Abstract
Key steps in the export of mRNA from the nucleus to the cytoplasm are the transport through the nuclear pore complex (NPC) and the subsequent remodeling of messenger RNA-protein (mRNP) complexes that occurs at the cytoplasmic side of the NPC. Crucial for these events is the recruitment of the DEAD-box helicase Ddx19 to the cytoplasmic filaments of the NPC that is mediated by the nucleoporin Nup214. Here, we present the crystal structure of the Nup214 N-terminal domain in complex with Ddx19 in its ADP-bound state at 2.5 Å resolution. Strikingly, the interaction surfaces are not only evolutionarily conserved but also exhibit strongly opposing surface potentials, with the helicase surface being positively and the Nup214 surface being negatively charged. We speculate that the positively charged surface of the interacting ADP-helicase binds competitively to a segment of mRNA of a linearized mRNP, passing through the NPC on its way to the cytoplasm. As a result, the ADP-helicase would dissociate from Nup214 and replace a single bound protein from the mRNA. One cycle of protein replacement would be accompanied, cooperatively, by nucleotide exchange, ATP hydrolysis, release of the ADP-helicase from mRNA and its rebinding to Nup214. Repeat of these cycles would remove proteins from a mRNP, one at a time, akin to a ratchet mechanism for mRNA export.
Keywords: crystal structure, immunofluorescence localization, mRNA export, mutagenesis, Nup214-helicase complex
Bidirectional traffic into and out of the nucleus occurs via nuclear pore complexes (NPCs) (1, 2). While transport of proteins across the nuclear envelope is reasonably well understood, the mechanism of nuclear export of mRNPs remains elusive (1, 3).
In yeast, the DEAD-box helicase Dbp5 localizes to the cytoplasmic side of the NPC by binding to Nup159, and, by removing proteins from mRNA (4–7), plays an essential role in remodeling of mRNP complexes. Mutants of Dbp5 that interfere with Dbp5's NPC localization or with its mRNP remodeling activity result in mRNA accumulation in the nucleus (6, 7). Targeting to the NPC appears to be conserved throughout evolution, because the human Dbp5 homolog, Ddx19, binds to Nup214, the Nup159 homolog (8, 9).
DEAD-box helicases are composed of an ≈350-residue core region, consisting of 2 globular domains termed domain 1 and 2 that contain 9 evolutionarily conserved sequence motifs, including the sequence DEAD, the eponymous motif of the entire family. The human Ddx19 protein encompasses 479 residues and contains the conserved core region, which is complemented by an N-terminal extension (NTE) of 94 residues (Fig. 1A). In the cell, Ddx19 was found to localize to the cytoplasm and to the nuclear envelope (8, 10).
Fig. 1.
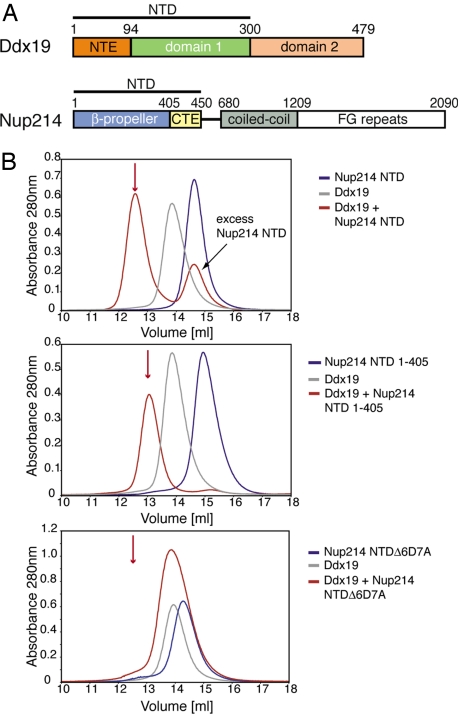
The 6D7A loop of the Nup214 NTD is essential for Ddx19 binding. (A) Domain organization of Nup214 and Ddx19. For Ddx19, the N-terminal extension (NTE, orange) and the 2 RecA-like domains (domain 1 and 2, green and light orange) are indicated. For Nup214, the β-propeller domain (light blue) and its C-terminal extension (CTE, yellow), the coiled-coil domain (gray), and the C-terminal unstructured region containing numerous FG-repeats (white) are indicated. The Nup214 NTD is composed of the β-propeller domain followed by the CTE. The bars above the domain structures mark the fragments of the orthorhombic crystal form. (B) Gel filtration profiles of full-length wild-type Ddx19 incubated with the Nup214 NTD, Nup214 NTD 1–405, or Nup214 NTD Δ6D7A before injection.
