Abstract
All DNA repeats known to undergo expansion leading to human neurodegenerative disease can form one, or several, alternative conformations, including hairpin, slipped strand, triplex, quadruplex, or unwound DNA structures. These alternative structures may interfere with the normal cellular processes of transcription, DNA repair, replication initiation, or polymerase elongation and thereby contribute to the genetic instability of these repeat tracts. We show that (CCTG)·(CAGG) repeats, in the first intron of the ZNF9 gene associated with myotonic dystrophy type 2, form slipped-strand DNA structures in a length-dependent fashion upon reduplexing. The threshold for structure formation on reduplexing is between 36 and 42 repeats in length. Alternative DNA structures also form in (CCTG)58·(CAGG)58 and larger repeat tracts in plasmids at physiological superhelical densities. This represents an example of a sequence that forms slipped-strand DNA from the energy of DNA supercoiling. Moreover, Z-DNA forms in a (TG)·(CA) tract within the complex repeat sequence 5′ of the (CCTG)n·(CAGG)n repeat in the ZNF9 gene. Upon reduplexing, the presence of the flanking sequence containing the Z-DNA-forming tract reduced the extent of slipped-strand DNA formation by 62% for (CCTG)57·(CAGG)57 compared with 58 pure repeats without the flanking sequence. This finding suggests that the Z-DNA-forming sequence in the DM2 gene locus may have a protective effect of reducing the potential for slipped-strand DNA formation in (CCTG)n·(CAGG)n repeats.
Keywords: alternative DNA structure, DNA repeat diseases, DNA repeat expansion, repeat instability
Expansion of the (CCTG)·(CAGG) tetranucleotide repeat tract in the first intron of the zinc finger protein 9 (ZNF9) gene on chromosome 3 is a causative factor in myotonic dystrophy type 2 (DM2) (1, 2). DM2 causes symptoms including myotonia, proximal muscle weakness and atrophy, cataracts, cardiac conduction abnormalities, and frontal balding. Although similar in pathology, DM2 is a much less severe disease than myotonic dystrophy type 1 (DM1) (3, 4). The prevailing paradigm is that both the DM1 and DM2 repeat expansions lead to a pathogenic effect of the RNA containing the (CUG)n and (CCUG)n repeat expansions (5, 6), respectively. The mean expansion in DM2 is 5,000 repeats, with a range between 75 and 11,000 repeats (3, 7). The exact threshold length is difficult to determine because of the somatic instability of the repeat tract (1, 7, 8). A complex, polymorphic flanking sequence consisting of (TG)14–25(TCTG)4–11 immediately precedes the DM2 (CCTG)n repeat tract. The polymorphisms at this locus include sequence interruptions of (TCTG) and (GCTG) in the (CCTG)n repeat tract (9). The loss of the sequence interruptions may predispose the tract to repeat expansion, as suggested for FRAXA, DM1, and Friedreich ataxia (10–12). The left-handed Z-DNA-forming (TG)n tract in this flanking sequence is evolutionarily conserved (9) and thus, may have some biological role.
Alternative, non-B-DNA structure formation within unstable DNA repeat sequences associated with human disease may contribute to their genetic instability (see refs. 13–16 for review). (CTG)·(CAG) repeats associated with DM1, Huntington's disease, and several spinocerebellar ataxias can form hairpins and slipped-strand structures that may block replication and lead to replication restart or DNA repair activity during which repeat lengths may change. (CGG)·(CCG) repeats associated with fragile X syndrome also form hairpins and slipped strands, whereas the Friedreich ataxia (GAA)·(TTC) repeat forms triplex structures, which are strong blocks to replication. The spinocerebellar ataxia type 10 (ATTCT)·(AGAAT) repeat does not form any hairpin or slipped-strand structure that would stall a polymerase. Rather, it forms unwound structures that can function as DNA unwinding elements (DUEs) and initiate aberrant replication (17). Expansion of this repeat has been associated with the proximity of an active replication origin in human cells (18). In contrast, little is known about the DM2 (CCTG)·(CAGG) repeats, although a single strand of (CAGG)26 can form hairpins, as determined by chemical probe analysis (19). RNA transcripts of (CCTG)·(CAGG) repeats can form hairpins similar to those of the (CTG)·(CAG) repeats (20).
We show that the DM2 (CCTG)n·(CAGG)n repeats form slipped-strand structures, and they form in duplex DNA from the energy of negative DNA supercoiling. In addition, the (TG)n tract flanking the DM2 (CCTG)·(CAGG) repeats forms Z-DNA in supercoiled DNA. Significantly, the presence of this flanking sequence, including the Z-DNA-forming tract, reduced the extent of slipped-strand DNA formation in the (CCTG)·(CAGG) repeat tract, suggesting a biological role for Z-DNA. Z-DNA formation, and the concomitant relaxation of negative supercoiling, may exert a protective effect against the formation of slipped-strand DNA in this DM2 locus.
Results
(CCTG)·(CAGG) Repeats Form Slipped-Strand Structures.
