Abstract
Reliable studies of enzymatic reactions by combined quantum mechanical /molecular mechanics (QM(ai)/MM) approaches, with an ab initio description of the quantum region, presents a major challenge to computational chemists. The main problem is the need for very large computer time to evaluate the QM energy, which in turn makes it extremely challenging to perform proper configurational sampling. One of the most obvious options for accelerating QM/MM simulations is the use of an average solvent potential. In fact the idea of using an average solvent potential is rather obvious and has implicitly been used in Langevin dipole / QM calculations. However, in the case of explicit solvent models the practical implementations are more challenging and the accuracy of the averaging approach has not been validated. The present study introduces the average effect of the fluctuating solvent charges by using equivalent charge distributions, which are updated every m steps. Several models are evaluated in terms of the resulting accuracy and efficiency. The most effective model divides the system into an inner region with N explicit solvent atoms and an external region with two effective charges. Different models are considered in terms of the division of the solvent system and the update frequency. Another key element of our approach is the use of the free energy perturbation (FEP) and/or linear response approximation (LRA) treatments that guarantees the evaluation of the rigorous solvation free energy. Special attention is paid to the convergence of the calculated solvation free energies and the corresponding solute polarization. The performance of the method is examined by evaluating the solvation of a water molecule and a formate ion in water and also the dipole moment of water in water solution. Remarkably, it is found that different averaging procedures eventually converge to the same value but some protocols provide optimal ways of obtaining the final QM(ai)/MM converged results. The current method can provide computational time saving of 1000 for properly converging simulations relative to calculations that evaluate the QM(ai)/MM energy every time step. A specialized version of our approach that starts with a classical FEP charging and then evaluates the free energy of moving from the classical potential to the QM/MM potential appears to be particularly effective. This approach should provide a very powerful tool for QM(ai)/MM evaluation of solvation free energies in aqueous solutions and proteins.
Keywords: QM/MM, ab initio, solvation, free energy, polarization
I. Introduction
QM/MM approaches have provided a general scheme for studies of chemical processes in proteins.1-12 Significant progress has been made with calibrated semiempirical QM/MM approaches2, 7, 10, 11 that include careful evaluations of the relevant activation free energies by free energy perturbation approaches that date back to the 80's13. These studies exploit the rapid evaluation of the semiempirical energies and sample the phase space of the QM atoms and the surrounding MM atoms. However, the current challenge is to move to an ab initio representation with a QM/MM treatment, since such QM(ai) representations have been shown to provide “chemical accuracy” in studies of gas phase reactions of small molecules. Here, we use the term ab initio mainly to differentiate form semiempirical methods. Therefore, QM(ai) may also refer to density functional theory, which uses empirical parameters in common implementations. Unfortunately, it is at present extremely challenging to evaluate the potential of mean force (PMF) for enzymatic reactions by QM(ai)/MM approaches14, 15 due to the requirement of very extensive sampling, which results in the extremely computationally expensive repeated evaluation of the QM energies.
The recent realization of the importance of the proper sampling of QM(ai)/MM surfaces led to several advances16-25. A major direction of these advances have been based on different adaptations20-25 of our idea16, 17 of using a classical potential as a reference for the QM/MM calculations. Other strategies have also been quite promising19, 26-28. Nevertheless there is clearly a need for more “mainstream“ approaches that can be used in standard implementations and aid in obtaining converging QM(ai)/MM free energies.
Here, we consider a simple and powerful treatment that can be viewed as a variable time step approach (from the QM/MM perspective) that appears to provide a powerful way to evaluate electrostatic free energies by QM/MM approaches. This approach is formally equivalent to approaches that add the average potential to the solute Hamiltonian and is thus a mean field approximation. Obviously, adding the average potential is an old idea that was implicitly implemented in the QM/Langevin Dipole (QM/LD) model29-31. It is also implemented implicitly in continuum models32-35. Furthermore, an averaging approach was implemented recently in an instructive work of Yang and coworkers26. However, while the addition of the average potential to the semiempirical Hamiltonian is very simple29, it requires specialized implementation in standard commercial QM(ai)/MM codes (that are designed to handle external point charges). Furthermore, the use of the average potential may not reproduce the average energy obtained by using the instantaneous potential in each time step, as in the common case when the solvent fluctuations are significant.
In some respects our approach is close in spirit to the work of Aguilar and coworkers36-39 who examined important aspects of this issue. However, our work focuses on examining the relationship between the QM/MM approaches for the evaluation of free energies that do not involve any averaging and of different averaging strategies. This issue presents a general problem that, to the best of our knowledge, has not yet been addressed. We also pay special attention to the contribution of the solute polarization that reflects the response to the potential from the surrounding solvent. Obviously, it is quite simple to estimate this effect by classical approximations30, but the uncertainties in such approximations can be quite significant in the challenging case of charged transition states in proteins.
The present approach maps the effect of the fluctuating MM environment during m time steps, on a set of mL point charges (where L is the number of atoms within a relatively small cutoff radius) and two additional point charges that represent the rest of the environment. This seemingly simple implementation is found to be very effective, leading to computational time saving of a factor of 1000 in QM(ai)/MM calculations of solvation free energies where the solute structure is fixed. Furthermore, the present study casts an interesting light on the relationship between the solvent fluctuations and the convergence of QM(ai)/MM calculation. This insight will be exploited in QM(ai)/MM free energy calculations that allow the solute to fluctuate.
II. Methods
Our starting point is the QM/MM energy of our system that is expressed here as:
| (1) |
where R and r are the solute (S) and solvent (s) coordinates, respectively and where ΨS and ϕs are the wavefunction of the solute and solvent, respectively and Q is the vector of solute residual atomic charges that depends on the potential created by the solvent (Us). Here the first term, , is the energy of the polarized gas phase Hamiltonian with a solute wavefunction that reflect polarization by the solvent. The second term is the solute-solvent electrostatic interaction. The third term, EνdW (R,r), is the solute-solvent van der Waals interaction and the last term, EMM(r), is the solvent potential surface. One can study different types of problems with above Etot, and here we will take the evaluation of the solvation free energy as an example. Our starting point is the free energy perturbation (FEP) adiabatic charging (AC) approach29, 40 where we can use a potential in the form:
| (2) |
where E′ denotes energy of system without electrostatic solute-solvent interaction and where λk changes from zero to one in n+1 steps.
We can use the standard FEP equation
| (3) |
where β=1/(kBT); kB is the Boltzmann constant and T is the absolute temperature. The <>Ek stands for the average obtained during the propagation of the configurations using the potential of Ek.
Eq. (3) can be effectively approximated by using the linear response approximation (LRA) treatment 41:
| (4) |
where ΔGcav is the solvation free energy of the nonpolar neutral form of the solute. This term consist of two parts describing hydrophobic and van der Waals free energy of cavity, which are not included in the first two terms of Eq. (4). They were described in Ref. 31 and implemented in ChemSol 2.0 program,31 which was used to obtain ΔGcav in this study. In the case of homogenous solutions the second LRA term of Eq (4) is zero as the propagation of the configurations using the E′ potential, which excludes the solute-solvent electrostatic interaction, does not lead to proper orientation of the solvent molecules.