The human Nup214 gene, also known as CAN, was first identified as a target for chromosomal translocations involved in leukemogenesis. Nup214 localizes to the cytoplasmic side of the NPC as part of the cytoplasmic filaments (11–13). The polypeptide chain of Nup214 contains 2,090 residues and can be divided into 3 distinct regions: a 450-residue N-terminal domain (NTD), a long, flexible linker that connects the NTD to a centrally located coiled-coil domain, and a C-terminal ≈1,000-residue region, which contains numerous FG-repeats (Fig. 1A). The localization of Ddx19 to the cytoplasmic side of the NPC has been shown to be dependent on the Nup214 NTD (8). Previously, we solved the structure of the human Nup214 NTD that revealed a β-propeller domain and suggested several possible protein–protein interaction surfaces, including the 45-residue C-terminal extension (CTE) and the conserved interblade connector 6D7A (14).
To gain deeper insight into the Nup214-dependent recruitment of Ddx19 to the cytoplasmic side of the NPC, we determined the structure of the Nup214 NTD in complex with the ADP-bound state of Ddx19. Moreover, we present an extensive biochemical analysis of the evolutionarily conserved Ddx19-Nup214 interaction and demonstrate that the interblade connector 6D7A that forms a conserved surface loop of the Nup214 β-propeller is essential for the interaction with Ddx19. We identify a hot-spot residue of Ddx19, arginine 259, that is critical for complex formation with Nup214 in vitro and for the localization of Ddx19 to the nuclear envelope in vivo.
Results
Nup214 Interacts with Ddx19 via Its Conserved 6D7A Loop.
To identify the interface between Ddx19 and Nup214, we tested several putative protein interaction sites on the Nup214 NTD surface for Ddx19 binding. We designed Nup214 variants, including Nup214 NTD 1–405 in which the CTE is deleted and Nup214 NTD Δ6D7A in which a flexible linker replaces the highly conserved surface loop 6D7A. The recombinant proteins were expressed, purified to homogeneity, and monitored for complex formation with Ddx19 using size exclusion chromatography (SEC) (Fig. 1B). While the CTE-deletion mutant Nup214 1–405 was capable of complex formation, the loop-deletion mutant Nup214 NTD Δ6D7A resulted in a protein that was incapable of binding to Ddx19 (Fig. 1B).
Crystal Structure of the Nup214 NTD·Ddx19 Complex.
Crystals appeared in the presence of ADP in the monoclinic space group P21 with 2 complexes in the asymmetric unit. The structure resolved the Nup214 NTD bound to the N-terminal RecA-like domain (domain 1) of Ddx19 bound to ADP at 3.2 Å resolution. However, no electron density was observed for the smaller C-terminal RecA-like domain (domain 2) of Ddx19. To optimize the diffraction data quality, we crystallized the Nup214 NTD in complex with only the N-terminal domain (NTD) of Ddx19, comprised of the NTE and domain 1 (residues 1–300), bound to ADP (Fig. 1A). Crystals of this complex belong to the orthorhombic space group P212121 with 1 complex in the asymmetric unit and diffracted to 2.5 Å resolution. The final model contains residues 8 to 428, and 69 to 300 of Nup214 and Ddx19, respectively. The structure was refined to an Rcryst factor of 19.9% and an Rfree factor of 24.2%. For details of the crystallographic statistics, see Table S1.