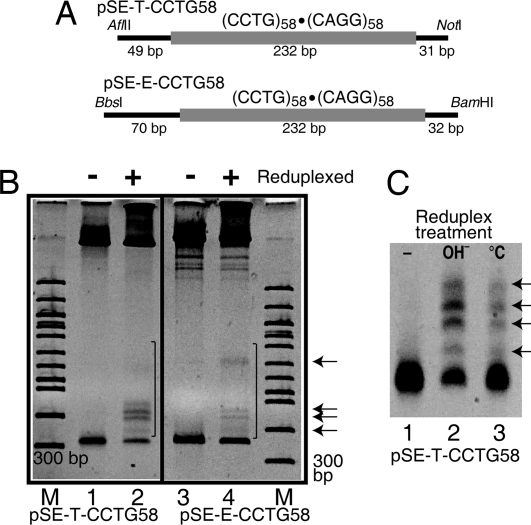
DNA containing slipped-strand structures migrates with slower mobility than the corresponding duplex DNA due to bends resulting from multiple 3-way junctions (10, 21, 22). Slipped-strand structures formed in a (CCTG)58·(CAGG)58 tract in plasmids pSE-T-CCTG58 and pSE-E-CCTG58 following an alkaline reduplexing procedure (21) (Fig. 1). To ensure that the slowly migrating fragments were not due to heteroduplexes formed from strands with different repeat lengths, plasmids were not linearized before the reduplexing treatment. The single strands of the plasmids remain catenated upon denaturation increasing the likelihood that they renature with their complimentary strand (21). In Fig. 1B, DNAs in lanes 1 and 3 were not subjected to reduplexing treatment. DNAs in lanes 2 and 4 were reduplexed and show anomalous, slowly migrating bands (indicated by the black bars and arrows) not present in the control samples. A very similar pattern of anomalous bands was formed for both plasmids analyzed, as indicated by the arrows. The reproducible pattern of anomalous bands is characteristic of slipped-strand DNA structure formation (21, 22).
Fig. 1.
Slipped-strand structures in (CCTG)58·(CAGG)58. (A) Map of fragments containing the (CCTG)58·(CAGG)58 repeats. The CCTG repeats are indicated by the thick, lightly shaded line. (B) Nondenaturing 5% polyacrylamide gels showing the presence of slipped-strand DNA structures upon reduplexing the plasmid. Lanes 1 and 2, pSE-T-CCTG58 cut with AflII and NotI to generate a 312-bp fragment with 80 bp of flanking DNA. Lanes 3 and 4, pSE-E-CCTG58 cut with BamHI and BbsI to generate a 334-bp fragment with 102 bp of flanking DNA. Lanes 1 and 3, control, no reduplexing. Lanes 2 and 4, alkaline reduplexed. Anomalous, slowly migrating bands are indicated by the black bars. Arrows indicate major products present in both plasmids. M, 100-bp DNA ladder (NEB). (C) Comparison of alkaline and temperature-based reduplexing for pSE-T-CCTG58. Lane 1, control, no reduplexing; lane 2, alkaline reduplexing, lane 3, temperature reduplexing. All lanes contain 1 μg of DNA.
A second protocol, which involves temperature controlled denaturation and renaturation (23), was also used to form slipped-strand structures. Both the alkaline- and temperature-based methods resulted in a similar pattern of anomalous bands for pSE-T-CCTG58 (Fig. 1C). The alkaline method resulted in a higher proportion of slipped-strand structures, but it also resulted in a greater amount of nicked plasmid, and plasmid that remained single stranded after renaturation (51).
Threshold Length for the (CCTG)·(CAGG) Slipped-Strand Structure Formation.
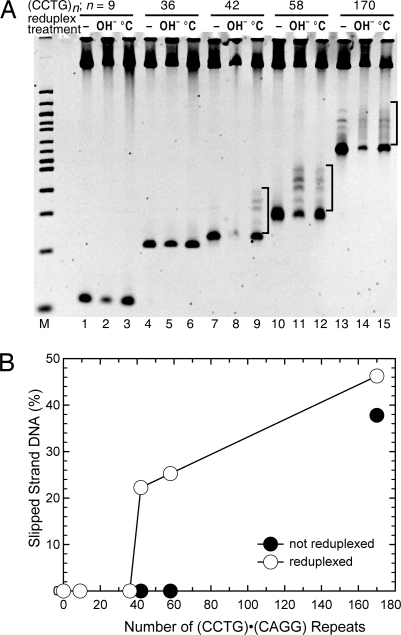
The formation of slipped-strand structures as a function of repeat tract length is shown in Fig. 2. Structures did not form in plasmids containing 9 or 36 repeats, using either the alkaline or temperature reduplexing protocol (Fig. 2A, lanes 1–6), however, structures were formed in plasmids containing 42, 58, or 170 repeats (lanes 7–15). This indicates a threshold for structure formation of between 36 and 42 repeats. Interestingly, this is just above a premutation allele pool for DM2 full mutations of 22–33 (CCTG) repeats (8). In addition, the percentage of structure formed in the population increased with repeat length from 22% for 42 repeats to 47% for 170 repeats (Fig. 2B).
Fig. 2.
Length threshold for (CCTG)·(CAGG) slipped-strand DNA formation. (A) (CCTG)n·(CAGG)n repeats, where n = 9 (lanes 1–3), n = 36 (lanes 4–6), n = 42 (lanes 7–9), n = 58 (lanes 10–12), and n = 170 (lanes 13–15) were analyzed for slipped-strand structures on a nondenaturing 5% polyacrylamide gel. –, control samples with no reduplexing; OH−, alkaline reduplexing; °C, temperature reduplexing. Slipped-strand structures appear as 4 prominent, slowly migrating bands when n = 42, 58, and 170, and are denoted by brackets. (B) Slipped-strand DNA structures are quantitated as the percentage of the DNA species migrating above the linear fragment (not including the larger linear plasmid band at the top of the gel). Results are from a representative temperature reduplexing experiment.
Superhelical-Density-Dependent Formation of Alternative DNA Structures in the DM2 Repeat Sequences.
Significantly, anomalously migrating bands formed spontaneously in the ‘control sample’, which was not subjected to reduplexing treatment at a level of 38% in pSE-T-CCTG170 containing 170 (CCTG)·(CAGG) repeats (Fig. 2A, lane 13; Fig. 2B). The slower migrating products were not due to the presence of larger repeat lengths (results not shown). Moreover, these slipped-strand structures could not be removed from a preparation of naturally supercoiled DNA by heating and quick cooling, as described in Materials and Methods. This protocol has worked well to remove cruciforms (24) and slipped-strand structures associated with shorter repeats, as shown below. In addition, the basic pattern of anomalously migrating bands was identical in the non reduplexed and reduplexed samples, indicating that the same population of slipped-strand structures formed in supercoiled DNA as following either of the reduplexing protocols. A greater degree of heterogeneity exists after reduplexing, as indicated by the “smearing” in the lane. This is a significant demonstration of the superhelical density-dependent formation of slipped-strand DNA.
