Abstract
Enzymes that hydrolyze complex carbohydrates play important roles in numerous biological processes that result in the maintenance of marine and terrestrial life. These enzymes often contain noncatalytic carbohydrate binding modules (CBMs) that have important substrate-targeting functions. In general, there is a tight correlation between the ligands recognized by bacterial CBMs and the substrate specificity of the appended catalytic modules. Through high-resolution structural studies, we demonstrate that the architecture of the ligand binding sites of 4 distinct family 35 CBMs (CBM35s), appended to 3 plant cell wall hydrolases and the exo-β-d-glucosaminidase CsxA, which contributes to the detoxification and metabolism of an antibacterial fungal polysaccharide, is highly conserved and imparts specificity for glucuronic acid and/or Δ4,5-anhydrogalaturonic acid (Δ4,5-GalA). Δ4,5-GalA is released from pectin by the action of pectate lyases and as such acts as a signature molecule for plant cell wall degradation. Thus, the CBM35s appended to the 3 plant cell wall hydrolases, rather than targeting the substrates of the cognate catalytic modules, direct their appended enzymes to regions of the plant that are being actively degraded. Significantly, the CBM35 component of CsxA anchors the enzyme to the bacterial cell wall via its capacity to bind uronic acid sugars. This latter observation reveals an unusual mechanism for bacterial cell wall enzyme attachment. This report shows that the biological role of CBM35s is not dictated solely by their carbohydrate specificities but also by the context of their target ligands.
Keywords: carbohydrate protein binding, enzyme targeting, plant cell wall degradation, protein cell attachment, X-ray crystallography
A generic feature of enzymes that hydrolyze complex carbohydrates is their modular structure. The most common noncatalytic modules are the carbohydrate binding modules (CBMs), which are grouped into sequence-based families (1). The general function of CBMs is to promote the interaction of the enzyme with the target substrate, thereby increasing the efficiency of catalysis (2–4). CBMs are prevalent in plant cell wall degrading enzymes where they direct the cognate catalytic modules to their target substrate within these complex composite structures (1). The release of sugars from the recalcitrant carbohydrates of plant cell walls provides essential nutrients for microbial ecosystems that support marine and terrestrial life. Indeed, the ability of mammalian herbivores to access their major source of nutrients depends on the plant cell wall degrading activity of intestinal microorganisms (5). Within an industrial context the microbial enzymes that catalyze this process are integral to the exploitation of lignocellulose as an environmentally sustainable substrate for biofuel production (6).
In addition to their contribution to plant cell wall degradation, there are increasing numbers of examples of CBMs playing a prominent role in other biological systems. For example, bacterial pathogens also deploy CBM-containing enzymes as virulence factors (7, 8), whereas deficiencies in CBM-containing human proteins, such as phosphatases that regulate glycogen synthase (9, 10), can lead to serious diseases. CBMs are also located in bacterial enzymes that are components of protection systems. Thus, soil bacteria, such as Amycolatopsis orientalis, detoxify and metabolize the fungal polysaccharide chitosan by secreting an endo-chitosanase (Csn) and the CBM-containing exo-β-d-glucosaminidase, CsxA, which together depolymerize the polymer (11, 12).
As a general rule, the ligand specificity of bacterial CBMs reflects the substrates attacked by the cognate catalytic modules (13–15). An intriguing divergence from this general rule is provided by 4 family 35 CBMs (CBM35s) appended to CsxA and 3 different plant cell wall degrading enzymes (detailed in Results). These modules, despite being appended to enzymes with different substrate specificities, display extensive sequence similarity indicating that the CBMs recognize highly related ligands. Here, we show that all 4 CBM35s display specificity for Δ4,5-anhydrogalacturonic acid (Δ4,5-GalA), although 2 of the proteins also interact with glucuronic acid (GlcA). X-ray crystallographic data reveal that the ligand binding site is highly conserved in the 4 CBM35s. Subtle modifications to the ligand binding site in 2 of the CBM35s enable these modules to discriminate between Δ4,5-GalA, a signature molecule for plant cell wall degradation, and GlcA, which is presented on the surface of several bacteria. By targeting Δ4,5-GalA, the CBM35s direct the plant cell wall hydrolases to accessible regions of the plant that are either undergoing biological degradation or remodeling. In contrast, the CBM35 appended to the defense enzyme CsxA appears to act as an adhesin molecule that anchors the glucosaminidase to the surface of the bacterium, rather than targeting the substrate chitosan. This report shows how CBMs that display conserved ligand specificities can exhibit divergent biological functions.
Results
Uronic Acid Recognition by CBM35.