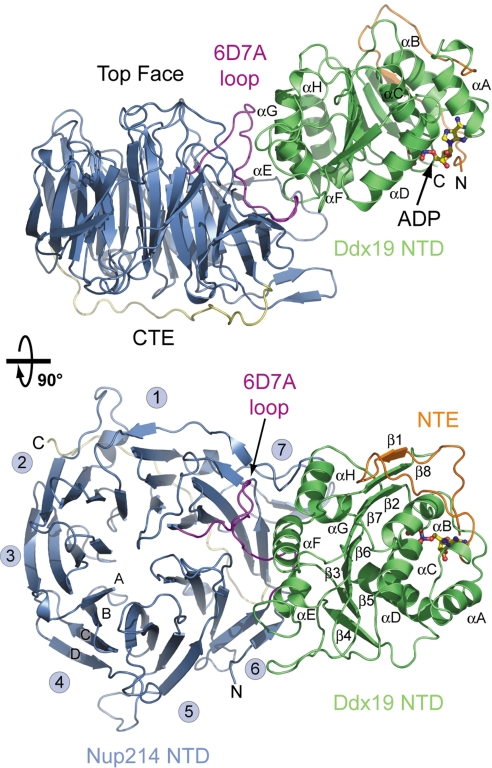
As previously described, Nup214 forms a 7-bladed β-propeller domain that is extensively decorated by long interblade connector loops, and contains a C-terminal extension (CTE) that binds to the bottom face of the β-propeller (14) (Fig. 2). Our structure only resolves domain 1 and 26 residues of the NTE of Ddx19. The polypeptide chain of domain 1 of Ddx19 folds into the canonical RecA-like domain found in other DEAD-box helicases, such as MjDEAD, and in eIF4A (15, 16) (Fig. 2). This globular domain is formed by an extensive, centrally located β-sheet composed of 7 parallel β-strands (β2–β8), which is sandwiched by 4 α-helices on each side (αA–αD on one side, and αE–αH on the other) (Fig. 2). In our structure, the unique N-terminal extension (NTE) is primarily unstructured, but the 26 residues that are resolved (residues 69 to 94) form an extensive loop, and supplement the β-sheet core of domain 1 with an additional anti-parallel β-strand, β1. Moreover, the N-terminal part of the NTE contributes to the nucleotide-binding pocket. Strikingly, the binding of Ddx19 to the extensively decorated Nup214 NTD results in only minor conformational changes in the Nup214 NTD, as indicated by a root-mean square deviation (RMSD) of ≈0.6 Å over all atoms between the isolated and Ddx19-bound Nup214 NTD structures.
Fig. 2.
Overview of the Nup214 NTD·Ddx19 NTD structure. Ribbon representation of the Nup214 NTD·Ddx19 NTD complex (Upper). A 90° rotated view is shown in Lower. For the Nup214 NTD, the β-propeller domain (blue), the 6D7A loop (magenta), the C-terminal extension (CTE; yellow), and the blade numbers are indicated. For the Ddx19 NTD, the N-terminal RecA-like domain (green) and the unique N-terminal extension (NTE; orange) is indicated. The ADP molecule bound to the Ddx19 NTD is shown in ball-and-stick representation.
The Nup214 NTD binds with its side to the N-terminal RecA-like domain (domain 1) of Ddx19. The interaction between the 2 proteins is primarily mediated by the interblade connector loop 6D7A with minor contributions from residues of the β-strand 6E, which forms the double Velcro closure, and the 1CD, 5D6A, and 6BC loops of the Nup214 β-propeller domain. In Ddx19, the 4 helices αE–αH and their interhelical loops contribute to the interface (Fig. 2).
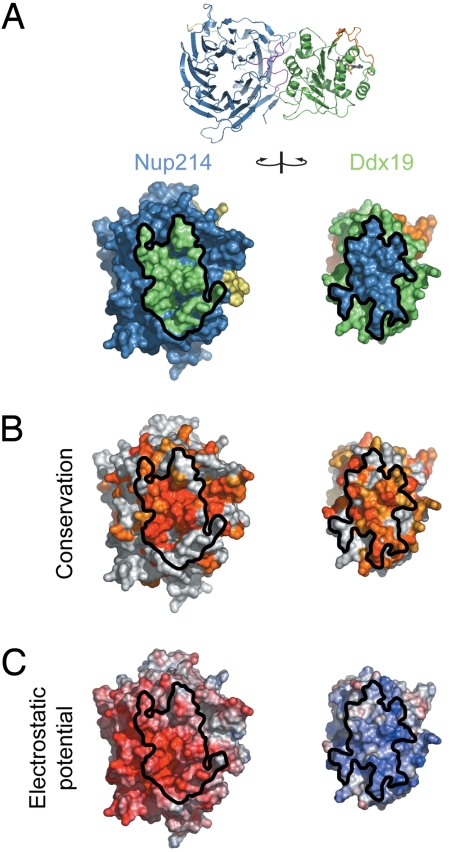
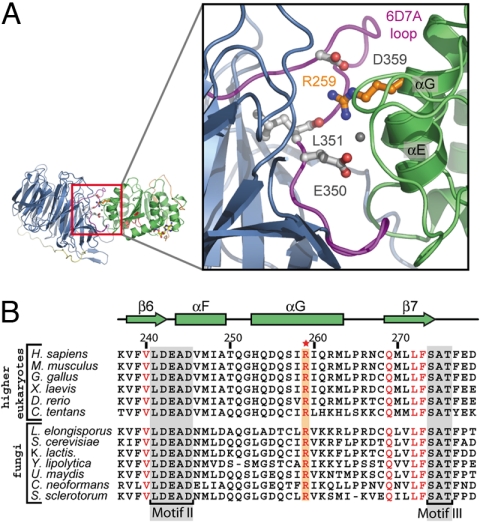
Strikingly, the association between the 2 proteins is predominantly mediated by surfaces with strongly opposing electrostatic potential that are evolutionarily highly conserved (Fig. 3 and Fig. S1). While the binding surface of Nup214 features a highly negative surface potential, the binding surface of Ddx19 is highly positively charged. In fact, the highest positive surface potential of domain 1 of Ddx19 is buried at the interface with Nup214. Importantly, 22 of 51 residues of Ddx19 are charged and form numerous salt bridges at the interface, dominating the association of the 2 proteins via electrostatic interactions (Fig. 3C and Fig. S2). For example, R259 of Ddx19 is located in the center of the interface and forms a critical salt bridge with D359, a water-mediated