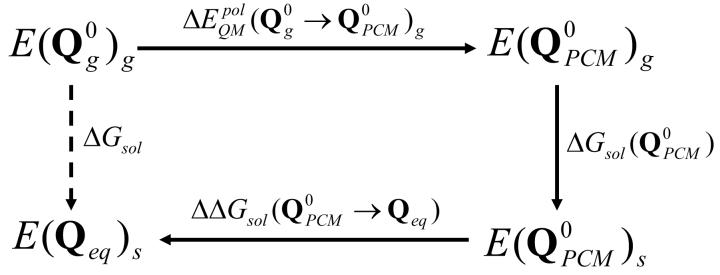
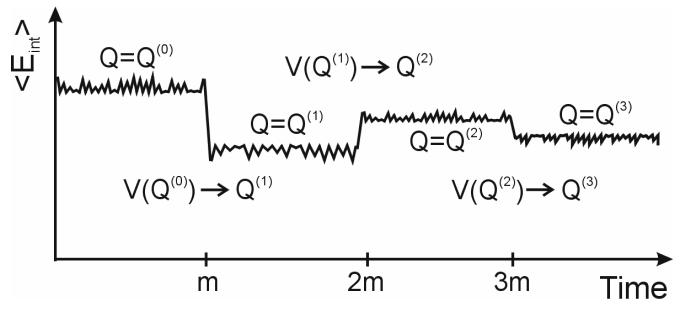
Alternatively, we can combine the approaches of Eqs. (3) and (4) into more efficient procedure presented in the following. This procedure use the cycle of Fig. 1 where we first run a classical MM simulation and use the FEP/AC approach29 to evaluate the free energy of charging the solute to a given charge distribution (e.g. partial charges obtained with the PCM solvation model (, where “0” in superscript designates a constant value)), and then evaluate the change in the free energy allowing the charge to “equilibrate” with the solvent potential. The vector of equilibrated QM/MM residual atomic charges is designed by Qeq. Thus the QM/MM solvation free energy can also be written as:
| (5) |
where the term is the solvation free energy of the solute, the atomic charges of which have been obtained from PCM model. The can be obtained using classical adiabatic charging approach based on FEP and therefore can be replaced by . The term in Eq. (5) is the polarization energy, which is given by:
| (6) |
and representing the polarization energy of a molecule in the PCM solvation model. The last term of Eq. (5), namely the , can be expressed using the LRA approach as:
| (7) |
where Etot(Q) is the QM/MM surface with the fluctuating charge.In case of both terms of Eq (7), the calculation of the Etot (Q) and is performed at exactly the same solute and solvent coordinates, and those systems differ only by the set of residual charges of the solute. Therefore, the average <> term becomes the difference in the polarization energies and the interaction energies of both systems. Calculation of each <> terms of Eq (7) requires performing QM calculations as the configurations are propagated to obtain solute polarization enegies included in Etot(Q). Therefore one can expect that calculation of Eq (7) might be expensive.
Figure 1.
Energy scheme. is calculated by the classical adiabatic charging approach.
Since 〈Q〉 and are similar, the LRA treatment can be approximated by assuming that both terms are equal (this corresponds to a single step FEP with a very small (E0−En) in Eq. (3)). Thus we can substitute the last term of Eq (5) with an approximated Eg (7), and write:
| (8) |
Of course, we can also use Eq. (7) without approximating it any further.
The main time consuming step in the evaluation of Eq. (8) is still the evaluation of the LRA term 〈 〉E(Q) term which is the expensive even for studies of solvation free energies. Thus, we move to the principle point of the present paper, which is the acceleration of the evaluation of Etot.
Obviously Etot can be approximated by using fixed solute charge and writing:
| (9) |
where Q̄ is some estimate of the solute charge in solution (from PCM or from QM results obtained at few solvent configurations). We can also replace by its classical approximation:
| (10) |
where q are the solvent residual charges, whereas i and j are indexes of the solute and solvent atoms, respectively.
can be estimated from LD or PCM calculations or even from a few QM/MM calculations at some solvent configurations. It is also possible to use the classical approximations considered in Ref. 30. Unfortunately, the approximations used for are very problematic since the solute wavefunction fluctuates during the solvent fluctuations. Here the obvious alternative is to evaluate the polarization by using the averaged potential from the solvent. This can be done by adding the solvent averaged potential to the solute Hamiltonian which in the simplest semiempirical formulation will look like29, 30:
| (11) |
where F is the Fock matrix. However, in the ab initio code the corresponding treatment is more complicated. Thus, although an innovative approach that add the average solvent potential to the solute Hamiltonian was recently presented26, most widely used codes treat the solvent potential by considering the solvent residual charge as an external charge, and our aim is to exploit this general feature in combining MM and QM programs.
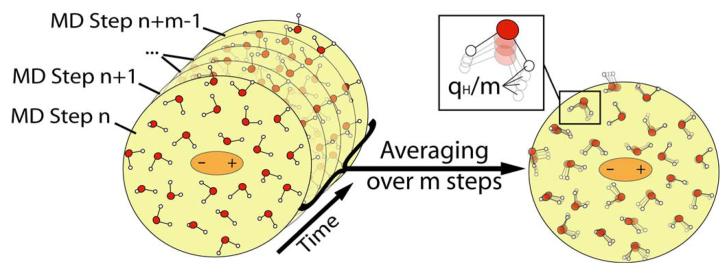
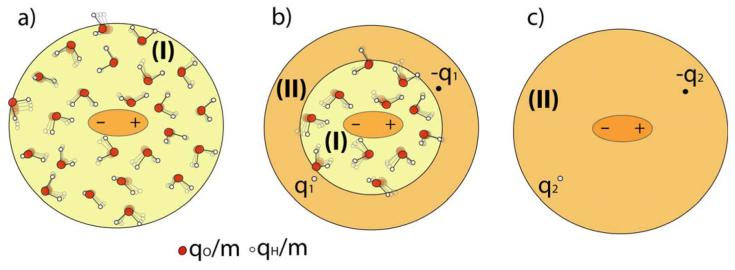
Our approach, which is technically similar to the approach of Aguilar and coworkers36, is demonstrated schematically in Fig. 1. In this strategy, we constrain the QM atoms (the solute atoms), evaluate the QM charges, Q(1), where (1) designates the first step and run m MM/MD steps allowing the solvent molecule to move in the potential . After m MD steps, we have m snapshots of solvent coordinates which can be used in two ways. The first is to evaluate the average potential from the m snapshots but this would require a special implementation. The second option is to scale the charge of each solvent atom by 1/m and to send m×N solvent atoms with the scaled solvent charges to the QM program. This approach (Fig. 2 and Fig. 3b) is very simple but unfortunately it generates m×N external charges to be included into the Hamiltonian within the QM program. This can be too expensive and inconvenient as will be shown in the following sections. Thus, we introduce the approximation described in Fig. 3b. In this treatment we divide the solvent into two regions. In the first region (region I), we convert the Next solvent atoms to m×Next external charges (scaled by 1/m), while in region II, we represent the average solvent field coming from N-Next solvent molecules, by two point charges (q and −q) using:
| (12) |
where EO is electric field gradient at point O (the geometrical center of QM system) and OR is pointing along EO to charge q.