CBMs generally display ligand specificities that reflect the substrates hydrolyzed by the appended catalytic modules (see ref. 1 for review). It is intriguing, therefore, that 4 CBM35s, which exhibit a high level of amino acid sequence identity (>38%), are components of enzymes where the cognate catalytic modules display highly divergent substrate specificities. The enzymes containing these CBMs are as follows: Rhamnogalauronan acetyl esterase Rgae12A (Cthe_3141) from Clostridium thermocellum, a PL10 pectate lyase (Pel10) from an environmental isolate (16), the exo-β-d-glucosaminidase CsxA from A. orientalis (11), and the xylanase CjXyn10B from Cellvibrio japonicus (17). The 4 CBM35s are designated, henceforth, as Rhe-CBM35 (Rgae12A), Pel-CBM35 (Pel10), Chi-CBM35 (CsxA), and Xyl-CBM35 (CjXyn10B).
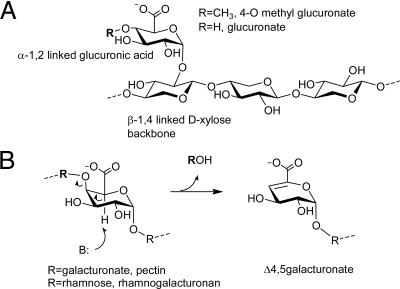
Although a previous study showed that Xyl-CBM35 bound to an unknown component of xylans (13), the specificities of the other CBM35s have not been investigated. Here, isothermal titration calorimetry (ITC) shows that all of the CBM35s appended to plant cell wall degrading enzymes bind to polygalacturonic acid predigested with a pectate lyase, but the proteins do not interact with the intact polysaccharide. The CBMs do not bind to pectin degraded with a hydrolytic polygalacturonase (Table 1), hinting that Δ4,5-GalA, the product of the β-elimination action (Fig. 1), is the target ligand. Crystallographic data revealed that Chi-CBM35 also bound to the reaction products released from homogalacturonan by a pectate lyase. These data suggest that all 4 CBM35s recognize Δ4,5-GalA, as the anhydro sugar is located at the nonreducing end of the oligosaccharides generated by pectate lyase action (18). These CBM35s do not bind to GalA or a range of neutral sugars or their corresponding oligosaccharides (Table S1). ITC shows that Xyl-CBM35 and Chi-CBM35 also bind GlcA and Rhe-CBM35 interacts weakly with the uronic acid, whereas Pel-CBM35 displays no affinity for this sugar. Pel-CBM35 bound to calcium and, consistent with previous studies on Xyl-CBM35, all of the CBM35s derived from the plant cell wall degrading enzymes required the metal ion to bind their carbohydrate ligands (Table 2). GlcA and Δ4,5-GalA recognition by the CBMs is dominated by enthalpic forces with the change in entropy making an unfavorable contribution to ligand binding (Table 1). This thermodynamic signature is typical of the binding of proteins to soluble carbohydrates (reviewed in ref. 1).
Table 1.
Thermodynamics of the binding of CBM35s to uronic acids
| CBM35 | Ligand | KA, M−1 | ΔG, kcal mol−1 | ΔH, kcal mol−1 | TΔS, kcal mol−1 | n |
|---|---|---|---|---|---|---|
| Xyl-CBM35 | GlcA | 4.5 (±0.0) × 104 | −6.3 (±0.0) | −9.6 (±0.0) | −3.3 (±0.0) | 1.1 (±0.0) |
| Xyl-CBM35 | Δ4,5-GalA* | 2.4 (±0.0) × 105 | −7.34 (±0.0) | −10.40 (±0.0) | −3.06 (±0.0) | 1.0 (±0.0) |
| Pel-CBM35 | GlcA | NB | — | — | — | — |
| Pel-CBM35 | Δ4,5-GalA | 4.7 (±0.4) × 104 | −6.4 (±0.0) | −8.2 (±0.2) | −1.8 (±0.2) | 1.1 (±0.0) |
| Pel-CBM35 | OligoGalA† | NB | — | — | — | — |
| Rhe-CBM35 | GlcA | 2.3 (±0.1) × 103 | −4.6 (±0.0) | −6.3 (±0.2) | −1.9 (±0.2) | 1.0 (±0.0) |
| Rhe-CBM35 | Δ4,5-GalA | 9.5 (±0.2) × 104 | −6.8 (±0.0) | −10.0 (±0.1) | −3.20 (±0.1) | 1.1 (±0.0) |
| Chi-CBM35 | GlcA | 1.9 (±0.0) × 104 | −5.8 (±0.0) | −8.2 (±0.1) | −2.4 (±0.1) | 1.2 (±0.0) |
NB, no binding detected.
*Δ4,5-GalA is the digestion product produced by the action of CjPel10A on polygalacturonic acid which generates predominately the disaccharide Δ4,5-GalAα 1,4Gal and the trisaccharide Δ4,5-GalAα 1,4Galα 1,4Gal.
†The ligand comprised polygalacturonic acid hydrolyzed with polygalacturonase. The products are: GalA, GalAα 1,4Gal, and GalAα 1,4Galα 1,4Gal.
Fig. 1.
