig. S5A).
Fig. 4.
miR-145 directly targets c-Myc and inhibits its expression. (A) Western blot shows suppression of the c-Myc protein by miR-145, but not the mutant miR-145 (miR-145-mt). Cells were transfected with vector alone or miR-145 or miR-145-mt for 24 h before harvesting for protein extraction. (B) Blockade of miR-145 by anti-miR145 (Anti-145) causes up-regulation of c-Myc. HCT-116 cells were transfected with scrambled oligo or anti-miR-145 oligo and were harvested 24 h later for protein or RNA extraction. The same transfected cells were separately used for Western blot or TaqMan real-time PCR. (C) Suppression of the c-Myc downstream target genes eIF4E and CDK4 by miR-145 in HCT-116 cells. (D) Effect of miR-145 on cell cycle. HCT-116 cells were transiently transfected with vector control or miR-145 expression vector. Two days later, cells were harvested for cell cycle analysis. (E) A putative miR-145 binding site in the c-Myc 3′-UTR and mutant miR-145 in which the seed sequences were mutated. (F) Effect of miR-145 or mutant miR-145 on the luciferase activity of Luc-c-Myc-UTR and Luc-c-Myc-UTR-d in which the putative miR-145 binding site was deleted. For luciferase assays, 293T cells were transfected with vector or miR-145 expression vector and then harvested for lysis of cells 2 days after transfection. Values in F are means ± SE of 3 separate experiments and normalized to their respective controls as 100%. **, P < 0.01.
To further determine the effect of miR-145 on c-Myc, we then examined c-Myc downstream target genes, including the mRNA cap-binding protein eIF4E, and CDK4, both of which are direct target genes of c-Myc (21, 22). miR-145 suppressed the expression of eIF4E and CDK4 (Fig. 4C) and it also suppressed the miR-17∼92 cluster (Fig. S5B), which has been shown to a direct target of c-Myc (23). These results suggest that miR-145 can indirectly suppress these downstream genes by silencing c-Myc. Given that c-Myc plays a role in the regulation of the cell cycle, and c-Myc is able to increase S phase cell population (24), we performed cycle analysis of the miR-145 transfected cells. miR-145 reduced the S phase cell population by ≈9% (Fig. 4D). Together, these results suggest that c-Myc is a target for miR-145.
To determine whether miR-145 directly targets c-Myc, we constructed a luciferase reporter carrying c-Myc 3′-UTR with a putative miR-145 binding site (Fig. 4E). We detected a reduction of luciferase activity by ≈50% in the miR-145 transfected cells as compared with vector control (Fig. 4F). This suppression of the luciferase activity was specific to miR-145 because the mutated miR-145 at the seed sequence lost its suppressive activity (Fig. 4F). We further showed that the miR-145 binding site in c-Myc 3′-UTR was required for this regulation; deletion of the binding site (Luc-c-Myc-UTR-d) abolished its suppression (Fig. 4F). Thus, miR-145 directly targets c-Myc, which is likely through interacting with this binding site.
miR-145 Suppresses Tumor Cell Growth both in Vitro and in Vivo.
Given the ability of miR-145 to silence c-Myc, miR-145 is expected to have an impact on tumor cell growth. Growth inhibition assays for cells transfected with miR-145 supported this notion, because miR-145 caused a significant decrease in cell growth for both HCT-116 and MCF-7 cells (Fig. S6 A and B). In addition, soft agar assays indicated that miR-145 suppressed anchorage-independent growth (Fig. S7 A–D). Therefore, it appears that miR-145-mediated c-Myc suppression could in part be attributed to the tumor suppressive role of miR-145.
To further define the tumor suppressive role of miR-145, we infected HCT-116 cells with the lentiviral vector expressing miR-145 (Fig. S8). Real-time PCR analysis revealed an ≈6-fold higher of the miR-145 level in the miR-145-infected cells than vector control (Fig. S9A), and this difference between the miR-145-infected cells and vector control cells is approximately equivalent to the fold difference between the normal colon tissue and matched tumor tissue samples (Fig. 1A), suggesting its clinical relevance. As shown in Fig. S10A, tumor growth with miR-145 was significantly slower than that with the vector control. In agreement with the tumor growth curve, the tumor weight for miR-145-infected cells was significantly lower than that of the vector control (Fig. S10 B and C ). We randomly selected 1 tumor each for vector control and miR-145, respectively, and found that the level of miR-145 of the tumor derived from the miR-145-infected cells (Fig. S9B) was higher than vector control; this difference was similar to that found when the cells were injected into mice (Fig. S9A), suggesting that the level of ectopically expressed miR-145 remains constant during the period of tumor growth in mice. We also extracted protein from 3 tumors derived from vector controls or miR-145 infections. Western blot analysis confirmed that the miR-145 tumors expressed lower levels of c-Myc than the vector control tumors (Fig. S10D ). Consistent with the Western blot results, we found that c-Myc mRNA was also down-regulated in the miR-145 tumor (Fig. S9C). These results support a role for the miR-145-mediated silencing of c-Myc in the tumor growth inhibition, although given the nature of miRNA targeting, it is also possible that other miR-145 targets could be responsible for the observed inhibition phenotype.
