Abstract
Evolutionary transitions in individuality (ETIs) underlie the watershed events in the history of life on Earth, including the origins of cells, eukaryotes, plants, animals, and fungi. Each of these events constitutes an increase in the level of complexity, as groups of individuals become individuals in their own right. Among the best-studied ETIs is the origin of multicellularity in the green alga Volvox, a model system for the evolution of multicellularity and cellular differentiation. Since its divergence from unicellular ancestors, Volvox has evolved into a highly integrated multicellular organism with cellular specialization, a complex developmental program, and a high degree of coordination among cells. Remarkably, all of these changes were previously thought to have occurred in the last 50–75 million years. Here we estimate divergence times using a multigene data set with multiple fossil calibrations and use these estimates to infer the times of developmental changes relevant to the evolution of multicellularity. Our results show that Volvox diverged from unicellular ancestors at least 200 million years ago. Two key innovations resulting from an early cycle of cooperation, conflict and conflict mediation led to a rapid integration and radiation of multicellular forms in this group. This is the only ETI for which a detailed timeline has been established, but multilevel selection theory predicts that similar changes must have occurred during other ETIs.
Keywords: evolution, multicellularity, multilevel selection, transitions in individuality, Volvox
The history of life on Earth has involved a number of evolutionary transitions in individuality (ETIs), in which groups of once-autonomous individuals became new individuals. Through the transfer of fitness from the individuals making up the group to the group itself, a new entity was formed with a single fitness and a single evolutionary fate. In this way, groups of interacting molecular replicators became single-celled organisms, prokaryotic cells became a primitive eukaryote, groups of single-celled organisms became multicellular organisms, and groups of multicellular organisms became social individuals (as in the social insects). In many cases such transitions have opened up entire new adaptive landscapes leading to vast radiations as completely new ways of being alive became available (e.g., cellular life, eukaryotes, plants, and animals). Understanding how and why groups of individuals become new kinds of individuals is a major challenge in explaining the history of life.
The transition from unicellular to multicellular life is the paradigm case of the integration of lower-level individuals (cells) into a new higher-level individual—the multicellular organism. This transition has occurred dozens of times independently, for example in the red algae, brown algae, land plants, animals, and fungi (reviewed in ref. 1). Among the best-studied ETIs is the origin of multicellularity in the green alga Volvox and its relatives (the volvocine algae), which have been developed as a model for the developmental genetics of multicellularity and cellular differentiation (2, 3).
The volvocine algae include 3 families of haploid, facultatively sexual eukaryotes totaling ≈50 species in the Chlorophycean order Volvocales. Together with unicellular relatives in the genera Chlamydomonas and Vitreochlamys, members of this group span a range of sizes and levels of complexity from unicells to macroscopic multicellular organisms with cellular differentiation (examples are shown in Fig. 2). The smallest multicellular forms are in the family Tetrabaenaceae (Basichlamys and Tetrabaena) and are made up of 4 Chlamydomonas-like cells held together in a common extracellular matrix. Members of the Goniaceae include Gonium and Astrephomene. Gonium ranges from 8 to 32 cells arranged in a flat or slightly curved plate. Astrephomene consists of 32–64 cells arranged on the perimeter of a spheroid, with 2–4 sterile somatic cells at the posterior pole. The diverse Volvocaceae include 7 genera (Eudorina, Pandorina, Platydorina, Pleodorina, Volvox, Volvulina, and Yamagishiella) ranging from 16 cells up to 50,000 cells. Members of this family are spheroidal (with the exception of Platydorina, a secondarily flattened sphere) and may have complete, partial, or no division of labor between reproductive and somatic cells. This diversity of intermediate grades of organization provides a unique window on the evolutionary transition from single cells to fully differentiated multicellular individuals such as Volvox.
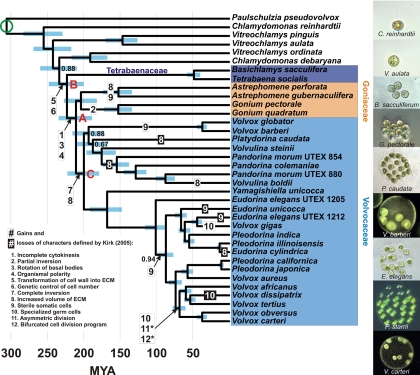
Fig. 2.