Ligands targeted by the CBM35 domains described in this work. (A) α-1,2 linked GlcA moiety in glucuronoxylan is shown. (B) Δ4,5GalA (4,5anhydrogalactosyl) moiety produced by the action of lyases (β-eliminases) on pectin (a polysaccharide of α-1,4 linked galacturonides) or rhamnogalacturonan is revealed.
Table 2.
Affinity of wild type and mutants of Pel-CBM35 for metal and carbohydrate ligands
| Protein | Ligand | KA, M−1 |
|---|---|---|
| Apo Pel-CBM35* | Calcium | 3.2 × 104 |
| Pel-CBM35 + EDTA + calcium | Δ4,5-GalA | 1.7 × 104 |
| Xyl-CBM35 + EDTA + calcium | Δ4,5-GalA | 1.8 × 105 |
| Rhe-CBM35 + EDTA + calcium | Δ4,5-GalA | 4.3 × 104 |
| Pel-CBM35 D32A/K21N | Δ4,5-GalA | 4.1 × 103 |
| Pel-CBM35 D32A/K21N | GlcA | 2.7 × 103 |
| Pel-CBM35 D32N | GlcA | Weak binding, <103 |
| Pel-CBM35 D32N | Δ4,5-GalA | 1.1 × 104 |
| Pel-CBM35 K21N | GlcA | NB |
| Pel-CBM35 K21A | Δ4,5-GalA | 2.6 × 104 |
EDTA was at 2 mM and calcium was at 10 mM. NB, no binding detected.
*Apo Pel-CBM35 was treated with Chelex-100 to remove any bound divalent metal ions.
Δ4,5-GalA, appearing as a result of pectate degradation or remodeling, is a signature molecule for plant cell wall damage. Thus, the likely function of the CBM35s located in the plant cell wall hydrolases is to target the cognate enzymes to regions of the plant that are particularly accessible to enzyme attack (see Discussion). It is also evident that Xyl-CBM35 binds to xylan but does not interact with unsubstituted xylooligosaccharides (13). To explore the unusual basis for xylan recognition, the polysaccharide was digested with xylanases and arabinofuranosidases to completion, and the products were fractionated (see SI Materials and Methods). ITC experiments identified a negatively charged oligosaccharide fraction containing GlcA (identified by HPLC analysis of the component sugars) bound to Xyl-CBM35. Thus, the capacity of Xyl-CBM35 to recognize xylan is likely to reflect the presence of GlcA side chains on the polymer. The protein, however, did not recognize 4-methyl-d-glucuronic acid (MeGlcA) (Table S1). Recent studies have shown that the ratio of GlcA and MeGlcA in xylans depends on the balance between the rate of glucuronoxylan synthesis and the rate at which the GlcA side chains in nascent glucuronoxylan are methylated (19). It has been proposed that in model plants, such as Arabidopsis, the rate of GlcA methylation is lower than glucuronoxylan synthesis, explaining the presence of significant quantities of the unmethylated uronic acid (20). Although the biological rationale for targeting the unmethylated uronic is unclear, we speculate that regions of the glucuronoxylans containing GlcA rather than MeGlcA may have a more open structure. Thus, the Xyl-CBM35 may be directing its appended catalytic modules to regions of the hemicellulose that are particularly susceptible to enzyme degradation.
Chi-CBM35 Is a Bacterial Adhesion Molecule.
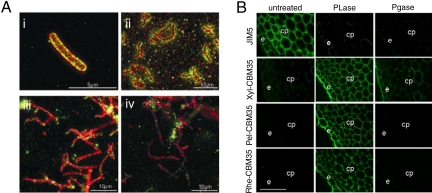
The data described above indicate that Chi-CBM35 does not target CsxA to its substrate chitosan, which lacks uronic acid. GlcA, however, is a component of some bacterial cell walls (21, 22) and thus Chi-CBM35 may anchor CsxA to the A. orientalis cell wall through an interaction with the exposed surface polysaccharides. This view is supported by in vivo localization studies revealed that both the catalytic module and CBM of CsxA are closely associated with the perimeter of A. orientalis cells (Fig. 2Ai). In contrast, Csn, a protein known to be secreted into the extracellular milieu (12), did not colocalize with A. orientalis cells, but rather was found in diffuse halos around the cells and associated with extracellular foci that were likely to be chitosan particles (Fig. 2Aii). Washing the A. orientalis cells with GlcA, but not glucose, removed endogenous CsxA from the cell surface of the bacterium (Fig. 2 Aiii and Aiv).
Fig. 2.