To further examine the superhelical density dependent formation of alternative DNA secondary structures in the DM2 (CCTG)·(CAGG) repeat, DNA topoisomer populations of increasing superhelical density were prepared for pSE-U-CCTG58 and pSE-U-CCTG170, which contain 58 or 170 (CCTG)·(CAGG) repeats, respectively, cloned into the EcoRI site of pUC8.
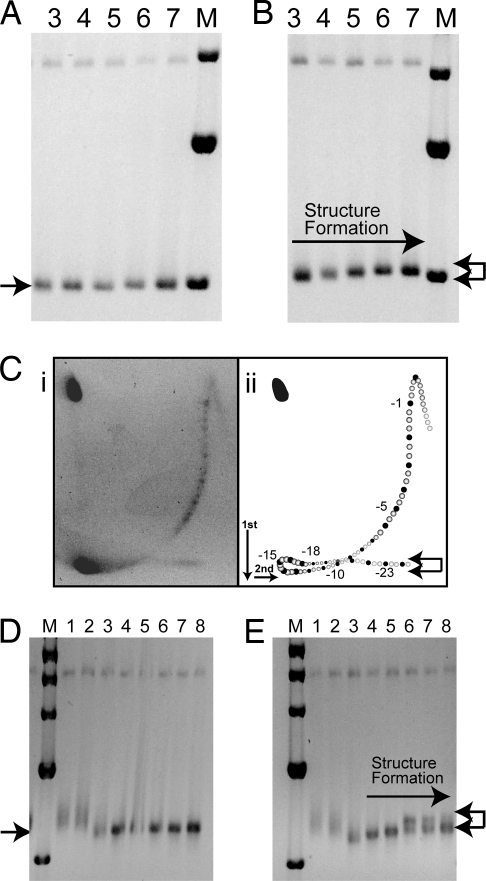
Any preexisting alternative DNA secondary structures in plasmids containing (CCTG)58·(CAGG)58 were removed as described in Materials and Methods, and all topoisomers migrated with the same mobility with respect to the marker (1-kb linear ladder) (Fig. 3A). In Fig. 3B, the same topoisomers were heated for 5 min at 80 °C and then allowed to slowly cool to room temperature to promote DNA structural transitions. The gels were run at 4 °C to prevent any subsequent structural transitions. The plasmid DNA underwent a structural transition as evident by the retardation in migration of the topoisomers populations with increasing superhelical density. The arrows in Fig. 3B show the positions of supercoiled DNA without (bottom arrow) and with a structural transition (top arrow). A 2D gel of topoisomers (from ΔL = +2 to ΔL < −25) treated to promote structure formation is shown in Fig. 3Ci. Distinct topoisomers can be seen, although the topoisomer pattern appears smeared due to the presence of plasmid molecules with a nonintegral number of helical turns that result from plasmid molecules containing deletions or additions of the 4-bp repeat unit (for discussion, see ref. 25). Such heterogeneity in plasmids containing unstable repeats is common (21, 22, 26), and is evident in the width of some of the linear restriction fragments in Figs. 1–3. Because the 2-D gel pattern represents a mixture of plasmids with 58 ± 3 (CCTG)·(CAGG) repeats, additional spots occur between the major spots corresponding to 58 (CCTG)·(CAGG) repeats. (In fact, 4 additional spots would result from 4-, 8-, and 12-bp deletions and additions). The filled circles in Fig. 3Cii represent the predominant topoisomers containing (CCTG)58·(CAGG)58. The open circles represent other spots present in the gel. From this analysis, we estimate that the DNA secondary structural transition is occurring at L − Lo = −15, or σ = −0.054. At higher superhelical densities, (L − Lo < −18) additional (or different) transitions may occur, as observed for (GAA)·(TTC) and (ATTCT)·(AGAAT) repeats (17, 27). The arrows shown in Fig. 3B are duplicated in Fig. 3Cii on the idealized 2D gel. This 2D gel pattern is consistent with the changes shown in the topoisomer populations in Fig. 3B. No retardation in the first dimension is observed in plasmid without the repeat (17). Analysis of 170 (CCTG)·(CAGG) repeats in pSE-U-CCTG170 showed similar results in the 1D analysis of topoisomer populations where a clear transition was visible after heating to 80 °C and slow cooling (compare Fig. 3D with Fig. 3E).
Fig. 3.
Alternative DNA structure formation in long (CCTG)·(CAGG) tracts at high superhelical densities. (A) The 1.75% agarose gel (in TAE buffer) shows individual topoisomer populations (lanes 3, 4, 5, 6, and 7) of pSE-U-CCTG58 (with 58 CCTG repeats) of increasing negative supercoiling. Plasmids were treated to remove any preexisting secondary structures, as described in Materials and Methods. (B) The topoisomer populations, as in A, after heating and slow cooling to promote DNA secondary structure formation. The arrow pointing to the right indicates increasing levels of DNA secondary structure formation as indicated by the reduced electrophoretic mobility. The left-facing arrows at the right of the gel indicate positions of fully supercoiled DNA, with no DNA secondary structures (bottom), and DNA containing secondary transitions (top). (C) A 2D agarose gel shows alternative DNA structure formation (i). Panel ii shows an annotated representation of the gel pattern. The filled spots represent the positions of plasmid containing 58 (CCTG) repeats, while intermediate spots are those representing plasmids containing 58 ± 3 (CCTG) repeats. The upper and lower arrows denote plasmids with structures or no structures, respectively. (D) Agarose gel showing individual topoisomer populations of pSE-U-CCTG170 (with 170 CCTG repeats), containing increasing levels of negative supercoiling. Plasmid has been treated to remove any preexisting secondary structures. (E) The same topoisomer populations, as in D, after heating and slow cooling to promote DNA secondary structure formation. Arrows as described above.