salt bridge with E350, and an additional hydrogen bond with the backbone carbonyl of L351 (Fig. 4). All of the residues that R259 of Ddx19 interacts with are located within the 6D7A loop of the Nup214 β-propeller domain. However, in addition to the salt bridges formed by R259, several additional salt bridges are formed in the interface, providing the molecular basis for our biochemical data (see below) that the 2 proteins can be dissociated in high-salt conditions (Fig. S2). These electrostatic interactions appear to be further reinforced by additional hydrophobic van der Waals contacts. In total, a surface area of ≈1900 Å2 is buried at the interface between the 2 proteins. Interestingly, such intermolecular associations governed primarily by electrostatic interactions were also observed in other nucleoporin structures, namely the Nup58/45 tetramer and the Seh1·Nup85 hetero-octamers (17, 18).
Fig. 3.
Surface properties of the Nup214 NTD-Ddx19 NTD interaction. (A) Surface renditions of the NTDs of Nup214 and Ddx19 in an open book representation colored according to the participation of the various domains as in Fig. 2. Surfaces that mediate the association between the 2 proteins are indicated in green (Ddx19) and blue (Nup214). As a reference, a ribbon representation of the complex is shown in its original orientation. (B) Surface representation colored according to a multispecies sequence alignment (Fig. S1). The conservation at each position is mapped onto the surface and is shaded in a color gradient from light yellow (60% similarity) to dark red (100% identity). (C) Surface representation colored according to the electrostatic potential. The electrostatic potential is plotted onto the surface and colored in a gradient from red (−10 kBT/e) to blue (+10 kBT/e). The orientation of all surface representations is identical. Black lines indicate the interface borders.
Fig. 4.
The conserved arginine 259 of Ddx19 is a key residue for complex formation. (A) Details of the interaction between the NTDs of Nup214 and Ddx19. The ribbon representation is colored according to Fig. 2. The Inset illustrates the position of R259 and its interacting residues and is expanded on the right. (B) Multispecies sequence alignment of Ddx19 homologs. The red asterisk indicates the location of the invariant R259. The conserved sequence motifs II and III are highlighted in gray boxes. Invariant kBT residues outside of the conserved sequence motifs and R259 are illustrated in red. The residue numbering is relative to human Ddx19 and the secondary structure of Ddx19 is shown above the sequence alignment.
Domain 1 of Ddx19 provides a deep groove for the ADP cofactor that is partially covered by the unique NTE. The adenine ring of the ADP molecule is specifically recognized by several hydrogen-bond interactions with Q119 and the backbone carbonyl of R114. In addition, the adenine ring is tucked into a hydrophobic slot between F112 of the αA-αB loop and L70 of the NTE. Altogether, the binding of the ADP cofactor is similar to that observed in the structure of UAP56 (19). However, in contrast to UAP56, no Mg2+ ion is found to coordinate the pyrophosphate moiety of ADP.