Binding of CBM35s to their target ligands. (A Upper) Binding of Chi-CBM35 to the surface of A. orientalis. The confocal immunofluorescence image shows CsxA (red), Chi-CBM35 (green), and colocalization of the 2 proteins (orange) at the A. orientalis cell wall level (i). An epi-fluorescence image reveals the difference in cellular localization between a known secreted protein, Csn (green), and CsxA (red) (ii). (A Lower) Epi-fluorescence images of CsxA (red) and Chi-CBM35 (green) colocalization at the cell wall level. The bacterial DNA was stained in blue. Before heat fixation the mycelia of A. orientalis were washed twice for 10 min, with 100 mM glucose (iii) or 100 mM GlcA (iv). (Magnification: i, 5,000×; ii, 1,250×; iii and iv, 1,380×.) (B) Binding of CBM35s to cell walls of tobacco stems. Cell walls were either untreated or incubated with pectate lyase (PL) or polygalacturonase (PG) before incubation with the CBM35s. The regions corresponding to epidermis (e) and cortical parenchyma (cp) cell walls are indicated. (Scale bars, 10 μm.)
CsxA is predicted to be a secreted protein with a Gram-positive signal peptide cleavage site after amino acid 32 (11) but does not display any of the known Gram-positive cell wall immobilization motifs. Collectively, these observations provide compelling evidence that CsxA is a secreted protein that can be noncovalently attached to the cell wall of A. orientalis via a CBM-carbohydrate interaction. This cell wall association is most likely through an interaction of the CBM with GlcA, which is known to be a component of the glycolipids of other Actinomycetales (21, 22), although the biological ligand of the CBM may also be a related but unidentified uronic acid. By contrast immunohistochemical studies showed that none of the C. japonicus CBM35s bound to the cell wall of the host bacterium (Table S2).
CBM35 Targeting of Enzymes to Degraded Regions of the Plant Cell Wall.
By contrast to Chi-CBM35, the CBM35s appended to plant cell wall hydrolases are likely to play a role in substrate targeting. Indeed, the capacity of the CBM35s to bind to pectins, which have been depolymerized by pectate lyases, indicates that the role of the modules is to direct their cognate enzymes to those regions of the plant cell wall that are being actively degraded. To test this hypothesis, the capacity of the various CBM35s to bind to transverse sections of tobacco stems, treated with a pectate lyase and a polygalacturonase, was assessed. Xyl-CBM35, Pel-CBM35, and Rhe-CBM35 bind only very weakly or not at all to epidermal and cortical parenchyma cell walls of untreated sections. All 3 CBM35s, however, bind extensively to tobacco stem cell walls treated with pectate lyase, indicating that the proteins target the Δ4,5-GalA generated during pectin degradation (Fig. 2B). To ensure that the degradation of the pectins did not uncover “natural” ligands recognized by these proteins, plant cell walls were treated with a polygalacturonase. This enzyme hydrolyses the glycosidic bond in polygalacturonic acid (homogalacturonan) pectins and thus the nonreducing sugar generated comprises GalA and not the anhydro sugar Δ4,5-GalA moiety. Degradation of pectins was demonstrated by the complete loss of binding of a homogalacturonan antibody to tobacco cells treated with either the pectate lyase or polygalacturonase (Fig. 2 B). Pel-CBM35 and Rhe-CBM35, however, displayed no binding to the polygalacturonase-treated tobacco cell walls (Fig. 2B). Xyl-CBM35 did bind weakly to epidermal and cortical parenchyma cell walls after polygalacturonase treatment reflecting the exposure of xylan epitopes by pectic homogalacturonan removal (Fig. 2B). These data show that all 3 CBM35s target regions of the plant cell wall containing high concentrations of Δ4,5-GalA, although the Xyl-CBM35 can also direct enzymes to regions of these composite structures, presumably decorated xylans that contain GlcA.
Structure of Unliganded CBM35s.
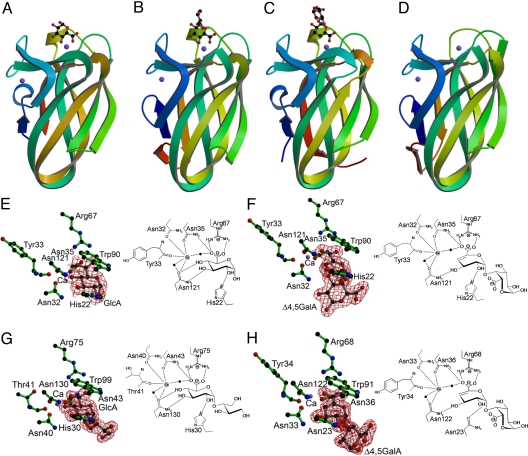
The crystal structures of the 4 CBM35s showed that the proteins were structurally homologous (Fig. 3). Indeed SSM (Secondary Structure Matching program; http://www.ebi.ac.uk/msd-srv/ssm/) analysis showed that Chi-CBM35 displays an rmsd of 1.1 Å with Pel-CBM35 over 121 Cα atoms, an rmsd of 0.8 Å with Rhe-CBM35 over 126 Cα atoms, and an rmsd of 1.1 Å with Xyl-CBM35 over 121 Cα atoms. In common with many CBM and lectin families, the 4 CBM35s display a jelly roll/β-sandwich fold comprising 2 antiparallel sheets consisting of 4 and 5 antiparallel β-strands, respectively. In all structures, except Rhe-CBM35, a metal ion is present, which has been modeled as calcium, based on its B-factor and coordination geometry exclusively with oxygen atoms. This calcium site is conserved in many lectin and several CBM families (reviewed in ref. 1). A second calcium is evident in all of the CBM35s, which is not structurally conserved in any other CBM family; the role of this metal in carbohydrate recognition is discussed below.