Z-DNA Formation in 5′ Human Flanking Sequence.
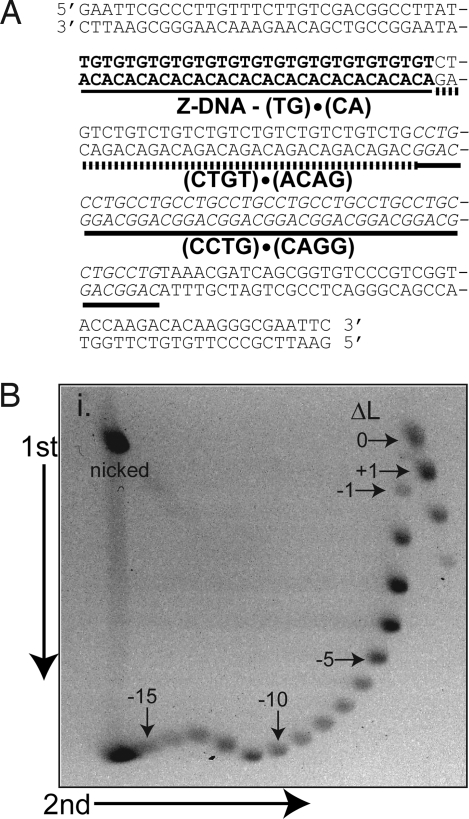
pSE-U-FS11 contains the sequence (TG)15(TCTG)8(CCTG)11 derived from a normal human DM2 allele cloned into the EcoRI site of pUC8 (Fig. 4A). A clear DNA secondary structural transition is evident in the 2D agarose gel containing a mixture of topoisomers (from ΔL = +3 to ΔL < −20) shown in Fig. 4B. This transition is not observed in plasmid without the repeat (17). Every 10 bp that forms Z-DNA is associated with relaxation of 1.78 turns of the helix (28). This particular sequence, (TG)15, has 1 additional T, 3′ to the repeat, which could also participate in Z-DNA formation. Relaxations of 2.3, 4.3, 5, and 5 superhelical turns are observed for topoisomers −12, −13, −14, and −15, respectively (Fig. 4B). This extent of relaxation is expected for a 31-bp Z-DNA transition (≈5.5 turns). Moreover, the transition begins at σ = −0.044 and is complete at σ = −0.052. This is consistent with previous reports of Z-DNA transitions for (TG)15 at σ = −0.045 to −0.055 (28–30).
Fig. 4.
Z-DNA formation at (TG)15 flanking the DM2 (CCTG)·(CAGG) repeat. (A) The sequence of the complex repetitive sequence at the ZNF9 locus cloned into pUC8, creating pSE-U-FS11. (B) Two-dimensional agarose gel analysis in a 1.75% agarose gel in TAE buffer shows a structural transition indicative of Z-DNA. Topoisomers were not treated to promote structure formation as Z-DNA forms spontaneously in supercoiled plasmid. Positions of selected isomers are indicated. A DNA secondary structural transition begins at topoisomer −12 as evidenced by the slower migration relative to that expected for this topoisomer.
Influence of the Z-DNA Forming Sequence on the Formation of Slipped-Strand DNA.
Z-DNA forms in the sequence flanking the (CCTG)n·(CAGG)n repeats at a lower superhelical density than that required for the formation of slipped-strand DNA. Therefore, Z-DNA formation may reduce the propensity for slipped-strand DNA formation. Alternatively, the transition between B-DNA and Z-DNA might potentially destabilize the helix in the flanking region, as occurs with DNA unwinding elements, and facilitate slipped-strand DNA formation (31). To investigate these possibilities, 2D agarose gel analysis was performed for pSE-T-FS31 containing (TG)20(TCTG)11(CCTG)31, in which the (CCTG) tract is below the transition threshold for structure formation of 36–42 repeats identified in Fig. 2. A transition characteristic of Z-DNA formation was observed; however, no additional transitions occurred that would suggest the formation of slipped-strand DNA within the (CCTG)31·(CAGG)31 tract (51). pSE-T-FS80 [containing (CCTG)80·(CAGG)80] was also analyzed using 2D gel electrophoresis, and a characteristic Z-DNA transition was also observed. Evidence for additional unwinding was difficult to assess, however, because of the heterogeneity caused by small expansions and deletions present within the DNA preparation of this long repeat.
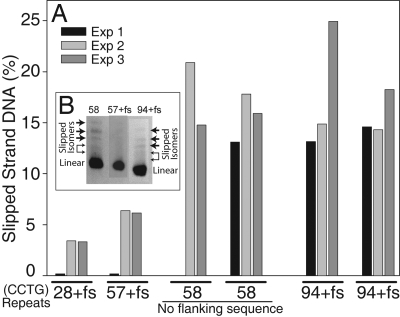
To ascertain whether the presence of the human flanking sequence, including the Z-DNA-forming tract, altered the propensity of slipped-strand DNA formation, the following experiments were performed. The percentage of slipped-strand DNA formed during the temperature-based renaturation protocol was analyzed in a series of plasmids containing 28, 57, and 94 (CCTG)·(CAGG) repeats within the human sequence (TG)18(TCTG)11 (CCTG)n·(CAGG)n. This was compared with 2 independent preparations of pUC8-U-CCTG58, containing (CCTG)58·(CAGG)58, without the human flanking sequences. The results, shown for 3 representative experiments in Fig. 5, demonstrate the following. A length dependence for slipped-strand formation was observed, as was observed for repeats lacking the flanking sequence (shown in Fig. 2). However, in these plasmids containing the flanking sequence, lower levels of slipped-strand DNA were detected. Significantly, with flanking human sequences, (CCTG)57·(CAGG)57 consistently formed on average 6.3% slipped-strand structures. The 2 preparations of (CCTG)58·(CAGG)58, without the flanking human sequences, formed on average 16.5% slipped-strand structures. Plasmid containing the (CCTG)94·(CAGG)94 tract with human flanking sequences formed, on average, 16.7% slipped-strand structures. The pattern of slipped-strand isomers was similar for 57 or 58 repeats, with or without the flanking human sequences (Fig. 5B). This demonstrates that a similar population of slipped-strand DNA isomers formed, albeit, at different levels. Also, a similar distribution of slipped-strand species was observed for 94 (CCTG) repeats (Fig. 5B). In summary, the presence of the flanking sequence containing the Z-DNA-forming tract reduced the extent of slipped-strand DNA formation in (CCTG)57 by 62% compared with the level formed in (CCTG)58 without the human flanking sequence.