Typically, the 2 RecA-like domains of DEAD-box helicases associate in tandem with the nucleotide bound between the 2 domains. The comparison of different DEAD-box helicases has revealed that nucleotide binding induces conformational changes that affect the association of the 2 domains and their spatial arrangement (15, 16, 19). Based on these conformational transitions, a model has been proposed in which the energy of ATP hydrolysis is used for the removal of proteins from the mRNP, resulting in the release of the free mRNA (19). Our findings that crystals of Nup214 NTD in complex with full-length Ddx19 showed no electron density for the second RecA-like domain of Ddx19, and that crystals were obtained only in the presence of ADP, suggest that the second RecA-like domain of Ddx19 changes its spatial alignment with respect to domain 1 in the ADP-bound state.
Interaction of Nup214 and Ddx19 Is Electrostatic in Nature.
Given the strongly opposing electrostatic potential of the surfaces that mediate the interaction between Nup214 and Ddx19, we examined the influence of high-salt buffer conditions on the stability of the Nup214 NTD·Ddx19 complex. Strikingly, we found that the apparent molecular weight of the complex decreased with increasing salt concentrations (Fig. S2). While the complex eluted with an apparent molecular mass of ≈235 kDa in a buffer containing 50 mM KCl, the apparent molecular mass decreased to ≈105 kDa when examined in a buffer containing 1 M KCl. Hence in agreement with the structural data, the association between the Nup214 NTD and Ddx19 is governed by electrostatic interactions that can be substantially weakened in high-salt conditions.
Further Characterization of the Nup214 Interacting Surface.
The 6D7A interblade connector loop encompasses 20 residues and connects the outermost β-strand 6D of blade 6 with the innermost β-strand 7A of blade 7 of the Nup214 β-propeller domain (Fig. S3). In yeast, it has been shown that 2 mutations within the 6D7A loop, V323E and I326E of Nup159 (the yeast homolog of Nup214), disrupt the interaction with Dbp5 (the yeast homolog of Ddx19) (9). However, while the yeast Nup159 and the human Nup214 display considerable sequence conservation in this loop, 1 of the 2 mutated Nup159 residues is a glutamate (E350) in the human Nup214 β-propeller domain (Fig. S3).
To address the differences between the yeast and human Nup214 homologs in helicase binding, we performed an alanine-scanning mutagenesis of the 6D7A loop. A series of 13 single alanine mutants covering the exposed residues of the 6D7A loop was generated (Table S2). In addition, we constructed the V353E mutant and the E350A/V353E double mutant, since these residues were found to be critical for complex formation in the yeast homolog. All 15 mutants were purified to homogeneity in milligram amounts, and their behavior on a gel filtration column was indistinguishable from that of the wild-type protein. The ability of all 15 mutants to bind to full-length Ddx19 was analyzed through the use of SEC. However, none of the 15 Nup214 NTD mutants tested markedly disrupted complex formation, suggesting that multiple residues of the 6D7A loop are involved in the binding of Ddx19.
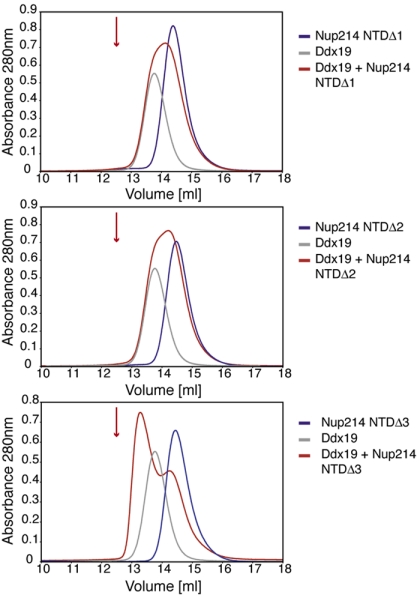
To identify a region of the 6D7A loop that is required for Ddx19 binding, we constructed 3 deletion mutants, Nup214 NTD Δ1, Δ2, and Δ3, covering residues 343 to 356 (Fig. S3B). For each mutant, a stretch of 5 consecutive residues was deleted from the solvent-exposed part of the 6D7A loop, as seen in the Nup214 NTD crystal structure (14). Again, the behavior of all 3 mutants on a gel filtration column was indistinguishable from that of the wild-type protein. We tested all 3 proteins for their ability to interact with full-length Ddx19 by SEC. While the Nup214 NTD Δ3 was capable of interacting with Ddx19, the complex peak displayed a substantial shift toward lower molecular weight, which indicates a weakened, more dynamic interaction between the 2 proteins (Fig. 5). The 2 remaining deletion mutants, Nup214 NTD Δ1 and Δ2, abolished complex formation with Ddx19 (Fig. 5). To determine whether the 6D7A loop was not only necessary, but also sufficient for complex formation, we tested a GST-6D7A-His6 loop fusion protein for complex formation with Ddx19. We found that the fusion protein was unable to bind to Ddx19. Our data demonstrate that the 6D7A loop of the Nup214 NTD is required for complex formation with Ddx19.