Fig. 3.
CBM35 structures. (A–D) Ribbon representations, color-ramped from the N-terminus (blue) to C-terminus (red), of Chi-CBM35 (in complex with GlcA) (A), Xyl-CBM35 (in complex with a GlcA containing disaccharide) (B), Rhe-CBM35 (in complex with Δ4,5-GalAα1,4Gal) (C), and Pel-CBM35 (D). The calcium ions are represented as blue spheres and ligands in ball-and-stick representation. Structures are shown in identical orientations. (E–H) Binding sites of Chi-CBM35 in complex with GlcA (E), Chi-CBM35 in complex with Δ4,5-GalAα1,4Gal (F), Xyl-CBM35 in complex with a GlcA containing disaccharide (G), and Rhe-CBM35 in complex with Δ4,5-GalAα1,4Gal (H). (E–H Left) The structures of the active site with the observed electron density for the ligand are shown. The electron density maps are shown as maximum likelihood weighted 2Fobs-Fcalc maps contoured at 1σ. Stereo views of these maps are given in Fig. S2. (E–H Right) Hydrogen bonding schematics with the calcium atoms are shown as gray circles and water molecules coordinating the calcium are shown as black circles.
Structure of CBM35-Ligand Complexes.
The mechanism of ligand recognition was revealed from the crystal structures of Xyl-CBM35 in complex with a GlcA-containing decorated xylooligosaccharide, Rhe-CBM35 in complex with a Δ4,5-GalA-containing galacturonooligosaccharide, and Chi-CBM35 bound to GlcA or Δ4,5-GalAα1,4Gal (Fig. 3). In each structure the carbohydrate ligand is housed in a shallow surface indentation in which O1 is solvent-exposed, which explains how the proteins are able to bind to the nonreducing end of pectin-derived oligosaccharides or the side chains of xylan. Thus, in Xyl-CBM35 the bound GlcA makes an α-linkage to another sugar that could not be identified with certainty from the electron density because of its disorder, whereas Δ4,5-GalA in complex with Rhe-CBM is α-1,4 linked to a d-GalA residue. The ligand binding site is highly conserved in the 4 CBM35s. Indeed, GlcA recognition by Xyl-CBM35 and Chi-CBM35 is identical (Fig. 3 E and G), and thus the Xyl-CBM35-GlcA complex will be used as a general reference (see Fig. S1 for an overlap of Xyl-CBM35 and Chi-CBM35 bound to uronic acid). The carboxylate at C6 of GlcA is a key specificity determinant, explaining why these proteins recognize GlcA and not glucose, although productive polar interactions are also made with O4, O3, and O2. The pyranose ring also makes extensive hydrophobic interactions with an adjacent tryptophan. The C6 carboxylate of the sugar makes bidentate hydrogen bonds with the 2 Nη atoms of Arg-75, whereas O6A also contacts a calcium ion that interacts with the protein through hydrogen bonds to the Oδ1 atoms of Asn-40 and Asn-43 and with the carbonyl of Thr-41. The inability of the R75A and N40A variants of Xyl-CBM35 and the equivalent mutants of Pel-CBM35 to bind either uronic acid (Table 2) demonstrates the importance of the arginine and calcium in carbohydrate recognition. O4 makes hydrogen bonds with the Oδ1 atoms of Asn-40 and Asn-130 and also makes a polar contact with the calcium ion. O3 interacts with Nδ2 of Asn-130 and O2 contacts with the Nε2 of His-30 (Fig. 3G). The pyranose ring makes extensive hydrophobic contact with Trp-99. The importance of these interactions is consistent with the observation that the N130A, W99A, and H30A variants of Xyl-CBM35 and the W89A, H22A, N35A, and N120A mutants of Pel-CBM35 displayed no binding to either GlcA or Δ4,5-GalA (Table S1). The conservation of these amino acids in the 4 CBM35s provides further support for their pivotal role in carbohydrate recognition. The Xyl-CBM35 residues Asn-130, Trp-99, Arg-75, and Asn-43 are invariant in the 4 proteins; Asn-40 and His-30 are functionally conserved as Asn-40 is replaced by an aspartate in Pel-CBM35 and Asn-23 is structurally equivalent to His-30 in Rhe-CBM35. The weaker binding of Rhe-CBM35 to GlcA, compared with Xyl-CBM35 and Chi-CBM35, likely reflects the replacement of His-30 with Asn-23; the hydrogen bond between Asn-23 Oδ1 with O2 is weaker than the polar contact between His-30 and O2 (Fig. 3 G and H). Δ4,5-GalA binds at an identical site to GlcA in Chi-CBM35 (and by inference Xyl-CBM35 as its binding site is identical to Chi-CBM35; see Fig. S1), and the interactions at C2, C3, C5, and C6 are conserved (Fig. 3 E and F). With Xyl-CBM35, the loss of the polar interaction at C4 appears to be compensated by a tighter hydrophobic interaction between Trp-99 and the conjugated sugar ring in Δ4,5-GalA compared to the saturated pyranose ring in GlcA.