Chronogram showing estimated divergence times among volvocine algae. Colored boxes identify the 3 multicellular families; ingroup species not highlighted in this manner are unicellular (Paulschulzia pseudovolvox, the outgroup, represents a separate origin of multicellularity). Blue bars are the central 95% of estimates from 300 Bayesian posterior trees. Bayesian posterior probabilities <0.95 are shown. The green circle indicates the calibration estimated in the broad-scale analyses. Red letters indicate nodes referred to in the text. Character state changes are those supported by hypothesis tests in ref. 5. We have retained Kirk's (4) original numbering for these steps. *, steps 11 and 12 may have had 2 separate origins in the clade including V. africanus and V. carteri.
On the basis of the development of V. carteri, Kirk (4) inferred a series of evolutionary changes involved in the transition to differentiated multicellularity. Most of the inferred intermediate stages in this transition are approximated by extant forms. Herron and Michod (5) reconstructed Kirk's steps in a phylogenetic framework, revealing a complex picture in which phylogeny does not strictly mirror ontogeny; some traits have multiple independent origins and reversals from derived to ancestral states. No previous work has rigorously estimated the timing of these changes.
Volvox has previously been thought to represent a recent experiment in multicellularity, having diverged from unicellular relatives only 50–75 million years ago (Ma). This estimate of divergence time was based on a single gene with a single fossil calibration, and no subsequent study has independently estimated the age of the multicellular volvocine algae. Nevertheless, the recent origin of multicellularity in this group has been widely accepted and has been integrated into thinking in this area. For example, the genetic simplicity underpinning development in these organisms has been thought to be a result of the relatively short time since they became multicellular (3, 6).
Results and Discussion
We estimated divergence times within the volvocine algae using a multigene data set with multiple fossil calibrations. Our results show, in contrast to all previous thought, that Volvox diverged from its closest unicellular relatives ≈234 Ma (95% Bayesian credibility interval 209–260 Ma) (Fig. 2) and that most of the developmental changes involved in the transition to multicellularity took place during an early, rapid radiation soon after this divergence. The 3 main lineages of multicellular volvocine algae, represented by the 3 modern families, were established by 200 Ma (179–222) (Fig. 2, node A).
We now consider the timing of morphological and developmental changes in more detail, using the series of 12 ontogenetic changes (Fig. 2) enumerated by Kirk for Volvox development (4). For the sake of continuity, we have retained Kirk's original numbering for the changes he identified. In addition to estimating dates for these changes, we compare traits of extant species with those reconstructed for ancestral species. Although the changes identified by Kirk address an important subset of morphological and developmental characters involved in the transition to multicellularity, it is important to keep in mind that our comparisons address only these 12 characters. The extant and ancestral species we compare may have differed in terms of characters we do not address.
Within 33.9 million years (My) (26.2–42.6) after the divergence from unicellular ancestors, a number of developmental changes related to the evolution of multicellularity took place (steps 1 and 3–8 in Fig. 2). The initial shift from a unicellular to a multicellular lifestyle occurred by 223 Ma (200–248; Fig. 2, node B), when the daughter cells of a Chlamydomonas-like ancestor became embedded in a common extracellular matrix (ECM) (Kirk's step 5). Around the same time, the number of daughter cells produced by a mother cell shifted from environmental to genetic control (step 6). Together, these changes led to a body plan morphologically and developmentally similar to the modern Tetrabaenaceae (Basichlamys and Tetrabaena).
By 211 Ma (189–234), the ancestors of Goniaceae and Volvocaceae had evolved a combination of characters found in modern Gonium (Fig. 2, node A). Cytokinesis became incomplete, leaving daughter cells connected by cytoplasmic bridges (step 1). The basal bodies of peripheral cells rotated to orient the flagella to beat in parallel (step 3), resulting in more efficient locomotion and establishing a center-to-edge polarity (step 4).
By 200 Ma (179–222), the ancestors of Volvocaceae had evolved a body plan consisting of a transparent sphere with the cells on the periphery, as in modern Volvulina, Yamagishiella, and Eudorina (Fig. 2, node C). Complete inversion established a spherical body plan with the flagella on the exterior (step 7), changing the center-to-edge polarity into an anterior–posterior polarity. The volume of ECM increased dramatically, leading to a hollow appearance with the cells on the periphery of the sphere (step 8).