Fig. 5.
A 9-residue region in the 6D7A loop of the Nup214 NTD is essential for Ddx19 binding. Gel filtration profiles of full-length wild-type Ddx19 incubated with the deletion mutants Nup214 NTDΔ1, Δ2, or Δ3 before injection. Gel filtration profiles of Ddx19 are colored in gray, the Nup214 NTD variants in blue, and the elution profile resulting from incubating Ddx19 with Nup214 proteins in red. The red arrow indicates the expected elution volume of the Nup214·Ddx19 complex (see Fig. 1B Top).
Further Characterization of the Ddx19 Interacting Surface.
To detect Ddx19 peptides that are involved in Nup214 binding, we probed a Ddx19 peptide array with radioactively labeled Nup214 NTD. This approach identified several consecutive Ddx19 peptide fragments that fall into 2 major regions when mapped onto the surface of a Ddx19 homology model (Fig. S4). The major interaction surface is found within domain 1, occupying approximately half of the surface area. A second, substantially smaller surface patch is found within domain 2, which is localized close to the surface cleft between the 2 domains (Fig. S4). These data suggest that the primary interaction between Ddx19 and Nup214 is mediated via the surface of domain 1.
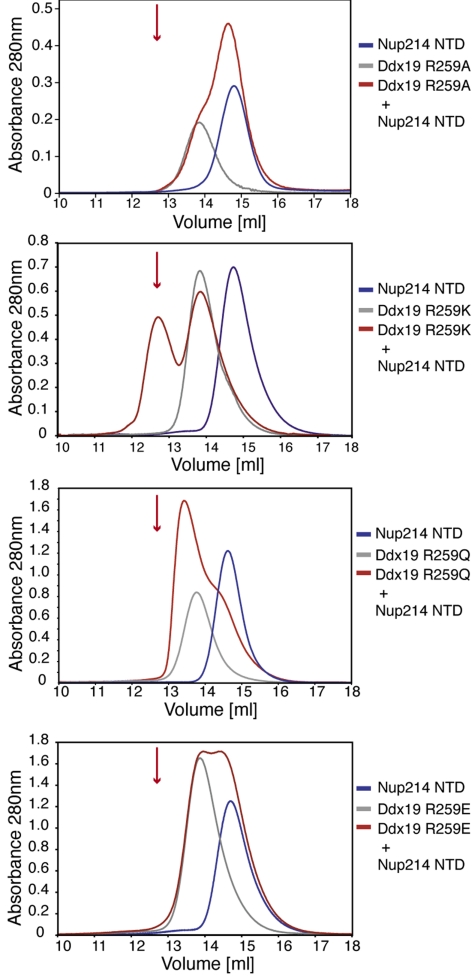
To identify Ddx19 residues that are critical for mediating complex formation, we performed alanine-scanning mutagenesis. We mutated 60 Ddx19 residues that were dispersed over the entire Ddx19 sequence and selected based on their predicted surface localization, their charge, and their conservation (Figs. S1 and S5, and Table S3). The surface localization and conservation was determined based on a Ddx19 homology model (Fig. S5). We were able to purify 54 of these mutants to homogeneity, and their capability to bind to the Nup214 NTD was measured by SEC. The behavior of the 54 individual mutants on a gel filtration column was indistinguishable from that of the wild-type protein, demonstrating that the introduced mutations did not interfere with the folding of the protein. Notably, we identified a single mutant of Ddx19, R259A, that abolishes the interaction between the 2 proteins (Fig. 6).
Fig. 6.