Rhe-CBM35 displays a marked preference for Δ4,5-GalA, binding ≈30-fold more tightly than to GlcA. In contrast, Pel-CBM35 recognizes the pectin-derived ligand but displays no affinity for GlcA. The mechanism for this difference in specificity is subtle. Of the 4 CBMs, only Pel-CBM35 contains a lysine at position 21, which forms an ion pair with Asp-32. This interaction is predicted to prevent the aspartate from making a hydrogen bond with the critical O4 moiety in GlcA. In the other 3 CBM35s the equivalent amino acids to Lys-21 and Asp-32 are both asparagine residues. Replacing Asp-32 and Lys-21 in Pel-CBM35 with asparagine residues restores GlcA recognition without compromising affinity for Δ4,5-GalA (Table 2).
Discussion
This report shows that despite displaying significant structural and biochemical similarities, there is considerable diversity in the biological functions of the 4 CBM35s. Although 3 of the CBM35s target the plant cell wall, notably Δ4,5-GalA, the capacity of Chi-CBM35 to function as a bacterial cell wall attachment module is perhaps the most striking feature of this protein ensemble. Proteins can be retained on the surface of eukaryotic and prokaryotic cells through a variety of mechanisms that do not feature carbohydrate recognition. Thus, the observation that Chi-CBM35 tethers CsxA to the surface of A. orientalis by binding to a carbohydrate ligand is an unusual biological role for a CBM and carbohydrate binding proteins in general. It should be noted, however, that a recent report has shown that a family of carbohydrate binding modules that bind to a wide spectrum of β-linked plant structural polysaccharides (CBM37), which are unique to Ruminococcus albus, anchor their cognate enzymes to the surface of the bacterium (23). The target ligand for Chi-CBM35 in the cell wall of A. orientalis is most likely an uronic acid such as GlcA. Although the ITC-derived affinity of Chi-CBM35 for the uronic acid (KD ≈ 100 μM) is modest, this solution affinity does not reflect the binding strength on a membrane surface where clustering of the ligands can result in a marked increase in apparent affinity. The biological rationale for this function appears to be to keep the enzyme in proximity to the bacterium. By presenting the enzyme on the surface of the bacterium, only the cationic fragments of chitosan, which pose the most immediate threat to the bacterium, are degraded by CsxA. Furthermore, the monosaccharide products of hydrolysis would be in the direct vicinity of the bacterium promoting rapid transport into the bacterial cell for further metabolism. The cell adhesion role of CBMs, reported here and by Ezer et al. (23), which was previously unconsidered, may prove to factor prominently in the function of these protein modules in the future.
The capacity of the 3 CBM35s, which are components of plant cell wall degrading enzymes, to bind Δ4,5-GalA indicates that they play a role in targeting the hydrolases and esterases to regions of their composite substrate that are accessible to biological degradation. Pectins are arguably the most accessible region of the plant cell wall and indeed cloak other polysaccharides, such as xyloglucan, occluding them from enzyme attack (24). Thus, by targeting enzymes to regions rich in Δ4,5-GalA, the CBM35s are directing their cognate catalytic modules to areas of the cell wall that are particularly accessible to further biological degradation. The origin of the pectate lyases that generate the Δ4,5-GalA molecules is unclear. It is possible that these enzymes are derived from saprophytic microorganisms that contribute to the plant cell wall degrading process. It is apparent, however, that plant genomes encode extensive families of pectate lyases (25), and thus it is possible that the anhydro sugar is released by endogenous enzymes. Although Δ4,5-GalA was not evident in the tobacco stem sections, suggesting that pectate lyases were not expressed in these tissues, it is possible that these enzymes are only synthesized in specific cell types or differentiation states. It should be emphasized, however, that Xyl-CBM35 also displays significant affinity for GlcA, a side chain present in several xylans (19), and thus this plasticity in uronic acid recognition provides the enzyme with a flexible targeting strategy that is tailored to the plant biomass presented to the bacterium. It is possible that Xyl-CBM35 initially directs the xylanase toward regions of cell walls that are being actively degraded but, as xylan structures are revealed, the enzyme is shuttled onto the hemicellulosic polysaccharide affording the enzyme access to its target substrate. It is interesting to note that the gene encoding the CBM35-containing xylanase is a component of a C. japonicus operon that also directs the synthesis of 2 other enzymes that contribute to xylan degradation; a GH62 arabinofuranosidase (Abf62A) and a CE1 ferulate/acetyl esterase (CE1A) (17, 26). Significantly, Abf62A and CE1 contain a CBM35 whose sequence is identical to Xyl-CBM35 and will therefore display specificity for Δ4,5-GalA and GlcA. Thus, the targeting of regions of plant cell walls that are being actively degraded is not restricted to a single xylanase, but is a feature of an enzyme ensemble that attacks decorated xylan structures.