Figure 2.
A schematic representation of the averaging of the solvent potential over m steps of a MD simulation.
Figure 3.
Different models for the evaluation of the average solvent charges: model M1 (a) involves an averaging of the explicit solvent molecules; models M2 and M3 (b) average the explicit molecules in region (I) while representing the average potential of the molecule in region II by two charges; model M4 (c) represents the solvent effect by two charges.
The validity of the averaging approach is related to the approximation
| (13) |
where < >m designates an averaged over m MD steps. However as will be seen below it is not essential to satisfy Eq. (13).
The validity of the use of effective charges simply depends on the magnitude of region I. In this study, we consider four solvent representation models that reflect different sizes of region I and region II. Model M1 and model M4 are single layer models consisting only of region I or region II respectively. Models M2 and M3 are dual layers models and the corresponding sizes of region I and II are presented in Table 1. At any rate, our task is to explore the range of validity of the above approximations and this will be done in the next section.
Table 1.
The different solvent representations used in this work.
| Model name |
Radius of regions (I) and (II) in Å |
Number of explicit water molecules solvating different systems |
Comment | ||
|---|---|---|---|---|---|
| rI | rII | H2O | HCOO− | ||
| Model M1 | 16 | n/a | 565 | 563 |
One layer, Fig. 3a |
| Model M2 | 10 | 16 | 137 | 143 |
Two layers, Fig. 3b |
| Model M3 | 6 | 16 | 28 | -* |
Two layers, Fig. 3b |
| Model M4 | n/a | 16 | 0 | 0 |
One layer, Fig. 3c |
This model was not used for this system.
The validation of the above approximations will be conducted for two different types of solutes: a polar molecule - water and an HCOO− ion. All the solute atoms will be solvated by explicit water molecules placed in a sphere with a radius of 16 Å, which corresponds to 565 and 563 water molecules respectively. All solvent molecules are represented by the ENZYMIX force filed42. In the simulation model the sphere of the explicit water molecules is surrounded by a surface region whose average polarization and radial distribution are determined by the surface constrained all-atom solvent (SCAAS) model40, 43, 44. The surface region is embedded in a bulk continuum region with a dielectric constant of 80. The long range interactions are treated by the local reaction field (LRF) approach45.
The MD simulations presented here were performed using the MOLARIS package42. In every case we first relaxed the system in a 10 ps long simulation of 1 fs time steps. All QM calculations were performed using the Gaussian03 package. The B3LYP exchange-correlation potential46, 47 was used with 6-31+G** and aug-cc-pVDZ basis sets48. The Merz-Kollman scheme49 with default atom radii was used to determine charges on atoms to be later used in the MD simulations.
The combined QM/MM calculations were facilitated by developing new extensions of MOLARIS that store solvent trajectories and calculate charges to be later used to define the external average potential in the QM calculation. The Gaussian03 - Molaris communication as well as free energy mapping was facilitated by application of the Gaussian Output Tools package50 and other Perl scripts. All benchmarked calculations presented here were performed on dual Intel Xeon 3.06 GHz machines with 2Gb of memory running the Red Hat Enterprise Linux 5 operating system.
III. Results and discussion
III.1 A solvated water molecule
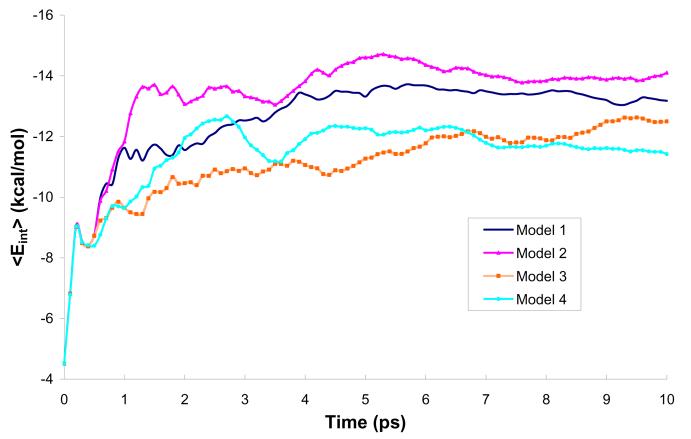
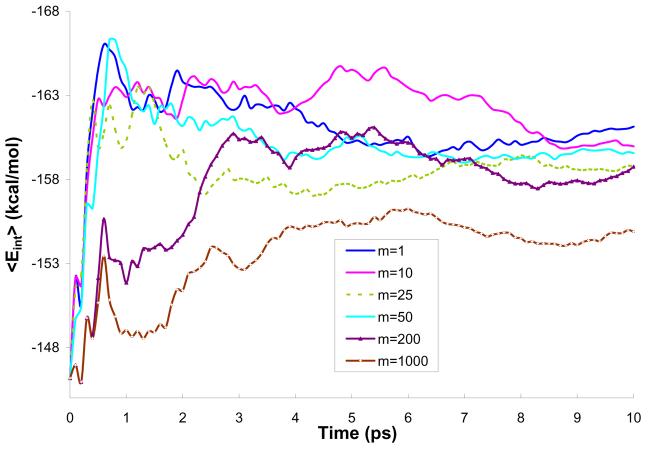
In the first stage of our study, we evaluated the performance of the four different models (M1-M4) for representing the solvent effects on the solute while using the standard QM/MM approach (m=1). This step was needed because the high m values (m>50) are only accessible for models with a reduced number of explicit water molecules (models M2-M4). Fig. 4 examines the averaged solute-solvent interaction energy (<Eint>) during a simulation of a solvated H2O molecule in water using models M1-M4. In the case of the reference model (model M1), the value of <Eint> converged after about 5ps; here we define the convergence by the reaching of a constant value. The other models converged in a similar time (see however below). The main point that emerged from the Fig. 4 is that model M2 (where about 75% of explicit water molecules are represented by two charges) provided an excellent approximation: the plot of <Eint> is practically parallel to that of model M1. The shift of ca. 0.5 kcal/mol originates in the beginning of the simulation (first 1.5 ps) and can be easily corrected by not taking the first 2ps of the QM/MM simulation (additional equilibration - see Table 2) into account. As seen from the Fig. 4 even the simplest models (M3 and M4) perform quite well, but they need much longer simulation times to converge. The longer simulation times can only be reached by increasing m. However, as will be shown below, the converged results are practically the same as ones obtained with the reference model M1. The rationale for using models M2-M4 is clarified in Table 3, where we evaluate the timing of one QM calculation using each model for large values of m. In this study, models M2 and M4 are tested most extensively. Apparently, model M2 provides a good balance between calculation time and the simplification of the solvent representation. Model M4 has the big advantage of short calculation times (despite the fact that it might require more steps to converge), while maintaining good accuracy. At the same time, its simplicity may seem somewhat artificial for those who may prefer more complete models.
Figure 4.
Convergence of the average interaction energy during 10ps simulations (m=1) using different models of solvent representation.
Table 2.