Arginine 259 of Ddx19 is crucial for binding to Nup214 NTD. Gel filtration profiles of the Nup214 NTD (blue), the Ddx19 mutants R259A, R259Q, R259K and R259E (gray), and the elution profile resulting from incubation of Nup214 NTD with Ddx19 (red) before injection are indicated. The red arrow indicates the expected elution volume of the Nup214·Ddx19 complex (see Fig. 1B Top).
Based on this finding, we analyzed additional mutants of R259, R259K, R259Q, and R259E, as well as 5 alanine mutants of residues surrounding R259 (L222A, D255A, Q256A, I258A, and R262A). The R259K mutant and the 5 surrounding mutants had no effect on the ability of Ddx19 to bind to the Nup214 NTD. However, the R259Q mutant was only capable of forming a weak complex with the Nup214 NTD, while the R259E mutant abolished complex formation (Fig. 6). These observations are consistent with an interface between the 2 proteins, in which R259 plays a dominant role by providing multiple key salt bridges and other polar interactions in the center of the interface. Overall, our biochemical data are in excellent agreement with our crystallographic data on the interaction between Nup214 and Ddx19.
Immunofluorescence Localization of R259 Mutants.
Ddx19 is dispersed throughout the cytoplasm and enriched at the nuclear envelope, whereas Nup214 is exclusively localized at the cytoplasmic face of the NPC (8, 12). To identify the region of Nup214 that is required for the anchoring of the protein to the NPC, we analyzed the localization of a series of HA-tagged Nup214 fragments in HeLa cells (Fig. S6A). These fragments were designed, taking the previous (14) and the present structural information of the Nup214 NTD into consideration. We found that full-length Nup214, and Nup214 fragments that lack the NTD (Nup214 ΔNTD), the FG-repeats (Nup214 ΔFG), or both regions (Nup214 ΔNTD ΔFG) localize at the nuclear envelope (Fig. S6B). In contrast, a Nup214 fragment that encompasses only the NTD is dispersed throughout the cell, and does not localize to the nuclear envelope. We conclude that only the coiled-coil domain of Nup214 (Nup214 ΔNTD ΔFG, residues 450 to 1209) is involved in the anchoring of the protein to the NPC.
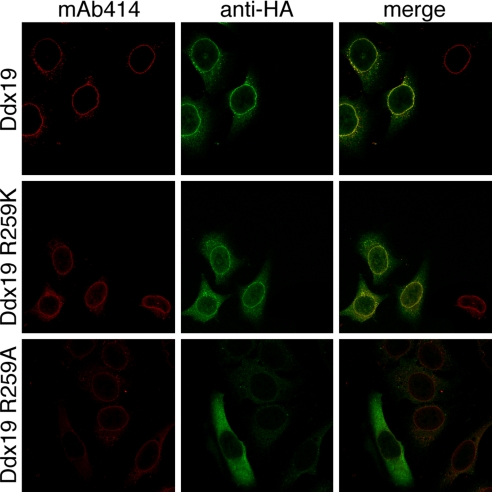
In yeast, it has been demonstrated that the deletion of the β-propeller domain of Nup159 prevents Dbp5 recruitment to the nuclear envelope (9). To test whether mutations of R259 not only disrupt the interaction between Nup214 and Ddx19 in vitro, but also inhibit the localization of Ddx19 at the nuclear envelope, we transiently transfected HeLa cells with wild-type Ddx19, and with the R259A and R259K mutants. While the wild-type Ddx19 and the R259K mutant were recruited to the NPC as predicted by our biochemical experiments, the R259A mutant failed to colocalize at the nuclear envelope (Fig. 7). Our data therefore demonstrate that R259 of Ddx19 is critical for the interaction between the 2 proteins not only in vitro, but also in vivo.
Fig. 7.
In vivo localization of Ddx19 and Ddx19 mutants. HeLa cells were transfected with Ddx19 and Ddx19 mutants containing a C-terminal HA-tag and analyzed with confocal microscopy. The monoclonal antibody mAb414 was used as a reference for nuclear envelope staining (red). The cellular localization of HA-tagged proteins was detected with an anti-HA antibody (green). The merged image reveals the colocalization of wild type Ddx19 and Ddx19 R259K with the nuclear envelope, while Ddx19 R259A displays no detectable nuclear envelope staining.