This report demonstrates how nature has exploited convergent binding specificity for divergent function. It is evident, therefore, that the context of the ligands recognized by CBMs is central to their biological function. In addition to providing insights into the biological role of CBMs as enzyme delivery molecules and host cell adhesins, respectively, this report also highlights the potential exploitation of these protein modules within an industrial setting. The novel specificities displayed by the family 35 CBMs will not only contribute to the toolbox of biocatalysts required to generate biofuels from lignocellulose, but also represent valuable probes that can be deployed in the analysis of plant cell wall architecture.
Materials and Methods
Gene Cloning, Protein Expression, and Purification.
Cloning of the genes encoding the CBM35s into Escherichia coli expression vectors and the subsequent description of the purification of the proteins are described in SI Materials and Methods.
Mutagenesis.
Site-directed mutagenesis was carried out by using the PCR-based QuikChange site-directed mutagenesis kit (Stratagene) by using appropriate plasmids as templates and primers pairs that are listed in Table S3.
Carbohydrate and Metal Binding Studies.
Ligand binding studies are described in SI Materials and Methods. The uronic acids used in these experiments were obtained as follows: GlcA, GalA, di- and tri-GalA were purchased from Sigma, whereas Δ4,5-GalAα1,4Gal and Δ4,5-GalAα1,4Galα1,4Gal were obtained by treating homopolygalacturonic acid (Sigma) with the pectate lyase CjPel10A to completion (monitored by no further increase at A257 nm). Xylooligosaccharides decorated with GlcA were obtained by digesting oat spelt xylan (Sigma) to completion, as monitored by HPLC (27), with the xylanase CjXyn10B, the arabinofuranosidase CjAbf51A, and the GH43 enzyme ddAXH (Megazyme). The products were purified by size exclusion chromatography, as described by Proctor et al. (27), and then by ion exchange using Dowex-100 resin. The smallest charged oligosaccharide fraction (designated Xyl-GlcA) that bound to Xyl-CBM35 was retained for further use in ITC and crystallography experiments.
Immunofluorescence Microscopy.
A. orientalis cells.
A. orientalis cultured in media supplemented with 0.2% vol/vol glucosamine and 0.8% wt/vol chitosan (6% N-acetylated) were harvested, and the mycelium was fixed by heat on Superfrost/Plus slides (Fisher). Bacteria were permeabilized with 50% methanol at −20 °C for 5 min and in PBSP (PBS with 50 mM glycine, 0.06% saponine, 0.06% Tween 20, 0.5% Nonidet P-40, and 0.5% TritonX-100) for 10 min. Smears were rinsed in PBSD (PBS with 0.01% saponine and 0.01% Tween 20) for 5 min at room temperature, and nonspecific binding sites were blocked with PBSB (PBSD with 2% normal goat serum, 2% BSA, and 0.45% fish gelatin) for 30 min. Incubation of the bacteria with antibodies and fluorescent microscopy are detailed in SI Materials and Methods.
Plant cell walls.
A 3-stage immunolabeling technique described previously (28) was used to assess CBM35 binding to tobacco stem sections. Specific details and pre-enzyme treatments are described in SI Materials and Methods.
Crystallization and Data Collection.
Details of protein crystallization, data collection, and structure determination are provided in SI Materials and Methods (Table S4).
Supplementary Material
Acknowledgments.
Work at the University of Victoria and Université de Sherbrooke was supported by Natural Sciences and Engineering Research Council of Canada Discovery grants (A.B.B., B.G.T., and R.B.), and work in Newcastle and York was supported by the Biotechnology and Biological Sciences Research Council. A.L.v.B is supported by doctoral fellowships from Natural Sciences and Engineering Research Council and the Michael Smith Foundation for Health Research. A.B.B. is a Canada Research Chair in Molecular Interactions and a Michael Smith Foundation for Health Research Scholar. T.M.G. holds a Sir Henry Wellcome postdoctoral fellowship, and G.J.D. has a Royal Society Wolfson Research Merit Award. Work in Lisbon was supported by Fundação para a Ciência e Tecnologia through grants SFRH/BD/23784/2005 (to M.A.C.) and PTDC/BIAPRO/69732/2006 and C.D. was supported by a Marie-Curie Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808972106/DCSupplemental.