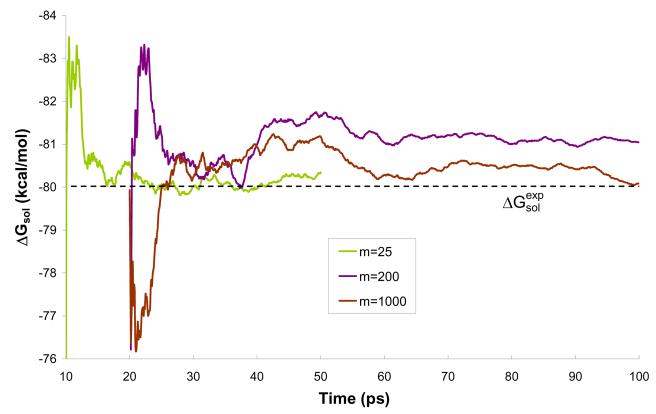
Average polarization energy (<Epol>), average solute-solvent electrostatic interaction energy (<Eint>) and free energy of solvation (ΔGsol of Eq. (7)) obtained during the last 8 ps of 10 ps long simulations of water molecule in a water solution. The best estimates of the free energy of solvation (ΔG sol * also of Eq. (7)) are obtained by performing 50 and 100 ps simulations for m=(25,50) or m=(100-1000), respectively. The free energies of solvation include the ΔGcav term of 0.9 kcal/mol.
| m | Solvent representation models | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model M1 | Model M2 | Model M3 | Model M4 | ||||||||||||
| <EPol> | <EInt> | ΔGsol | <EPol> | <EInt> | ΔGsol | ΔGsol* | <EPol> | <EInt> | ΔGsol | ΔGsol* | <EPol> | <EInt> | ΔGsol | ΔGsol* | |
| 1 | 1.6 | −14.0 | −6.1 | 1.6 | −14.0 | −6.1 | -* | 1.4 | −13.2 | −5.9 | -* | 1.0 | −11.7 | −5.3 | -* |
| 10 | 1.4 | −12.8 | −5.8 | 1.4 | −13.3 | −5.9 | -* | 1.4 | −13.1 | −5.8 | -* | 1.1 | −12.4 | −5.7 | -* |
| 25 | 1.4 | −13.0 | −5.7 | 1.9 | −17.0 | −7.5 | −6.1 | 1.1 | −11.2 | −4.9 | −5.9 | 1.4 | −14.3 | −6.3 | −6.1 |
| 50 | 1.5 | −14.2 | −6.2 | 1.4 | −13.5 | −6.1 | −6.3 | 1.5 | −14.4 | −6.4 | −6.2 | 1.4 | −15.4 | −6.8 | −6.3 |
| 100 | -** | -** | -** | 1.5 | −14.5 | −6.4 | −5.8 | 1.3 | −13.6 | −6.2 | −6.4 | 1.1 | −12.8 | −6.0 | −6.1 |
| 200 | -** | -** | -** | 1.1 | −11.7 | −5.3 | −6.0 | 1.3 | −13.9 | −6.2 | −6.2 | 1.2 | −14.0 | −6.5 | −6.3 |
| 500 | -** | -** | -** | 1.1 | −12.6 | −5.7 | −6.3 | 1.1 | −12.5 | −5.5 | −6.0 | 0.8 | −10.9 | −4.9 | −6.2 |
| 1000 | -** | -** | -** | 1.3 | −14.0 | −6.5 | −6.4 | 1.0 | −11.7 | −5.3 | −5.9 | 0.8 | −12.3 | −5.8 | −6.4 |
not included in this study
cannot be completed due to insufficient memory of the available computer
Table 3.
Total QM/MM simulation time (wall time, τ, in seconds) and average wall time required to complete 1 QM step (t) obtained for 10000 steps simulation of solvated a water molecule using solvent potential averaged over m steps. Difference solvent representations models are defined in Table 1.
| Solvent representation model | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model M1 | Model M2 | Model M3 | Model M4 | |||||
| m | τ | t | τ | t | τ | t | τ | t |
| 1 | 196941 | 22 | 192692 | 19 | 190123 | 19 | 189782 | 19 |
| 10 | 29816 | 30 | 21665 | 20 | 20568 | 19 | 20347 | 19 |
| 25 | 25980 | 62 | 10764 | 24 | 10103 | 19 | 8911 | 19 |
| 50 | 35502 | 170 | 7797 | 32 | 7092 | 29 | 5095 | 19 |
| 100 | -* | -* | 7536 | 61 | 4385 | 31 | 3184 | 19 |
| 200 | -* | -* | 9925 | 170 | 3125 | 36 | 2256 | 19 |
| 500 | -* | -* | 19436 | 893 | 2695 | 65 | 1657 | 19 |
| 1000 | -* | -* | 34803 | 3296 | 3119 | 168 | 1465 | 19 |
cannot be completed due to insufficient memory of the available computer
After validating our approach for obtaining V̄m, we moved to the examination of the performance of Eq. (12) for different values of m, (note that our final focus will be on solvation free energies rather than Eq. (13).). This was done for the test case of a solvated water molecule in water where we evaluate major contribution to solvation free energy (solute-solvent electrostatic interaction energy (Eint) and solute polarization energy(Epol)) and dipole moment as a function of the simulation time.
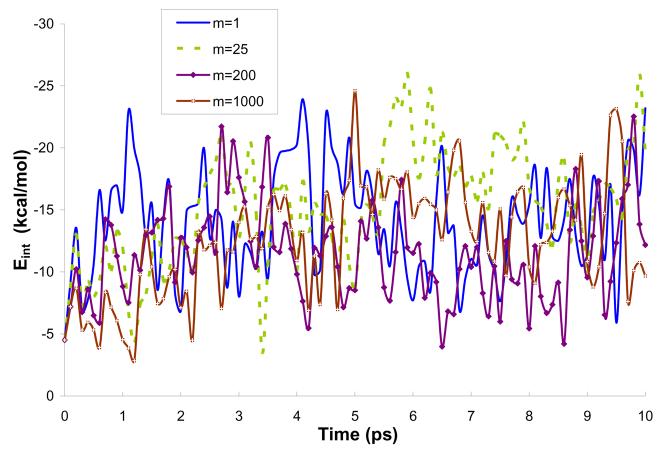
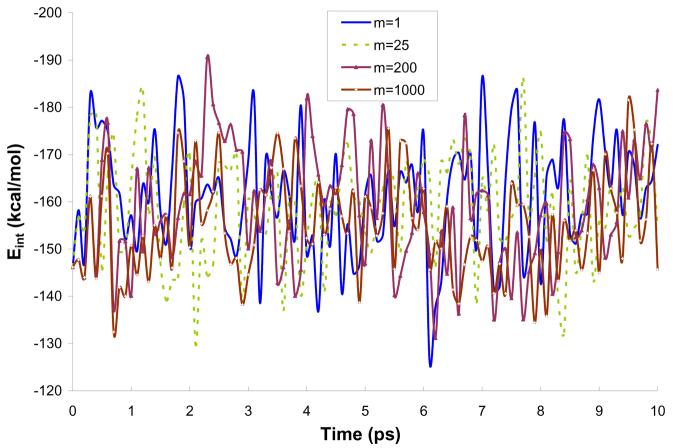
Fig. 5 describes the interaction energy for the different m's as a function of time, while Figs. 6 and 7 represent, respectively, the average solute-solvent electrostatic interaction energy and the solute polarization energy as a function of time. The plots represent 10 ps simulations using model M2. It can be seen from Fig. 5 that the amplitude of Eint during the simulation is similar for the m's studied (for the clarity of the plot only selected m's were presented in Fig. 5). This is due to fact that m only defines how often the charges on the solute atoms are updated according to the average solvent potential. However, since the reasonable charges obtained in PCM calculations are used as the starting point of MD simulation, the changes in charges are not significant and it does not seem to affect the solute-solvent interaction to a great extent.