The time span in which all of the above changes took place makes up <20% of the total time since the ancestors of the Volvocaceae diverged from single-celled ancestors. In this relatively short time, they evolved from single cells into tightly integrated (but undifferentiated) colonies with a developmental program (inversion) and a mode of locomotion that both required a high degree of coordination among cells.
Changes subsequent to the divergence of Goniaceae and Volvocaceae (Fig. 2, node A; 189–234 Ma) are mostly related to cellular specialization, and the associated dates cannot always be restricted to a usefully narrow range. The first step in a reproductive vs. somatic division of labor, the origin of sterile somatic cells, happened between 134 and 227 Ma in the lineage leading to Astrephomene, between 30.7 and 216 Ma in the lineage leading to Volvox barberi and V. globator, and between 76.6 and 116 Ma in the lineage leading to Pleodorina and the remaining species of Volvox. The specialized germ cells found in the genus Volvox originated between 65.0 and 98.7 Ma in the lineage leading to V. aureus and V. carteri and within the last 51.9 My in the lineage leading to V. gigas.
Although the striking transition in complexity from unicellular to multicellular in volvocine algae has been broken down into a series of modest changes, our results on the timing of evolution in this group reveal that the transition did not occur by smooth, constant change. Rather, the changes occurred sporadically, with periods of rapid change and occasional reversals from derived to ancestral states interspersed among long periods of stasis. For example, the basic body plan common to modern Eudorina, Volvulina, and Yamagishiella—undifferentiated spheroids with cells arranged at the periphery—was established within a few tens of millions of years after the initial divergence from unicellular ancestors but has been stable in several lineages for 200 My (179–222; Fig. 2, node C). These results support the view that the various grades of organization in volvocine algae represent not transitional forms but rather alternative stable states (7), each well adapted to a particular environment.
Implications for Evolutionary Transitions in Individuality in General.
ETIs occur when individuals combine to form new individuals, as occurred with the origin of cells, of eukaryotes, of multicellular organisms, and of integrated societies. During each of these ETIs, fitness was transferred from the individuals making up the group to the group itself, forming a new individual with a single fitness and a single evolutionary fate (8–10). According to multilevel selection theory (MLS) this transfer of fitness requires the evolution of cooperation and conflict mediators, which enhance cooperation by restricting the opportunities for defection and reducing selection among the members of the group (10).
The question of how a high degree of integration and coordination among cells arose so quickly in the volvocine lineage can be addressed using the framework of MLS. Several of the key changes involved in the transition to differentiated multicellularity in the volvocine algae may be interpreted as components of cycles of cooperation, conflict, and conflict mediation predicted by MLS (5). According to this view, one such cycle took place soon after the initial transition to multicellularity, when conflicts over the production of ECM were resolved by the shift to genetic control of cell number, which limits the benefits of defection (11). The rapid pace of subsequent changes (steps 1, 3, 4, 7, and 8) suggests that the resolution of this conflict was a key innovation enabling the diversification, integration, and increase in complexity that followed.
The specific details of this or any other ETI are bound to be unique, but there are basic principles of MLS that are relevant to all such transitions (8, 10). These principles and the results of the present study suggest a strategy for understanding other ETIs, such as the origins of multicellularity in plants and animals. Because of the lack of extant intermediate forms, the primary source of evidence in these cases has been the fossil record, which is absent for the volvocine algae. However, the tempo and mode of change in the volvocine algae suggest that actual transitional forms were restricted to narrow temporal ranges. If this is true for other origins of multicellularity, fossil evidence of the early steps in these ancient transitions may be scant, and comparative genomics may be a more fruitful approach. The importance of MLS in ETIs gives us an idea of the kinds of changes we should be looking for in these other groups. The challenge for understanding the origins of multicellularity in the animals and plants is to identify the key innovations along with the underlying genes leading to cooperation among cells, the resulting conflicts, and, most importantly, the mechanisms by which these conflicts were mediated.
Methods
We ran 2 sets of analyses for divergence time estimation, 1 with a broad taxonomic scale but only a few representatives of volvocine algae (Fig. 1) and 1 with a narrower taxonomic scale but dense sampling within the volvocine algae (Fig. 2). We used the results of the broad-scale analyses to calibrate the basal divergence in the fine-scale analyses.