Fig. 5.
Slipped-strand DNA structures form at reduced levels in (CCTG)·(CAGG) repeats within the human flanking region containing a Z-DNA forming tract. (A) The percentage of slipped-strand DNA formation after the temperature renaturation protocol is shown for plasmids pSE-U-FS28 (28+fs), pSE-U-FS57 (57+fs), pSE-U-FS94 (94+fs) [containing (TG)18(TCTG)11(CCTG)n, with n = 28, 57, or 94, respectively, within the human gene sequence] and pSE-U-CCTG58 [containing (CCTG)58] (“fs” denotes flanking sequence). Data are shown for 3 representative experiments. (B) A region of an acrylamide gel shows the slipped-strand products for (CCTG)58 alone (58), and 57 or 94 (CCTG) repeats within the human flanking sequence (57+fs and 94+fs). The linear band is denoted and the slipped-strand isomers are indicated by arrows.
Discussion
All DNA repeats associated with neurodegenerative repeat expansion diseases analyzed to date can form 1 or more alternative DNA structures. For example, the (GAA)·(TTC) repeat in Friedreich ataxia forms triplex and parallel DNA structures (27, 32), the (ATTCT)·(AGAAT) repeats form unwound structures (17), and (CTG)·(CAG) and (CGG)·(CCG) repeats form slipped-strand structures (21, 22, 26). Here, we show that the (CCTG)n·(CAGG)n repeats associated with DM2 form slipped-strand DNA. Essentially identical patterns of slipped-strand DNA structures were observed after reduplexing treatment of 3 different plasmids containing (CCTG)58·(CAGG)58 using 2 different reduplexing procedures. This demonstrates that the population of slipped-strand structures is a function of the (CCTG)58·(CAGG)58 tract and not flanking DNA sequences. A threshold length at which DNA repeats form slipped strands was between 36 and 42 repeats, as determined using 2 different reduplexing protocols. This is similar to a threshold observed for DM1 (CTG)·(CAG) repeats (22). Moreover, this length is close to the high end of the premutation allele length of 22–33 identified by Bachinski et al. (8). Although a wide range of isomers of slipped-strand structures may be expected, as observed for (CTG)·(CAG) repeats (10, 21, 22), only a limited number of preferred structures form within (CCTG)n·(CAGG)n repeats as indicated by identification of the 4 prominent bands corresponding to slipped-strand isomers. This result was similar to that for a fragile X (CGG)·(CCG) repeat with sequence interruptions, in which 2 slipped isomers were observed (21).
We present a demonstration of the superhelical density dependent formation of slipped-strand DNA. Slipped-strand DNA formed spontaneously in the (CCTG)170·(CAGG)170 repeat tract in naturally supercoiled plasmid. While thermodynamically favorable, the supercoil-dependent formation of slipped-strand structures has not been reported for (CTG)·(CAG) or (CGG)·(CCG) repeats. The patterns of slipped-strand DNA isomers formed in supercoiled DNA and after reduplexing were similar, indicating that the same slipped-strand products were formed under both conditions. In plasmid containing (CCTG)58·(CAGG)58, a DNA structural transition occurred at a superhelical density of σ = −0.054. When an all-or-none structural transition occurs, for example with cruciform, Z-DNA, or intramolecular triplex formation, the first topoisomer, in which the structural transition occurs, migrates with reduced mobility. Successive topoisomers with decreasing linking number (i.e., more negative supercoiling), migrate faster in the gel for an all-or-none transition (see ref. 28). This is in contrast to the 2D gel pattern for DNA unwinding elements, including the (ATTCT) repeats and flanking A+T rich region at the SCA10 locus, in which a plateau of topoisomers indicative of continued unwinding is observed (17). Subsequent to an initial structural transition, topoisomers with increasing supercoiling in plasmids containing the (CCTG)n·(CAGG)n repeats continued to migrate with reduced mobility (Fig. 3), indicating continued structure formation with increasing superhelical density, as observed for (GAA)·(TTC) repeats (27). This is perhaps not surprising given the lengths of the repeat tracts involved (232 and 684 bp).
DNA supercoiling may be driving slipped-strand DNA formation by a mechanism that involves repeat unwinding, strand extrusion, and branch migration of a (CCTG) loop. Subsequent interaction of a (CAGG) hairpin loop and the (CCTG) loop would result in formation of a folded slipped-strand structure, as shown in Fig. S1.
Left-handed Z-DNA forms in alternating purine-pyrimidine tracts and is stabilized by negative DNA supercoiling and DNA–protein interactions in cells (33, 34). The degree of negative supercoiling necessary to stabilize Z-DNA depends on both the length and the composition of the Z-DNA-forming tract (28). The (TG)15·(CA)15 repeat in the normal human sequence flanking the DM2 (CCTG)n repeat forms Z-DNA at a physiological superhelical density of σ = −0.043, consistent with previous analyses of similar DNAs containing (TG)n repeats (see ref. 28 for review). This superhelical density is within the range detected in various human gene regions in human cells (35, 36), suggesting that this sequence has the potential to exist as Z-DNA in cells. The slipped-strand structure forms at a higher superhelical density than Z-DNA, and the (CCTG)n repeat did not alter the superhelical density at which Z-DNA formed. Moreover, the presence of the Z-DNA-forming tract in the flanking human sequence did not promote slipped-strand DNA formation in a premutation repeat length of (CCTG)31·(CAGG)31 (pSE-T-FS31), which is below the transition length threshold of 36–42. This result suggests that a flanking Z-DNA forming sequence may not stimulate structure formation in the adjacent (CCTG)·(CAGG) repeat tract, at least for this size of tract.