Figure 5.
Interaction energy during 10ps simulations of water molecule in water using model M2. The charge on the solute atoms were updated every mth step.
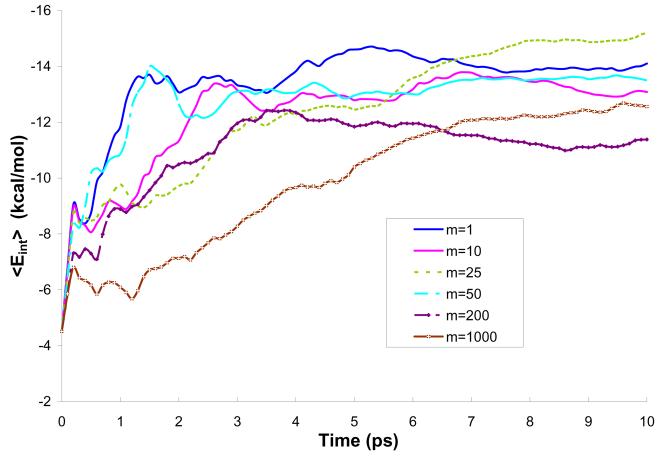
Figure 6.
Average solute-solvent interaction energy during 10 ps simulations of a water molecule in water solution (model M2).
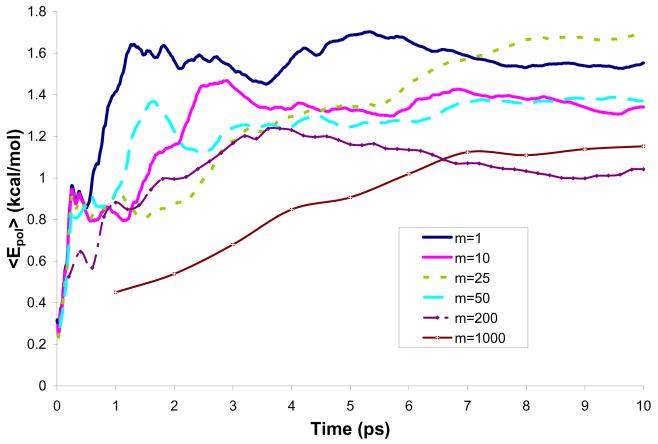
Figure 7.
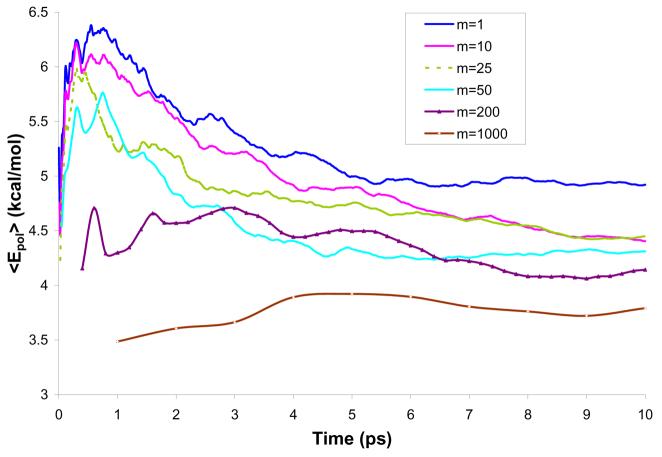
Average solute polarization energy during 10 ps simulations of water molecule in a water solution (model M2).
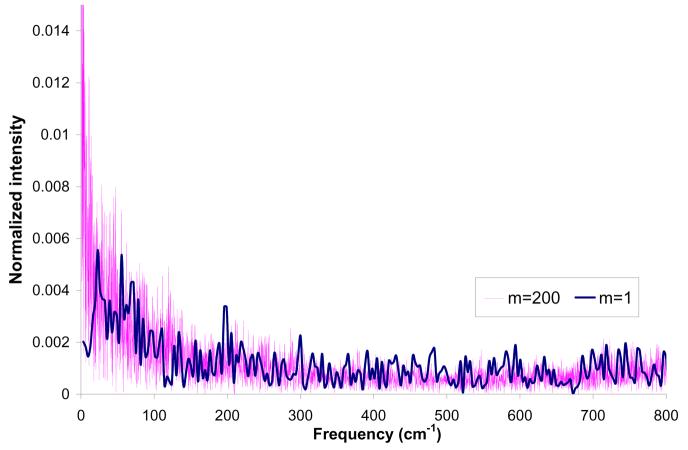
The plots of the averaged interaction energy, <Eint>, and the polarization energy, <Epol>, lead to the conclusion that all selected m's lead to convergence. At the same time it is apparent that even the m=10 simulation does not follow the m=1 simulation in an exact way. These results might look surprising at first glance. However, a closer look should start from the realization that the solvent molecules fluctuate on a wide frequency range and that some of these frequencies involve periods which are shorter than the time between different QM/MM evaluations for m>1. This point is clear from the Fourier transform of the fluctuating interaction energy (Fig. 8), which includes oscillations above 800 cm−1 (about 7 fs). Fortunately, the fast oscillations do not contribute to the final free energy. Thus we do not expect to have the same result for m=1 and m>1. In asking what the optimal value of m is, we have to realize that converging calculations require “equilibration” between the solute charge and the solvent potential. For example, we could use the approach described in Fig. 9. In this approach, we run long MD simulation with Q(0) and then take the resulting average potential, evaluate new Q and run another set of MD simulations. This procedure is repeated until convergence is reached. The plots demonstrating such convergence for different values of m and 3 models (M2-M4) are presented in Figs. S-1 - S-6 of the Supplementary Materials.
Figure 8.
Fourier transform of the fluctuating interaction energy as a function of the simulation length and the value of m obtained in simulation of water molecule in water using model M2.
Figure 9.
Schematic convergence.