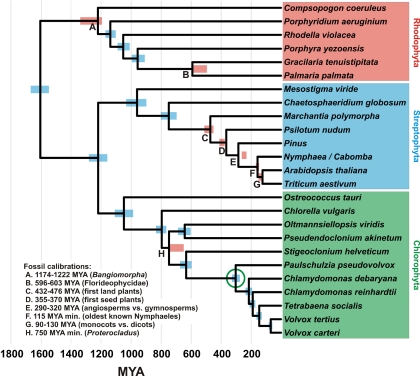
Fig. 1.
Chronogram showing estimated divergence times among photosynthetic eukaryotes. Blue bars are the central 95% of estimates from 300 Bayesian posterior trees. Red bars are the central 95% of dates inferred during fossil cross-validation. Bayesian posterior probabilities <0.95 are shown. References for the fossil calibrations are given in the text. The green circle indicates the most recent common ancestor of Paulschulzia pseudovolvox and Volvox carteri, the divergence used to calibrate the fine-scale analyses.
Broad-Scale Analyses.
We based our broad-scale phylogenetic analysis (Fig. 1) on a concatenated alignment of 5 chloroplast protein-coding genes [beta subunit of ATP synthase, P700 chlorophyll a-apoprotein A1 (psaA), P700 chlorophyll a-apoprotein A2 (psaB), photosystem II CP43 apoprotein (psbC), and large subunit of rubisco] and the nuclear small ribosomal subunit (18S) from representative red algae, green algae, and land plants, using the glaucophyte alga Cyanophora paradoxa as an outgroup [supporting information (SI) Table S1]. Two sequences in the alignment were chimeras of two different species: the operational taxonomic unit labeled “Pinus” in Fig. 1 consisted of chloroplast sequences from Pinus thunbergii and an 18S sequence from P. luchuensis; and that labeled “Nymphaea/Cabomba” consisted of chloroplast sequences from Nymphaea alba and an 18S sequence from Cabomba caroliniana.
We ran all analyses on 4 subsets of the full alignment, each with 2 partitioning strategies: (i) a partition for each codon position and 1 for 18S and (ii) a partition for protein-coding genes excluding third codon positions and 1 for 18S. In each case we used a separate substitution model for each partition, chosen using the Akaike information criterion in MrModelTest (12). Consensus trees and Bayesian posterior probabilities were based on the combined post-burn-in trees from 4 independent runs in MrBayes (13).
We estimated divergence times using Langley–Fitch (LF), penalized likelihood with an additive penalty (PL) (14), and nonparametric rate smoothing (NPRS) (15) algorithms in r8s (16) and Bayesian inference in MultiDivTime (17). For each analysis, divergence time estimates were based on a sample of 300 Bayesian posterior trees, filtered to be compatible with the 50% consensus tree (filtering was necessary to obtain reliable results from r8s, in which a node is defined by 2 descendants). The performance of each combination of data set, partitioning strategy, and algorithm for date estimation was evaluated by fossil cross-validation, in which analyses are rerun excluding each calibration in turn and the date for each calibration is inferred and compared to the fossil date.
All of the primary fossil calibrations used in this study were used for the same purpose in previous estimates of eukaryotic divergence times (18, 19). As in those studies, we included uncertainty in the ages of fossils by constraining each calibration to a range rather than to a point estimate. We constrained the divergence between red algae and (green algae + land plants) to a maximum of 3500 My, the age of the first known fossils (20, 21). Within the red algae, we constrained the deepest divergence to the age of the oldest known red algal fossil, Bangiomorpha pubescens (1174–1222 My) (22) and that between Gracilaria tenuistipitata and Palmaria palmata to the age of the Doushantuo Florideophyte fossils (595–603 My) (23, 24). Within the green plant lineage, we used the following constraints: the first appearance of spore tetrads indicating the emergence of land plants (432–476 My) (25), the oldest known seeds indicating the emergence of seed plants (355–370 My) (26), several fossils indicating the divergence between angiosperms and gymnosperms (290–320 My) (27), the earliest known fossil Nymphaeales indicating their divergence from eudicots (115 My minimum) (28), and the Early Cretaceous palynological diversification of angiosperms indicating the divergence of monocots and eudicots (90–130 My) (29). Within the green algae, we constrained the divergence between the Ulvophyceae and the Volvocales to the age of the cladophoracean fossil Proterocladus (750 My minimum) (30).