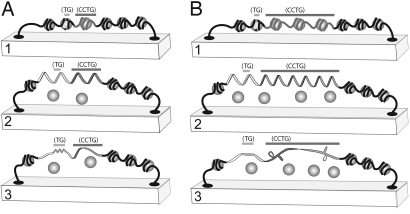
Z-DNA formation at the evolutionarily conserved Z-DNA forming tract in the ZNF9 gene suggests a biological role for Z-DNA. Z-DNA forming sequences can act as a torsional sink, absorbing and reducing negative supercoiling, preventing other alternative DNA structure formation. Competition for supercoiling energy can exist between 2 or more sequences that can form different supercoil-dependent alternative DNA structures (17, 37). For example, when adjacent to an A+T rich region, the formation of Z-DNA increased the negative supercoiling required to melt the A+T tract (30). Consistent with this result, slipped-strand DNA structures formed in (CCTG)·(CAGG) tracts at reduced levels when a flanking human sequence including the Z-DNA-forming region was present. The formation of slipped-strand DNA may lead to replication fork arrest, primer-template misalignment, or recombination during replication restart, all of which may be a causative factor in the DNA-directed expansions and deletions associated with neurodegenerative disease-associated repeats (14). Z-DNA formation would reduce the potential for formation of slipped-strand DNA, thereby protecting normal length, or some premutation length (CCTG)·(CAGG) repeats from structure formation and subsequent genetic instability (Fig. 6A). The energy required for the formation of most alternative DNA secondary structures decreases as the repeat length increases (28). As long repeat lengths (e.g., 170 repeats) form slipped-strand structures very easily, the protective effect of the Z-DNA tract is expected to have limits. As repeat tract lengths increase, the decrease in superhelical density resulting from Z-DNA formation will afford increasingly less protection against slipped-strand structure formation (Fig. 6B).
Fig. 6.
A protective effect for Z-DNA against the formation of slipped-strand DNA in the DM2 allele. (A) 1, Representation of a region of the DM2 allele with the DNA organized in nucleosomes (shaded circles) in the context of a topological domain. The Z-DNA-forming region is denoted by light shading and the (CCTG)·(CAGG) repeat by darker shading. 2, Unrestrained supercoiling is present in the DNA as a result of (or concomitant with) the loss of nucleosomes. 3, Z-DNA formation leads to relaxation of negative supercoiling in the topological domain, which prevents the supercoil-dependent formation of slipped-strand DNA structures. (B) 1 and 2, Representation as in A but with a (CCTG)·(CAGG) repeat expansion. 3, In the longer (CCTG)·(CAGG) tract, slipped-strand structures form easily and the relaxation of supercoils from Z-DNA may not be sufficient to prevent slipped-strand DNA formation. Formation, however, occurs at reduced levels compared with a (CCTG) tract lacking the flanking Z-DNA-forming sequence.
Z-DNA forming sequences have a variety of biological functions and consequences in mammalian cells (34). Z-DNA sequences have been associated with deletions (38, 39) and recombination hot spots in human chromosomes (40). Z-DNA may be particularly important in the regulation of genes requiring nuclear factor 1 binding, as demonstrated for the CSF1 promoter (34, 41–43). The vaccinia virus Z-DNA binding protein E3L is critical for pathogenic activity suggesting that 1 component of viral pathogenesis may be interference with the normal role of Z-DNA (44–46). The role of the evolutionarily conserved Z-DNA forming sequence in the DM2 gene is unknown. Both the 5′ and 3′ flanking regions of this ZNF9 gene are highly conserved in chimpanzees and gorillas (9), so this tract may have some regulatory or other biological function. While the Z-DNA tract may have a protective effect preventing slipped-strand DNA formation, if the Z-DNA region has a critical gene regulatory function, the formation of slipped-strand structures in long repeat lengths may reduce unrestrained supercoiling in the region, such that Z-DNA cannot form, thereby abrogating a biological role of Z DNA.
Materials and Methods
Plasmids.
Plasmids are described in Table 1 and were purified as described in ref. 21. All (CCTG)·(CAGG) repeats containing and lacking human repetitive flanking sequence were cloned into the EcoRI sites of pUC8 (2,665 bp), pTracer-SV40 (4,217 bp) (Invitrogen), or pEP-I-EGFP (6,692 bp) [a gift from H. J. Lipps, University of Witten, Witten, Germany (47)]. For plasmids containing (CCTG)·(CAGG) repeats and human flanking sequences, the DNA sequences were cut from plasmids containing PCR products of human alleles. For plasmids lacking human flanking sequences, repeats were generated as described, adding EcoRI linkers such that the (CCTG)·(CAGG) repeat is flanked by EcoRI sites (48).
Table 1.
Plasmids used for structural analysis
| Name | Vector base | Relevant sequences |
|---|---|---|
| pSE-U-FS11 | pUC8 | (TG)15(TCTG)8(CCTG)11 |
| pSE-U-FS28 | pUC8 | (TG)18(TCTG)11(CCTG)28 |
| pSE-U-FS57 | pUC8 | (TG)18(TCTG)11(CCTG)57 |
| pSE-U-FS94 | pUC8 | (TG)18(TCTG)11(CCTG)94 |
| pSE-U-FS31 | pUC8 | (TG)20(TCTG)11(CCTG)31 |
| pSE-U-CCTG58 | pUC8 | (CCTG)58 |
| pSE-U-CCTG170 | pUC8 | (CCTG)170 (±5 repeats) |
| pSE-T-CCTG9 | pTracer-SV40 | (CCTG)9 |
| pSE-T-CCTG36 | pTracer-SV40 | (CCTG)36 |
| pSE-T-CCTG42 | pTracer-SV40 | (CAGG)42 |
| pSE-T-CCTG58 | pTracer-SV40 | (CCTG)58 |
| pSE-T-CCTG170 | pTracer-SV40 | (CCTG)170 (±5 repeats) |
| pSE-E-CCTG58 | pEP-I-EGFP | (CCTG)58 |
| pSE-E-CCTG170 | pEP-I-EGFP | (CCTG)170 (±5 repeats) |
All repeat sizes were verified by using restriction digestion band size analysis and/or sequencing.