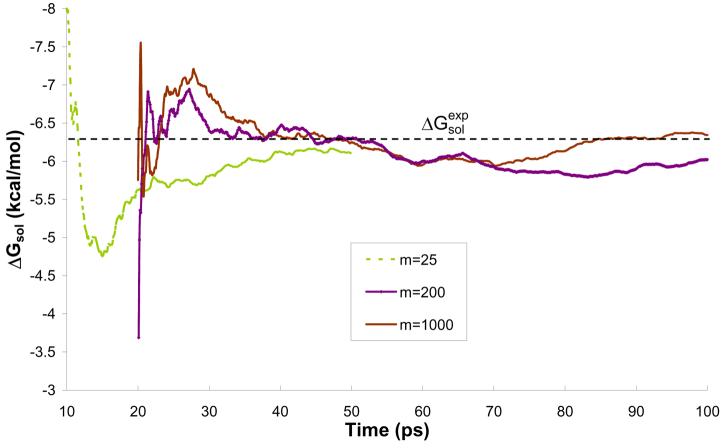
The convergence issue becomes very important when we evaluate solvation free energies by QM/MM models. Thus, we present the results for calculations of ΔGsol for different values of m and different solvent representations obtained after 10 ps simulation in Table 3. In the case of the model M1 and m=1 the result of −6.1 kcal/mol is converged within a 10ps simulation and is almost equal to experimental value of −6.3 kcal/mol51. When using the simpler solvent representation, model M2, the result is also converged and it the same as for model M1. For m=1, models M3 and M4 give results further from the experimental value (−5.9 and −5.3, respectively). These results are not converged, however, but because of the computational cost the longer simulation needed for full verification was not performed. Such longer simulations were performed for m>10. As an example let us consider the ΔGsol results obtained with model M2 for m=25 and m=200. In the 10 ps simulation, we obtained −7.5 and −5.3 kcal/mol, respectively, which is up to 20% off the correct value. However, these results do not reflect proper convergence and longer simulation had to be conducted. The ΔGsol results of 50 and 100 ps simulations conducted for m=25 and m=200, respectively, are in perfect agreement with the corresponding experimental result (Figs. S1-S3 and Table 2). Similar results were obtained for model M1, which performed poorly over most of tested m's in the 10 ps simulation. However, the free energy of solvation averaged over the 50 and 100 ps simulations reproduce the experimental value (Table 2). Interestingly, for all presented solvent representation models, m=50 give perfect agreement with the experimental result, but this is coincidental, since m=50 did not perform as well for other solvent representation models not presented in this study.
At this point, one should wonder what are our recommendations for choosing the optimal model for solvent representation and the value of m for calculating the free-energy of solvation? As was shown here, there is strong coupling between these parameters and the required time to obtain converged results. When using models M1-M3, increasing m will lead to an increase of the single QM step calculation time (Table 3). This trend is drastic for model M1 and might also become evident for model M3 for very large values of m. The single QM calculation time using model M4 does not depend on m, however, this model might require much more MD steps to converge. One also has to keep in mind that model M4 is only good for small molecules, as this model uses two charges to reproduce the electric field only in the geometrical center of the solute. Larger molecules might require more sophisticated approaches to reproduce the local polarization of the molecular fragments. Therefore, the selection of the value of m and the appropriate model should be based on the system being studied and the estimated total calculation time required for convergence. In the case of the water molecule the most efficient approach would be to perform 100 ps simulation using model M4 and m=1000. Comparing to the standard approach (model M1, m=1), it offers calculation time savings by 100 times less, while still giving the same result. As mentioned before, chemists might prefer to use more complete solvent potential models like M2 or M3, which performed well for values of m from 25 to 1000 offering computational time savings by factor of up to 73 in the presented case (M3, m=500).
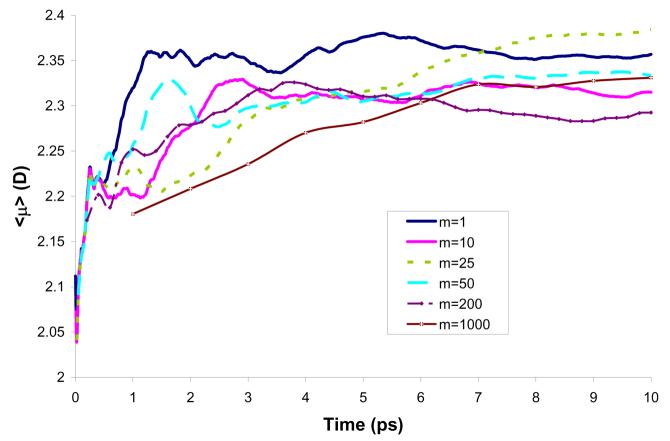
After exploring the energetic issue, it is useful to examine the convergence of the solute polarization and this was done in the present study in terms of the dipole moment of water. As seen in Fig. 10, the use of different values of m leads to the convergence of the dipole moment. The converged value of the dipole moment obtained with m=1 is 2.35D. For other values of m considered in this study the values obtained are within 2.28-2.38 D, which correspond to shifts of 0.48-0.58 D compared to the gas-phase result. Other studies employing broad range of computational techniques suggested the shifts of 0.4-0.8 D.52-54 Mendoza et al.38 have used a mean field approach with averaging potential over a different number of configurations and obtained dipole moments between 2.72 and 2.78 D. At any rate, our aim is not to reproduce of the dipole moment, but rather compare the results obtained by different m's.
Figure 10.
Average dipole moment of water molecule in water along a 10 ps simulation using model M2.
III.2 A solvated formate ion
After studying a polar molecule (namely H2O) we examined the performance of our approach in the case of solvated ion. As a test case, we consider the formate ion and the corresponding results are summarized in Table 4 and Figs. 12-15. The plot of the solute-solvent interaction energy, Eint, obtained in a number of simulations using m=1…1000 and model M2 is presented in Fig. 12. It can be seen from the figure that the amplitudes of Eint are on the same order of magnitude for all values of m considered. Also, the average solute-solvent interaction energy, <Eint>, appears to converge nicely (Fig. 13). More specifically, <Eint>, converges (within 2 kcal/mol) in all simulations with m<100. The convergence with m=1000 appeared to be much slower since the solute charge was updated only 10 times and thus did not reach satisfactory equilibration. Additional plots demonstrating the convergence of <Eint> in 50 ps and 100 ps simulations for m=25,50 and m≥100, respectively are presented in Figs. S-7 and S-8.
Table 4.
Average polarization energy (<Epol>), average solute-solvent electrostatic interaction energy (<Eint>) and free energy of solvation (ΔGsol of Eq. (8)) obtained during the last 8 ps of 10 ps simulations of HCOO− anion in water solution. t is the average time required for one QM step. The best estimates of the free energy of solvation (ΔGsol*) are obtained by taking average over the last 80ps of 100 ps simulations (available only for m≥100). Average energies and free energies are given in kcal/mol, a time is given in seconds. For all calculations using model M4 t=26. The free energies of solvation include the ΔGcav term of 1.5 kcal/mol.
| m | Solvent representation models | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model M1 | Model M2 | Model M4 | ||||||||
| <EPol> | <EInt> | ΔGsol | t | <Epol> | <EInt> | ΔGsol | ΔGsol* | t | ΔGsol* | |
| 1 | 4.1 | −156.2 | −78.6 | 28 | 4.7 | −160.9 | −80.6 | -* | 27 | -* |
| 10 | 4.4 | −158.7 | −79.7 | 38 | 4.1 | −157.8 | −79.3 | -* | 41 | -* |
| 25 | 4.3 | −159.1 | −79.9 | 74 | 4.3 | −159.3 | −79.9 | −80.1 | 45 | -* |
| 50 | 4.0 | −156.5 | −78.9 | 175 | 4.2 | −159.0 | −79.8 | −79.6 | 42 | -* |
| 100 | -** | -** | -** | -** | 4.2 | −157.0 | −78.8 | −79.8 | 73 | −81.7 |
| 200 | -** | -** | -** | -** | 4.1 | −157.6 | −79.2 | −80.9 | 174 | −81.5 |
| 500 | -** | -** | -** | -** | 4.4 | −163.7 | −82.2 | −80.3 | 918 | −81.5 |
| 1000 | -** | -** | -** | -** | 3.8 | −154.4 | −77.8 | −80.0 | 3379 | −81.7 |
not included in this study
cannot be completed due to insufficient memory of the available computer
Figure 12.