The best-performing analysis, and the one with which our fine-scale analyses were calibrated, was a 4-gene data set (psaA, psaB, psbC, and 18S), with a partition for each codon position and one for 18S, analyzed using the LF algorithm. The 95% Bayesian credibility intervals (BCIs) of dates inferred during fossil cross-validation overlapped the ranges used for fossil calibration in all but 2 cases: the inferred origin of seed plants was slightly too old (381–417 vs. 355–370 Ma; Fig. 1, node D), and the inferred divergence of angiosperms from gymnosperms was too young (236–268 vs. 290–320 Ma; Fig. 1, node E). No single fossil calibration had a large effect on estimated divergence times; the inferred date of the divergence used in the fine-scale analyses (see below) when each calibration was removed in turn ranged from 296 to 310 Ma. The maximum time constraint of 3500 Ma for the root of the tree had no effect on estimates of divergence times. Bayesian analyses recovered dates similar to those found using r8s, with overlapping 95% confidence intervals.
Fine-Scale Analyses.
Because analyses with large amounts of missing data were found to perform poorly in the broad-scale analyses, we based our inference of divergence times within the volvocine algae (Fig. 2) on Bayesian posterior trees from a supplementary analysis (“reduced data set”) in (5), which included only those taxa for which no gene sequences were missing. To check for the possibility that our results were affected by saturation of substitutions at third codon positions, we ran a second set of analyses on the same data set excluding third codon positions. Median divergence time estimates from the analysis excluding third codon positions were highly correlated with those from the analysis including all 3 codon positions (R2 = 0.991) and fell within the 95% BCI in every case but 1 (Vitreochlamys aulata vs. V. pinguis) on which none of our conclusions are based.
We calibrated divergence time estimates within the volvocine algae using the divergence of Paulschulzia pseudovolvox from V. carteri, for several reasons. First, this divergence is basal in the fine-scale tree, and estimates of its age were remarkably consistent across analyses and robust to removal of fossil calibrations. In addition, the branching order of Chlamydomonas reinhardtii, C. debaryana, and the multicellular volvocine algae (Tetrabaenaceae, Goniaceae, and Volvocaceae) differed between the 2 sets of analyses. This discrepancy is not surprising given the short branch lengths of the relevant internodes, but it implies that choosing either of these later divergences as a calibration would have biased the results of the fine-scale analyses in one direction or the other.
When the date of the basal divergence was delimited with the upper and lower bounds of the 95% BCI from the broad-scale analysis (282–333 Ma), r8s used only the lower end of the range, yielding artificially narrow confidence intervals (effectively failing to account for uncertainty in the calibration). To rectify this, we based our final estimates on 100 iterations in which each of the 300 volvocine trees was randomly assigned one of the 300 dates inferred for the divergence in question.
Robustness of Divergence Time Estimates.
Molecular methods of estimating divergence times have been criticized even to the point of suggesting that they should not be used (31), but in cases in which no relevant fossil record exists they are the only methods available. One of the harshest criticisms of such methods (31) can also be taken as a prescription for their proper use. We have carefully followed this prescription by using multiple genes, multiple fossil calibrations, and methods allowing for rate heterogeneity; by accounting for uncertainty both in the ages of fossils and in the phylogeny; by reporting confidence intervals that include all of these sources of error; and by evaluating our results using fossil cross-validation. Estimates of divergence times within the volvocine algae are based on a secondary calibration, but we have rigorously accounted for the uncertainty in this estimate. Although uncertainties remain, this study represents the best information currently available.
Supplementary Material
Acknowledgments.
We thank Darren Boss and Mike Sanderson for help with the r8s analyses, Adam Bjork and Joel Wertheim for helpful discussions, and Aurora Nedelcu and Deborah Shelton for their comments on earlier versions of the manuscript. In addition, we thank David Kirk for a helpful discussion on the nature of complete differentiation. This work was supported by the University of Arizona College of Science, the University of Arizona Department of Ecology and Evolutionary Biology, a graduate student award (to M.D.H.) from the Society of Systematic Biologists, and a grant-in-aid of research (to M.D.H.) from Sigma Xi.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811205106/DCSupplemental.