Slipped-Strand DNA Formation and Analysis.
Two different DNA reduplexing reactions were used to form slipped-strand structures in plasmid DNA. One method involves alkaline denaturation and renaturation at 68 °C (21). The second method of reduplexing used temperature denaturation (23). DNA in 100 mM NaCl, 50 mM Tris·HCl pH 7.9, and 1 mM DTT was incubated for 1 min at 100 °C, 10 min at 85 °C, 60 min at 70 °C, and slowly cooled to room temperature. Slipped-strand DNA was analyzed on 5% nondenaturing polyacrylamide gels in TBE buffer (90 mM Tris, 90 mM borate, 2.5 mM EDTA, pH 8.3). Gels were run at a constant voltage (150 V, 10–12 V/cm) at room temperature, stained with ethidium bromide, and photographed. DNA fragment sizes were calculated relative to migration of a 100-bp or 1-kb ladder (New England Biolabs), or a known plasmid restriction digest.
Gel Electrophoresis.
Supercoiled DNA topoisomers of varying superhelical density were prepared as described in ref. 17. A broad distribution of topoisomers was combined, precipitated, and resuspended in 10 μL of TEN buffer (10 mM Tris·HCl, pH 7.6, 1 mM EDTA, 50 mM NaCl). To eliminate preexisting alternative DNA structures, DNA was incubated for 5 min at 80 °C and immediately frozen in a dry ice/ethanol bath, as described previously for removing cruciforms (49, 50). To promote DNA secondary structure formation, DNA was allowed to slowly cool to room temperature after a 5-min incubation at 80 °C.
DNA was loaded onto a 1.5% or 1.75% (wt/vol) agarose gel in TAE buffer (40 mM Tris, 25 mM acetate, 1 mM EDTA, pH 8.3). Two-dimensional gel electrophoresis was performed at 4 V/cm for both dimensions, with addition of 30 μg/mL chloroquine in the second dimension, which was run at 90° with respect to the first dimension. Gels for slipped-strand DNA analysis were run at 4 °C, and gels for Z-DNA analysis were run at 20 °C in the first dimension. All gels were run at 20 °C in the second dimension.
Supplementary Material
Acknowledgments.
We thank Pooja Dalal, Lindsay A. Raghoobar, and Payal M. Raulji for experimental assistance. This work was supported by National Institutes of Health Grant AR48171 (to R.K.), the Muscular Dystrophy Association (R.K.), and National Institutes of Health Grant ES05508 (to R.R.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807699106/DCSupplemental.
References
- 1.Liquori CL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 2.Bachinski LL, et al. Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: A single shared haplotype indicates an ancestral founder effect. Am J Hum Genet. 2003;73:835–848. doi: 10.1086/378566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krahe R, Bachinski LL, Udd B. In: Genetic Instabilities and Neurological Disease. Wells RD, Ashizawa T, editors. Burlington, MA: Academic; 2006. pp. 131–150. [Google Scholar]
- 4.Vihola A, et al. Histopathological differences of myotonic dystrophy type 1 (DM1) and PROMM/DM2. Neurology. 2003;60:1854–1857. doi: 10.1212/01.wnl.0000065898.61358.09. [DOI] [PubMed] [Google Scholar]
- 5.Tapscott SJ. Deconstructing myotonic dystrophy. Science. 2000;289:1701–1702. doi: 10.1126/science.289.5485.1701. [DOI] [PubMed] [Google Scholar]
- 6.Ranum LP, Day JW. Myotonic dystrophy: RNA pathogenesis comes into focus. Am J Hum Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day JW, et al. Myotonic dystrophy type 2: Molecular, diagnostic, and clinical spectrum. Neurology. 2003;60:657–664. doi: 10.1212/01.wnl.0000054481.84978.f9. [DOI] [PubMed] [Google Scholar]
- 8.Bachinski LL, et al. Premutation allele pool in myotonic dystrophy type 2 (DM2) Neurology. 2008 Nov 19; doi: 10.1212/01.wnl.0000333665.01888.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liquori CL, et al. Myotonic dystrophy type 2: Human founder haplotype and evolutionary conservation of the repeat tract. Am J Hum Genet. 2003;73:849–862. doi: 10.1086/378720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson CE, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 11.Kunst CB, Leeflang EP, Iber JC, Arnheim N, Warren ST. The effect of FMR1 CGG repeat interruptions on mutation frequency as measured by sperm typing. J Med Genet. 1997;34:627–631. doi: 10.1136/jmg.34.8.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama Z, Izumi Y, Kameyama M, Kawakami H, Nakamura S. The effect of CAT trinucleotide interruptions on the age at onset of spinocerebellar ataxia type 1 (SCA1) J Med Genet. 1999;36:546–548. [PMC free article] [PubMed] [Google Scholar]
- 13.Cleary JD, Pearson CE. The contribution of cis-elements to disease-associated repeat instability: Clinical and experimental evidence. Cytogenet Genome Res. 2003;100:25–55. doi: 10.1159/000072837. [DOI] [PubMed] [Google Scholar]
- 14.Sinden RR, Pytlos MJ, Potaman VN. In: Human Nucleotide Expansion Disorders. Fry M, Usdin K, editors. Berlin: Springer; 2006. pp. 3–53. [Google Scholar]
- 15.Sinden RR, Pytlos-Sinden MJ, Potaman VN. Slipped strand DNA structures. Front Biosci. 2007;12:4788–4799. doi: 10.2741/2427. [DOI] [PubMed] [Google Scholar]
- 16.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 17.Potaman VN, et al. Unpaired structures in SCA10 (ATTCT)n·(AGAAT)n repeats. J Mol Biol. 2003;326:1095–1111. doi: 10.1016/s0022-2836(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 18.Liu G, Bissler JJ, Sinden RR, Leffak M. Unstable spinocerebellar ataxia type 10 (ATTCT)·(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol Cell Biol. 2007;27:7828–7838. doi: 10.1128/MCB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dere R, Napierala M, Ranum LP, Wells RD. Hairpin structure-forming propensity of the (CCTG·CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J Biol Chem. 2004;279:41715–41726. doi: 10.1074/jbc.M406415200. [DOI] [PubMed] [Google Scholar]