Interaction energy along 10ps simulations of HCOO− in water using model M2. Charge on the solute atoms updated every mth step.
Figure 15.
Free energy of solvation of the formate ion along 50 and 100 ps simulation. The calculations were performed using Eq. (4) and corrected by adding the difference between the ΔGsol of Eq. (7) and ΔGsol of Eq. (4).
Figure 13.
Average solute-solvent interaction energy along 10 ps simulation of HCOO− solvated by water (model M2).
The most remarkable difference between the convergence of <Eint> for H2O and HCOO− is that the convergence is much slower in the latter case. This reflects the fact that the initial orientation of the solvent molecules is far from optimal in the HCOO− case and more MD steps are required to orient them according to electric field of an ion. As shown by the plots of <Eint> along 50 and 100 ps simulations for m 25, 50 and m≥100 respectively the convergence within 1 kcal/mol is reached after 40 and 60 ps respectively.
In addition to the solute-solvent interaction energy, we studied the polarization energy of the HCOO− ion. As shown in Fig. 14, the average polarization energy, <Epol>, converges within a 10 ps simulation, for all the values of m considered. The converged value spread between 3.5 and 5.5 kcal/mol. The lowest value of ca. 3.5 kcal/mol correspond to m=1000 and is reflected in the low average solute-solvent interaction energy observed in this case.
Figure 14.
Average solute polarization energy along 10 ps simulation of HCOO− ion in water solution (model M2).
The converged result of <Eint> and <Epol> can be used to obtain the solvation free energy using Eq. (4). The convergence of this approach can be assessed from Fig. 15 (see, however, the comment below). However, the final converged value turn out to be −88.0 kcal/mol which represent a significant overestimation of the observed solvation energy (−80 kcal/mol43). A comparison of the classical ΔGsol evaluated by the LRA approach of Eq. (4) and by the FEP approach of Eq. (3) established that the overestimation is entirely due to the use of the LRA approximation for the entire charging process. Thus we used Eq. (8) and obtained the results summarized in Table 4. As seen from the table the calculated solvation free energy of HCOO− can converge to −80±1 kcal/mol and is in excellent agreement with the corresponding observed value. Since the QM/MM part of Eq. (8) converges much faster than the corresponding part of Eq. (4), we describe in Fig. 15 the convergence of Eq. (4) correcting, however, the final result to those obtained by Eq. (8).
Overall the QM/MM treatment of Eq. (4) converges after about 40 and 60 ps simulations for m=25,50 and m m≥100, respectively. On the other hand, the QM/MM part of Eq. (8) requires about 20 ps and the MM evaluation of is very inexpensive relative to the QM/MM part.
In order to assess the effect of the model used we also evaluate the solvation energies using model M4 (Table 4). The corresponding results are on average 1.5 kcal/mol lower than results obtained by the model M2 and even further from the experimental results.
Although the obtained free energy of solvation is in very good agreement with the experiment value, one has to keep in mind that the result can be inaccurate due to the QM method employed. The major contribution might be related to the size of the basis set used in QM calculations. We estimated these by conducting one test calculation (100 ps simulation with m=1000 and model M4) where we employed the larger aug-cc-pVDZ basis set rather than 6-31++G** used before. The larger basis set is expected to better reproduce the polarization effect as it provides more flexibility for the SCF procedure. Indeed the obtained free energy of solvation was decreased by only less than 1% (0.7 kcal/mol).
When discussing the performance issues one should consider the purpose of the calculation. For example, when trying to obtain solvation energies by Eq. (4) one should consider using the M1 model, averaging over m=25-50 solvent configurations and using accurate QM methods. In our test case of the HCOO− ion, the single QM calculation using model M1 and 50 solvent configurations was on average 6.35 times slower than calculation for m=1 using the same M1 model. However, since 50 times less QM calculations are required the overall speed-up is 7.8. For studies that require obtaining relative results for the number of systems, one could use models M2-M3 and m>100 which provided convergence within 100 ps simulation requiring 100-1000 QM steps. Our benchmark for model M2 and m=200 suggests that the single QM step is 6.5 times slower than for m=1, however, with averaging over m=200 the total speed-up using our accelerated approach is 30.6 times. On the other hand, for rough estimates of relative energies using the M4 model seem very attractive. In this case, the averaging even over 1000 steps takes as much time as single gas-phase calculation. Therefore, the total speedup using our accelerated approach can reach 1000 times in this case. This of course does not take into account the slower convergence observed in the case of HCOO− for m=1000, which in practice decreased the total speed-up to ca. 100 times, as a 10 times longer simulation was required.
IV. Concluding Remarks
The evaluation of the free energy changes in solutions and proteins by classical MM approaches usually requires very extensive averaging over the configurational space of the protein. This work establishes that the same requirement will hold for QM/MM calculations. Thus it is clear that simple minimization approaches of the type used in gas phase QM calculations would not be effective in evaluations of the activation energies of chemical reactions in proteins (e.g. see Ref. 55). Unfortunately, evaluating free energies of QM(ai)/MM surfaces is extremely challenging due to the need for extensive evaluation of the QM(ai) energies. Several innovative strategies for accelerating QM(ai)/MM sampling have already emerged16, 17,19, 26-28. Nevertheless there is clearly a need for more “mainstream“ approaches that can be used in standard implementations and help in obtaining converging QM(ai)/MM free energies.
The present study provides a practical and effective approach for performing QM(ai)/MM calculations of solvation free energies in solutions and in proteins. This is done by averaging the solvent potential over fixed a number of steps (m) in MM simulations, representing the effect of the fluctuating solvent charges by effective charges, and then updating the solute polarization by incorporating the effective charges in the solute Hamiltonian. The idea of using an average potential is obviously not new and has been used implicitly in our QM/LD models29-31. Furthermore, some important aspects of the current strategy have been used before. For example, the useful idea of representing the solvent potential by effective charges and a variable m value was used in Refs 36-39 and an average potential was used in Ref. 26. However, the main point in our work is the systematic examination of the convergence of the averaging approach in actual free energy calculations and the relationship between the uses of different m values and the full QM/MM simulations that do not use any updating. Moreover, we explored the validity of using simple charge distributions that lead to significant saving in computer time. Finally, the key point of our approach is the development of an accurate and effective approach for calculating QM(ai)/MM solvation free energies.