References
- 1.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boraston AB, Kwan E, Chiu P, Warren RA, Kilburn DG. Recognition and hydrolysis of noncrystalline cellulose. J Biol Chem. 2003;278:6120–6127. doi: 10.1074/jbc.M209554200. [DOI] [PubMed] [Google Scholar]
- 3.Hall J, et al. The noncatalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem J. 1995;309:749–756. doi: 10.1042/bj3090749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomme P, et al. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988;170:575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- 5.Hazlewood GP, Laurie JI, Ferreira LM, Gilbert HJ. Pseudomonas fluorescens subsp. cellulosa: An alternative model for bacterial cellulose. J Appl Bacteriol. 1992;72:244–251. doi: 10.1111/j.1365-2672.1992.tb01830.x. [DOI] [PubMed] [Google Scholar]
- 6.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 7.Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281:35263–35271. doi: 10.1074/jbc.M607620200. [DOI] [PubMed] [Google Scholar]
- 8.van Bueren AL, Higgins M, Wang D, Burke RD, Boraston AB. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat Struct Mol Biol. 2007;14:76–84. doi: 10.1038/nsmb1187. [DOI] [PubMed] [Google Scholar]
- 9.Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Rev. 1999;29:265–295. doi: 10.1016/s0165-0173(99)00003-x. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Stuckey JA, Wishart MJ, Dixon JE. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J Biol Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 11.Cote N, et al. Two exo-beta-D-glucosaminidases/exochitosanases from actinomycetes define a new subfamily within family 2 of glycoside hydrolases. Biochem J. 2006;394:675–686. doi: 10.1042/BJ20051436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai K, Katsumi R, Isobe A, Nanjo F. Purification and hydrolytic action of a chitosanase from Nocardia orientalis. Biochim Biophys Acta. 1991;1079:65–72. doi: 10.1016/0167-4838(91)90025-u. [DOI] [PubMed] [Google Scholar]
- 13.Bolam DN, et al. X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J Biol Chem. 2004;279:22953–22963. doi: 10.1074/jbc.M313317200. [DOI] [PubMed] [Google Scholar]
- 14.Czjzek M, et al. The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J Biol Chem. 2001;276:48580–48587. doi: 10.1074/jbc.M109142200. [DOI] [PubMed] [Google Scholar]
- 15.van Bueren AL, Morland C, Gilbert HJ, Boraston AB. Family 6 carbohydrate binding modules recognize the nonreducing end of beta-1,3-linked glucans by presenting a unique ligand binding surface. J Biol Chem. 2005;280:530–537. doi: 10.1074/jbc.M410113200. [DOI] [PubMed] [Google Scholar]
- 16.Solbak AI, et al. Discovery of pectin-degrading enzymes and directed evolution of a novel pectate lyase for processing cotton fabric. J Biol Chem. 2005;280:9431–9438. doi: 10.1074/jbc.M411838200. [DOI] [PubMed] [Google Scholar]
- 17.Kellett LE, et al. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990;272:369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran F, Nasuno S, Starr MP. Extracellular and intracellular polygalacturonic acid trans-eliminases of Erwinia carotovora. Arch Biochem Biophys. 1968;123:298–306. doi: 10.1016/0003-9861(68)90138-0. [DOI] [PubMed] [Google Scholar]
- 19.Pena MJ, et al. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauss H, Hassid WZ. Biosynthesis of the 4-O-methyl-D-glucuronic acid unit of hemicellulose B by transmethylation from S-adenosyl-L-methionine. J Biol Chem. 1967;242:1680–1684. [PubMed] [Google Scholar]
- 21.Chatterjee D, Aspinall GO, Brennan PJ. The presence of novel glucuronic acid-containing, type-specific glycolipid antigens within Mycobacterium spp. Revision of earlier structures. J Biol Chem. 1987;262:3528–3533. [PubMed] [Google Scholar]
- 22.Chatterjee D, Bozic C, Aspinall GO, Brennan PJ. Glucuronic acid- and branched sugar-containing glycolipid antigens of Mycobacterium avium. J Biol Chem. 1988;263:4092–4097. [PubMed] [Google Scholar]
- 23.Ezer A, et al. Cell-surface enzyme attachment is mediated by family-37 carbohydrate-binding modules, unique to Ruminococcus albus. J Bacteriol. 2008;190:8220–8222. doi: 10.1128/JB.00609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus S, et al. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8:60. doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrissat B, Coutinho PM, Davies GJ. A census of carbohydrate-active enzymes in the genome of. Arabidopsis thaliana. Plant Mol Biol. 2001;47:55–72. [PubMed] [Google Scholar]
- 26.Ferreira LM, et al. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a noncatalytic cellulose-binding domain. Biochem J. 1993;294(Pt 2):349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proctor MR, et al. Tailored catalysts for plant cell-wall degradation: Redesigning the exo/endo preference of Cellvibrio japonicus arabinanase 43A. Proc Natl Acad Sci USA. 2005;102:2697–2702. doi: 10.1073/pnas.0500051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCartney L, et al. Differential recognition of plant cell walls by microbial xylan-specific carbohydrate-binding modules. Proc Natl Acad Sci USA. 2006;103:4765–4770. doi: 10.1073/pnas.0508887103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.