Role of miR-145 in p53-Mediated Repression of c-Myc.
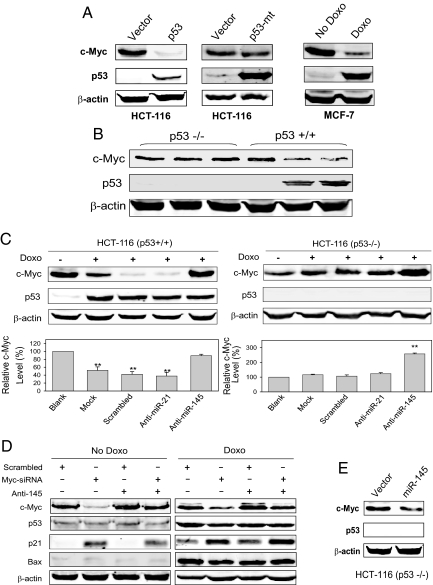
Having demonstrated that (i) miR-145 suppresses tumor cell growth both in vitro and in vivo, likely in part through direct silencing of c-Myc; and (ii) p53 induces miR-145, we next investigated whether p53 negatively regulates c-Myc through the induction of miR-145. As shown in Fig. 5A, ectopically expressed p53 substantially reduced c-Myc expression. However, the mutant p53 had no effect on c-Myc expression. We also showed that the Doxo-induced p53 repressed c-Myc in MCF-7 cells (Fig. 5A Right). The p53 status was critical for c-Myc repression after treatment with Doxo. In p53+/+ HCT-116 cells, Doxo induced p53, which in turn suppressed c-Myc. In contrast, in p53−/− HCT-116 cells no c-Myc suppression was observed (Fig. 5B). To better determine the relationship between the p53-mediated induction of miR-145 and repression of c-Myc, we performed time-course experiments and found that the c-Myc repression was mirrored by the miR-145 induction in a time-dependent manner, which only occurred in the p53+/+ HCT-116 cells (Fig. S11). Of note, the c-Myc repression had a small lag period compared with p53 and miR-145 induction.
Fig. 5.
Role of miR-145 in p53-mediated repression of c-Myc. (A) Effect of wild-type p53 and mutant p53 (R175H) and Doxo-induced p53 on c-Myc expression. HCT-116 cells were transduced with p53 or transfected with mutant p53 for 24 h and then harvested for extraction of protein. For induction of the endogenous p53, the experiment was carried out same as in Fig. 2B with 1 μg/ml Doxo. (B) HCT-116 (p53−/−) and HCT-116 (p53 +/+) were treated with Doxo at 0.5 or 1.0 μg/ml for 16 h. Over 50% repression was observed in the Doxo treated HCT-116 (p53+/+) cell. (C) Blockade of p53-mediated c-Myc repression by anti-miR-145. HCT-116 (p53−/−) and HCT-116 (p53 +/+) cells were first transfected with anti-miR-145 or control oligos. After 10 h, the cells were treated with Doxo at 1.0 μg/ml for 16 h before extraction of protein or RNA. Anti-miR-21 serves as an additional negative control. Blank without Doxo serves as a baseline of c-Myc for comparison between Doxo-treated and Doxo-untreated, or between anti-miR-145 and negative control (scrambled oligo or anti-miR-21). The mock control was performed by the same transfection experiments without oligos to confirm that the repression of c-Myc by Doxo/p53 is specific. Bottom: Relative c-Myc levels are averages ± SE of 3 separate experiments and normalized to the no Doxo control as 100%. The value for the anti-miR-145 lane in HCT-116 (p53 +/+) cells is 92%. **, P < 0.01 compared with blank controls. (D) Effect of c-Myc-siRNA and/or anti-miR-145 on c-Myc, p21 and Bax. HCT-116 cells transfected with scrambled oligo or c-Myc-siRNA for 24 h, followed by the treatment with or without Doxo (0.5 μg/ml) for 16 h. The c-Myc protein blot in the Right was exposed longer than the left 1 to detect various amounts of the c-Myc protein in different treatments. (E) miR-145 also suppresses c-Myc in p53−/− cells.