Conclusions
A key step in the transport of nuclear mRNPs to the cytoplasm is the transit through the NPC. This transit involves mRNP remodeling, whereby proteins bound to nuclear mRNA are dissociated and replaced by cytoplasmic mRNA binding proteins. The Nup214-associated helicase at the cytoplasmic side of the NPC has been implicated in this mRNP remodeling step (6, 7). Our Nup214·Ddx19 structure reveals that the interaction between the 2 proteins is mediated to a large extent via electrostatic interactions. In fact, we identified the highly conserved R259 in the N-terminal domain of Ddx19 as a key residue for the interaction with Nup214 in vitro and for nuclear envelope localization in vivo.
A salient feature of Ddx19 is the evolutionarily conserved, highly positive surface potential that suggests that Ddx19 is capable of directly interacting with mRNA. Strikingly, the highest positive surface charge is buried in the interface with Nup214. It is conceivable that the patch of these positively charged surface residues could competitively interact with mRNA. If this were the case, a plausible sequence of events might be as follows. As linearized mRNP approaches the Nup214-helicase-ADP complex, the ADP-helicase binds competitively to mRNA. Cooperatively, this could dissociate the ADP-helicase complex from Nup214, exchange ADP for ATP, strongly bind ATP-helicase to mRNA, and displace mRNA-bound protein; ATP hydrolysis would weaken the interaction with mRNA and reestablish binding of the ADP-helicase to Nup214, thereby completing 1 cycle of protein removal from a segment of mRNA. Each cycle would ratchet the mRNA toward the cytoplasm. It is conceivable that ATP hydrolysis and ADP/ATP exchange is enhanced by a specific activating protein and/or exchange factor, respectively.
The structure of the Nup214-helicase complex reported here, together with the elucidation of the structure of other nucleoporins at the cytoplasmic face of the NPC can be expected to improve our understanding of mRNA export and the accompanying remodeling of mRNP at the important interface between the NPC and the cytoplasm.
Methods
The details of molecular cloning, expression, purification, crystallization, X-ray diffraction data collection, structure determination, protein interaction analysis, and immunofluorescence experiments are described in SI Text and Tables S2–S4. In short, Nup214 NTD and Ddx19 variants were expressed by using a pET28a vector (pET28a-PreS) modified to contain a PreScission protease-cleavable N-terminal hexa-histidine tag (20). Recombinant protein was purified using several chromatographic techniques. Native X-ray diffraction data of Nup214·Ddx19 crystals were collected at the Advanced Light Source, Lawrence Berkeley National Laboratory. The structure was solved by molecular replacement using PHASER (21) and the atomic coordinates of the Nup214 NTD (14). A model was built with the program O (22) and refined with CNS (23) and Refmac (24). Data collection and refinement statistics are summarized in Table S1. The structure factors and atomic coordinates of the Nup214 NTD·Ddx19 and Nup214 NTD·Ddx19 NTD have been deposited to the Protein Data Bank with the accession codes 3FMO and 3FMP.
Supplementary Material
Acknowledgments.
We thank Daniel Andor, Martin Kampmann, Vivien Nagy, Alina Patke, Hang Shi, and Pete Stavropoulos for discussions and comments on the manuscript; D. King for mass spectrometry analysis; Stephanie Etherton for help with editing the manuscript; and Corie Ralston and the staff at beamlines 8.2.1 and 8.2.2 of the Advanced Light Source for support during data collection. The peptide array was synthesized by the Protein Center of the Rockefeller University. E.W.D. is a Dale F. and Betty Ann Frey Fellow of the Damon Runyon Cancer Research Foundation. A.H. is supported by a grant from the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3FMO and 3FMP).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813267106/DCSupplemental.
References
- 1.Hoelz A, Blobel G. Cell biology: Popping out of the nucleus. Nature. 2004;432:815–816. doi: 10.1038/432815a. [DOI] [PubMed] [Google Scholar]
- 2.Rout MP, et al. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 4.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 5.Weirich CS, et al. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 6.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 10.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Lindern M, et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suntharalingam M, Wente SR. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 14.Napetschnig J, Blobel G, Hoelz A. Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc Natl Acad Sci USA. 2007;104:1783–1788. doi: 10.1073/pnas.0610828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc Natl Acad Sci USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melcak I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 18.Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A Fence-Like Coat for the Nuclear Pore Membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci USA. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 21.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.