References
- 1.Bonner JT. The origins of multicellularity. Integr Biol. 1998;1:27–36. [Google Scholar]
- 2.Kirk DL. Germ-soma differentiation in Volvox. Dev Biol. 2001;238:213–223. doi: 10.1006/dbio.2001.0402. [DOI] [PubMed] [Google Scholar]
- 3.Kirk DL. Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge, UK: Cambridge Univ Press; 1998. [Google Scholar]
- 4.Kirk DL. A twelve-step program for evolving multicellularity and a division of labor. BioEssays. 2005;27:299–310. doi: 10.1002/bies.20197. [DOI] [PubMed] [Google Scholar]
- 5.Herron MD, Michod RE. Evolution of complexity in the volvocine algae: transitions in individuality through Darwin's eye. Evolution. 2008;62:436–451. doi: 10.1111/j.1558-5646.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 6.Rausch H, Larsen N, Schmitt R. Phylogenetic relationships of the green alga Volvox carteri deduced from small-subunit ribosomal RNA comparisons. J Mol Evol. 1989;29:255–265. doi: 10.1007/BF02100209. [DOI] [PubMed] [Google Scholar]
- 7.Larson A, Kirk MM, Kirk DL. Molecular phylogeny of the volvocine flagellates. Mol Biol Evol. 1992;9:85–105. doi: 10.1093/oxfordjournals.molbev.a040710. [DOI] [PubMed] [Google Scholar]
- 8.Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. San Francisco: Freeman; 1995. [Google Scholar]
- 9.Buss LW. The Evolution of Individuality. Princeton: Princeton Univ Press; 1987. [Google Scholar]
- 10.Michod RE. Darwinian Dynamics, Evolutionary Transitions in Fitness and Individuality. Princeton: Princeton Univ Press; 1999. [Google Scholar]
- 11.Michod RE. In: Genetic and Cultural Evolution of Cooperation. Hammerstein P, editor. Cambridge, MA: MIT Press; 2003. pp. 261–307. [Google Scholar]
- 12.Nylander JAA. MrModeltest 2.0. 2002 Available at www.ebc.uu.se/systzoo/staff/nylander.html.
- 13.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 14.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 15.Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 16.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 17.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 18.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 19.Berney C, Pawlowski J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc R Soc Lond B Biol. 2006;273:1867–1872. doi: 10.1098/rspb.2006.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schopf JW, Kudryavtsev AB, Agresti DG, Wdowiak TJ, Czaja AD. Laser-Raman imagery of Earth's earliest fossils. Nature. 2002;416:73–76. doi: 10.1038/416073a. [DOI] [PubMed] [Google Scholar]
- 21.Westall F, et al. Early Archean fossil bacteria and biofilms in hydrothermally-influenced sediments from the Barberton greenstone belt, South Africa. Precambrian Res. 2001;106:93–116. [Google Scholar]
- 22.Butterfield NJ. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. [Google Scholar]
- 23.Barfod GH, et al. New Lu-Hf and Pb-Pb age constraints on the earliest animal fossils. Earth Planet Sci Lett. 2002;201:203–212. [Google Scholar]
- 24.Xiao S, Zhang Y, Knoll AH. Three-dimensional preservation of algae and animal embryos in a Neoproterozoic phosphorite. Nature. 1998;391:553–558. [Google Scholar]
- 25.Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- 26.Gillespie WH, Rothwell GW, Scheckler SE. The earliest seeds. Nature. 1981;293:462–464. [Google Scholar]
- 27.Soltis PS, Soltis DE, Savolainen V, Crane PR, Barraclough TG. Rate heterogeneity among lineages of tracheophytes: integration of molecular and fossil data and evidence for molecular living fossils. Proc Natl Acad Sci USA. 2002;99:4430–4435. doi: 10.1073/pnas.032087199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friis EM, Pedersen KR, Crane PR. Fossil evidence of water lilies (Nymphaeales) in the Early Cretaceous. Nature. 2001;410:357–360. doi: 10.1038/35066557. [DOI] [PubMed] [Google Scholar]
- 29.Crane PR, Friis EM, Pedersen KR. The origin and early diversification of angiosperms. Nature. 1995;374:27–33. [Google Scholar]
- 30.Butterfield NJ, Knoll AH, Swett K. Paleobiology of the Neoproterozoic Svanbergfjellet Formation, Spitsbergen. Fossils and Strata. 1994;34:1–84. [Google Scholar]
- 31.Shaul S, Graur D. Playing chicken (Gallus gallus): methodological inconsistencies of molecular divergence date estimates due to secondary calibration points. Gene. 2002;300:59–61. doi: 10.1016/s0378-1119(02)00851-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.