To demonstrate the role of miR-145 in the p53-mediated repression of c-Myc, we determined whether this p53-mediated repression of c-Myc can be reversed by knockdown of miR-145. Although induction of p53 reduced the c-Myc level, this reduction was blocked by anti-miR-145 in p53+/+ HCT-116 cells (Fig. 5C Left). For example, Doxo/p53 caused reduction of the c-Myc level by ≈60%. However, when the cells were pretreated with anti-miR-145, the c-Myc repression was reversed to 8% (i.e., 92% of the no Doxo control), suggesting that anti-miR-145 can block this p53-dependent and miR-145-mediated c-Myc repression. The reason for incomplete blockade of the c-Myc repression is possibly due to other p53-mediated mechanisms involved. In p53−/− HCT-116 cells, Doxo caused no repression of c-Myc, as expected. Importantly, anti-miR-145 increased the c-Myc level by ≈250% (Fig. 5C Right), which was similar to the level in the p53+/+ cells without drug treatment (Fig. 4B). Similarly, anti-miR-145 was able to block the c-Myc repression in the Ly294002 or c-Myc-siRNA cells (Fig. S12). Moreover, anti-miR-145 increased S phase cell populations in general (Fig. S13 A and B). However, anti-miR-145 caused only a slight reduction of p53-induced apoptosis (Fig. S13C). We then examined the effect of c-Myc knockdown on the levels of p21 and Bax, both of which are direct targets of p53. As shown in Fig. 5D, c-Myc knockdown had no effect on Bax level, but p21 level was increased, which is likely because c-Myc can suppress p21 by partnering with Miz1 (25); when c-Myc is reduced, p21 is increased. Anti-miR-145 increased the c-Myc level in the presence or absence of c-Myc-siRNA; it had no effect on Bax (Fig. 5D). These results suggest that a lack of miR-145 impairs the p53-induced cell cycle arrest, but has little effect on p53-induced apoptosis or p53 targets such as Bax. Finally, to demonstrate that miR-145-mediated c-Myc suppression is a downstream event of p53, we ectopically expressed miR-145 in the p53 −/− cells and still detected reduction of the c-Myc level. (Fig. 5E), further suggesting that this miR-145-mediated c-Myc repression can be independent of p53, if sufficient miR-145 is present in the cell. Therefore, these results collectively suggest that miR-145 is an important component of the p53-mediated c-Myc repression regulatory network.
Discussion
Despite the overwhelming evidence suggesting that miRNAs could play a causal role in human malignancies, the mechanism underlying the deregulation of miRNAs and the miRNA-mediated gene silencing, which leads to cancer development, is still poorly understood. The available evidence suggests that p53 and c-Myc could be major key players in this complex network (26). Although the relationship between p53 and c-Myc and how they regulate each other has been a focus of research for a long time, we still do not have a clear picture as to how the cell balances the effect of “yin” (e.g., p53) and “yang” (e.g., c-Myc). We present evidence that p53 induces miR-145 expression and thus, miR-145 is a new member of this p53 regulatory network. More importantly, through the induction of miR-145, p53 is able to repress c-Myc expression at the posttranscriptional level. Therefore, our finding identifies a critical role of miR-145 as a tumor suppressor in the p53 regulatory network and thus provides new insight into the p53-mediated regulation of c-Myc.
As a master regulator for gene expression, p53 has been shown to directly or indirectly regulate numerous protein-coding genes. However, only a very limited number of miRNAs have been shown to be direct targets of p53. To date, the miR-34 family are the only known miRNAs that are directly regulated by p53 (27). Several groups have reported that the miR-34 family, including miR-34a, miR-34b, and miR-34c, are induced by DNA damage and oncogenic stress in a p53-dependent manner. Therefore, the identification of miR-145 as a direct p53 target expands the repertoire of p53-regulated genes. For example, through miR-145, p53 can indirectly suppress c-Myc, as we demonstrated in this study. Moreover, given that c-Myc and Miz1 can work together to suppress p21 (25), miR-145 is expeted to be able to potentiate the induction of p21 by p53 because suppression of c-Myc by miR-145 would relieve the repression of c-Myc on p21 (Fig. 5D). Finally, because silencing of c-Myc by miR-145 is a downstream event of p53 activity, p53 is able to specifically regulate different downstream pathways through induction of miR-145.
Given that miRNAs could play a “fine-tuning” role in gene regulation (28), identification of miR-145 as a component of this regulatory network highlights the complexity of gene regulation involving important genes such as p53 and c-Myc. Understanding the role of miR-145 may also help to explain in part why c-Myc usually has a short half life (29). Regulation of c-Myc is complex. In addition to the transcriptional regulation by various factors such as mitogens, other mechanisms such as ubiquitination also play an important role (30). Moreover, c-Myc can enhance p53 expression through p19Arf (31). Our study suggests another regulatory mechanism through p53 and miR-145. Evidently, such a sophisticated regulatory network may be necessary for the cell to maintain the normal growth and proliferation. Interruption of this balance, such as the down-regulation of miR-145, could lead to cell malignancy.