- 20.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 22.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n·(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wetmur JG. Excluded volume effects on the rate of renaturation of DNA. Biopolymers. 1971;10:601–613. doi: 10.1002/bip.360100402. [DOI] [PubMed] [Google Scholar]
- 24.Zheng G, Sinden RR. Effect of base composition at the center of inverted repeated DNA sequences on cruciform transitions in DNA. J Biol Chem. 1988;263:5356–5361. [PubMed] [Google Scholar]
- 25.Wang JC. Helical repeat of DNA in solution. Proc Natl Acad Sci USA. 1979;76:200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson CE, et al. Slipped-strand DNAs formed by long (CAG)·(CTG) repeats: Slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potaman VN, et al. Length-dependent structure formation in Friedreich ataxia (GAA)n·(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004;32:1224–1231. doi: 10.1093/nar/gkh274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinden RR. DNA Structure and Function. San Diego: Academic; 1994. [Google Scholar]
- 29.Vogt N, Rousseau N, Leng M, Malfoy B. A study of the B-Z transition of the AC-rich region of the repeat unit of a satellite DNA from Cebus by means of chemical probes. J Biol Chem. 1988;263:11826–11832. [PubMed] [Google Scholar]
- 30.Aboul-ela F, Bowater RP, Lilley DM. Competing B-Z and helix-coil conformational transitions in supercoiled plasmid DNA. J Biol Chem. 1992;267:1776–1785. [PubMed] [Google Scholar]
- 31.Sullivan KM, Murchie AI, Lilley DM. Long range structural communication between sequences in supercoiled DNA. Sequence dependence of contextual influence on cruciform extrusion mechanism. J Biol Chem. 1988;263:13074–13082. [PubMed] [Google Scholar]
- 32.LeProust EM, Pearson CE, Sinden RR, Gao X. Unexpected formation of parallel duplex in GAA and TTC trinucleotide repeats of Friedreich's ataxia. J Mol Biol. 2000;302:1063–1080. doi: 10.1006/jmbi.2000.4073. [DOI] [PubMed] [Google Scholar]
- 33.Rich A, Nordheim A, Wang AHJ. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 34.Rich A, Zhang S. Timeline: Z-DNA: The long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 35.Kramer PR, et al. Transcriptional state of the mouse mammary tumor virus promoter can affect topological domain size in vivo. J Biol Chem. 1999;274:28590–28597. doi: 10.1074/jbc.274.40.28590. [DOI] [PubMed] [Google Scholar]
- 36.Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- 37.Benham CJ. Statistical mechanical analysis of competing conformational transitions in superhelical DNA. Cold Spring Harbor Symp Quant Biol. 1983;47:219–227. doi: 10.1101/sqb.1983.047.01.027. [DOI] [PubMed] [Google Scholar]
- 38.Fuscoe JC, et al. V(D)J recombinase-like activity mediates hprt gene deletion in human fetal T-lymphocytes. Cancer Res. 1991;51:6001–6005. [PubMed] [Google Scholar]
- 39.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majewski J, Ott J. GT repeats are associated with recombination on human chromosome 22. Genome Res. 2000;10:1108–1114. doi: 10.1101/gr.10.8.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu R, et al. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Mulholland N, Fu H, Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Champ PC, Maurice S, Vargason JM, Camp T, Ho PS. Distributions of Z-DNA and nuclear factor I in human chromosome 22: A model for coupled transcriptional regulation. Nucleic Acids Res. 2004;32:6501–6510. doi: 10.1093/nar/gkh988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon JA, Rich A. Biological function of the vaccinia virus Z-DNA-binding protein E3L: Gene transactivation and antiapoptotic activity in HeLa cells. Proc Natl Acad Sci USA. 2005;102:12759–12764. doi: 10.1073/pnas.0506011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YG, Lowenhaupt K, Oh DB, Kim KK, Rich A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection. Proc Natl Acad Sci USA. 2004;101:1514–1518. doi: 10.1073/pnas.0308260100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langland JO, et al. Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J Virol. 2006;80:10083–10095. doi: 10.1128/JVI.00607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenke AC, et al. Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc Natl Acad Sci USA. 2004;101:11322–11327. doi: 10.1073/pnas.0401355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SH, Cai L, Pytlos MJ, Edwards SF, Sinden RR. Generation of long tracts of disease-associated DNA repeats. BioTechniques. 2005;38:247–253. doi: 10.2144/05382RR01. [DOI] [PubMed] [Google Scholar]
- 49.Zheng G, Kochel T, Hoepfner RW, Timmons SE, Sinden RR. Torsionally tuned cruciform and Z-DNA probes for measuring unrestrained supercoiling at specific sites in DNA of living cells. J Mol Biol. 1991;221:107–122. doi: 10.1016/0022-2836(91)80208-c. [DOI] [PubMed] [Google Scholar]
- 50.Oussatcheva EA, et al. Influence of global DNA topology on cruciform formation in supercoiled DNA. J Mol Biol. 2004;338:735–743. doi: 10.1016/j.jmb.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 51.Edwards SF. Houston: University of Texas Health Science Center; 2006. Non B-DNA stuctures and genomic instability associated with myotonic dystrophy type 2 (CCTG)·(CAGG) repeats. PhD dissertion. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.