Materials and Methods
Plasmids.
Constructs expressing miR-145, luciferase-c-Myc-3′-UTR, and miR-145 promoter were generated by standard methods as detailed in SI. An adenoviral vector expressing p53, Ad-p53-GFP (32), was obtained from Vector Biolabs; a mutant p53 (R175H) expression vector and Akt-DN were obtained from Addgene.
Transfection.
HCT-116 and MCF-7 cells were transfected by using DNAfectin reagent (Applied Biological Materials) following the manufacturer's protocol. Transfection of locked nucleic acid (LNA) anti-miR-145 (IDT) into HCT-116 cells was performed by using RNAfectin (Applied Biological Materials) in 6-well plates following the manufacturer's recommendations.
Transduction.
The lentiviral vector (System Biosciences) expressing miR-145 was packaged and used to infect MCF-7 or HCT-116 cells according to the manufacturer's protocol.
Luciferase Assay.
Cells were first transfected with appropriate plasmids in 12-well plates, and then were harvested and lysed for luciferase assay 24 h after transfection. Luciferase assays were performed by using a luciferase assay kit (Promega) according to the manufacturer's protocol. β-galactosidase or renilla luciferase was used for normalization.
PCR/RT-PCR and Real-Time RT-PCR.
PCRs were performed to amplify the putative miR-145 promoter according to the standard 3-step procedure. For RT-PCR, we isolated total RNA by using TRIzol reagent (Invitrogen) per the manufacturer's protocol and used 1 μg of RNA to synthesize cDNA by SuperScriptase III (Invitrogen) with random primers. To detect c-Myc mRNA, we used the SYBR green method with primers c-Myc-5.1 and c-Myc-3.1 (Table S2). Expression of miR-145 in cell lines or patient specimens was detected by TaqMan stem-loop RT-PCR method (33).
Supplementary Material
Acknowledgments.
We thank Dr. Bert Vogelstein (The Johns Hopkins University, Baltimore) for providing both the p53 wild type and the p53 null HCT-116 cell lines. This work was supported by Grant CA102630 from the National Cancer Institute and Grant BC052294 from the Department of Defense.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808042106/DCSupplemental.
References
- 1.Levy N, Yonish-Rouach E, Oren M, Kimchi A. Complementation by wild-type p53 of interleukin-6 effects on M1 cells: Induction of cell cycle exit and cooperativity with c-myc suppression. Mol Cell Biol. 1993;13:7942–7952. doi: 10.1128/mcb.13.12.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416:129–136. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 3.McNeil CM, et al. c-Myc overexpression and endocrine resistance in breast cancer. J Steroid Biochem Mol Biol. 2006;102:147–155. doi: 10.1016/j.jsbmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Ragimov N, et al. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene. 1993;8:1183–1193. [PubMed] [Google Scholar]
- 5.Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillai RS. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 8.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sempere LF, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 10.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Schepeler T, et al. Diagnostic and Prognostic MicroRNAs in Stage II Colon Cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 13.Shtilbans V, Wu M, Burstein DE. Current overview of the role of Akt in cancer studies via applied immunohistochemistry. Ann Diagn Pathol. 2008;12:153–160. doi: 10.1016/j.anndiagpath.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Roy HK, et al. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201–205. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 15.Mayo LD, Donner DB. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem Sci. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- 16.Ogawara Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277:21843–21850. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 17.Zhou BP, et al. HER-2/neu blocks tumor necrosis factor-induced apoptosis via the Akt/NF-kappaB pathway. J Biol Chem. 2000;275:8027–8031. doi: 10.1074/jbc.275.11.8027. [DOI] [PubMed] [Google Scholar]
- 18.Soussi T, Ishioka C, Claustres M, Beroud C. Locus-specific mutation databases: Pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- 19.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 20.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 21.Jones RM, et al. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermeking H, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubik D, Dembinski TC, Shiu RP. Stimulation of c-myc oncogene expression associated with estrogen-induced proliferation of human breast cancer cells. Cancer Res. 1987;47:6517–6521. [PubMed] [Google Scholar]
- 25.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 26.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 27.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobert O. miRNAs play a tune. Cell. 2007;131:22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 29.Jones TR, Cole MD. Rapid cytoplasmic turnover of c-myc mRNA: Requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: Ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia. 2006;8:630–644. doi: 10.1593/neo.06334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 32.Yu J, et al. Identification and classification of p53-regulated genes. Proc Natl Acad Sci USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.