Our study establishes the conditions for converging free energy calculations and provides several very effective ways of obtaining converged results which, to the best of our knowledge, has not been accomplished to date. At any rate our approach provides a time saving of up to 1000 in QM/MM calculations of solvation free energies while basically giving the same results that would be obtained in such calculations without averaging procedure. Of course if one would run only 100 MD steps overall, one will not save much time. Establishing this point is quite important since the argument that one is using a mean field approximation usually implies that the results are approximated. However we found here that the use of an average potential in free energy calculations give the same results as free energy calculations with the instant potential.
The present study has not considered the solvent polarization since it used the MOLARIS non-polarizable force field. However, we would like to clarify that we have been using polarizable force fields in QM/MM calculations since 19761 and have been considering the solvent polarization in many works (for review on polarizable force-fields see Ref. 56). Thus we can add the polarized solvent dipoles to the solvent average charges by using the polarizable MOLARIS force field. This detail is left, however, to subsequent works.
The approach used here for fixed solvent coordinates can be extended to the much more challenging (and arguably more important) case where both the solute and solvent are allowed to fluctuate. At present there are only a few approaches that can be used to address this issue in an efficient way20-25. Thus, it is important to explore practical approaches for evaluating the PMF in the solute-solvent configurational space without fixing the solute coordinate during the free energy calculations. The most obvious use is the refinement of our approach that use the EVB as a reference potential15. In this approach we perform MD simulation on both the EVB and corresponding QM/MM surface. Thus, we can obtain significant saving by reducing the expensive QM evaluations while using the average MM potential obtained between the number of QM evaluations. However we also explore other less involved strategies. We believe that the present strategy will provide a useful way of advancing in this direction.
Figure 11.
Free energy of solvation of water along 50 and 100 ps simulation of water molecule in water solution.
Acknowledgment
Author would like to thank Dr. Shina Kamerlin for her comments of the manuscript. This work was supported by the NIH grant GM24492. M.H. holds award from Foundation for the Development of the University of Gdansk (FRUG). The computational work was supported by University of Southern California High Performance Computing and Communication Center (HPCC).
Footnotes
Supporting Information
Figs. S-1 - S-8 are available as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
The affiliation should be the institution where the work was conducted.
References
- 1.Warshel A, Levitt M. Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 2.Shurki A, Warshel A. Structure/Function Correlations of Protreins using MM, QM/MM and Related Approaches; Methods, Concepts, Pitfalls and Current Progress. Advances in Protein Chemistry. 2003;66:249–312. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]
- 3.Gao J. Hybrid quantum and molecular mechanical simulations: an alternative avenue to solvent effects in organic chemistry. Acc. Chem. Res. 1996;29:298–305. [Google Scholar]
- 4.Bakowies D, Thiel W. Hybrid Models for Combined Quantum Mechanical and Molecular Approaches. J. Phys. Chem. 1996;100:10580–10594. [Google Scholar]
- 5.Field MJ, Bash PA, Karplus M. A Combined Quantum Mechanical and Molecular Mechanical Potential for Molecular Dynamics Simulations. J. Comp. Chem. 1990;11:700–733. [Google Scholar]
- 6.Friesner R, Beachy MD. Quantum Mechanical Calculations on Biological Systems. Curr. Op. Struct. Biol. 1998;8:257–262. doi: 10.1016/s0959-440x(98)80048-1. [DOI] [PubMed] [Google Scholar]
- 7.Monard G, Merz KM. Combined Quantum Mechanical/Molecular Mechanical Methodologies Applied to Biomolecular Systems. Acc. Chem. Res. 1999;32:904–911. [Google Scholar]
- 8.Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM. A QM/MM study of the Racemization of Vinylglycolate Catalysis by Mandelate Racemase Enzyme. J. Am. Chem. Soc. 2001;123:709–721. doi: 10.1021/ja002879o. [DOI] [PubMed] [Google Scholar]
- 9.Marti SA,J, Moliner V, Silla E, Tunon I, Bertran J. Theor. Chem. Acc. 2001;3:207–212. [Google Scholar]
- 10.Field M. Stimulating Enzyme Reactions: Challenges and Perspectives. J. Comp. Chem. 2002;23:48–58. doi: 10.1002/jcc.1156. [DOI] [PubMed] [Google Scholar]
- 11.Cui Q, Elstner M, Kaxiras E, Frauenheim T, Karplus M. A QM/MM Implementation of the Self-Consistent Charge Density Functional Tight Binding (SCC-DFTB) Method. J. Phys. Chem. B. 2001;105:569–585. [Google Scholar]
- 12.Lyne PD, Mulholland AJ, Richards WG. Insights into Chorismate Mutase Catalysis From a Combined QM/MM Simulation of the Enzyme Reaction. J. Am. Chem. Soc. 1995;117:11345–11350. [Google Scholar]
- 13.Warshel A, Sussman F, Hwang J-K. Evaluation of Catalytic Free Energies in Genetically Modified Proteins. J. Mol. Biol. 1988;201:139–159. doi: 10.1016/0022-2836(88)90445-7. [DOI] [PubMed] [Google Scholar]
- 14.Klahn M, Braun-Sand S, Rosta E, Warshel A. On Possible Pitfalls in ab Initio Quantum Mechanics/Molecular Mechanics Minimization Approaches for Studies of Enzymatic Reactions. J. Phys. Chem. B. 2005;109(32):15645–15650. doi: 10.1021/jp0521757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosta E, Klahn M, Warshel A. Towards Accurate Ab Initio QM/MM Calculations of Free-Energy Profiles of Enzymatic Reactions. J. Phys. Chem. B. 2006;110(6):2934–2941. doi: 10.1021/jp057109j. [DOI] [PubMed] [Google Scholar]
- 16.Muller RP, Warshel A. Ab Initio Calculations of Free Energy Barriers for Chemical Reactions in Solution. J. Phys. Chem. 1995;99:17516–17524. [Google Scholar]
- 17.Strajbl M, Hong G, Warshel A. Ab-initio QM/MM Simulation with Proper Sampling: “First Principle” Calculations of the Free Energy of the Auto-dissociation of Water in Aqueous Solution. J. Phys. Chem. B. 2002;106:13333–13343. [Google Scholar]
- 18.Olsson MHM, Hong G, Warshel A. Frozen Density Functional Free Energy Simulations of Redox Proteins: Computational Studies of the Reduction Potential of Plastocyanin and Rusticyanin. J. Am. Chem. Soc. 2003;125:5025–5039. doi: 10.1021/ja0212157. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Liu H, Yang W. Free energy calculation on enzyme reactions with an efficient iterative procedure to determine minimum energy paths on a combined ab initio QM/MM potential energy surface. J. Chem. Phys. 2000;112(8):3483–3492. [Google Scholar]
- 20.Rod TR. U, Quantum mechanical free energy barrier for an enzymatic reaction. Phys. Rev. Lett. 2005;94(13):138302. doi: 10.1103/PhysRevLett.94.138302. [DOI] [PubMed] [Google Scholar]
- 21.Liu WW,RH, Doren DJ. Density and temperature dependences of hydration free energy of Na+ and Cl− at supercritical conditions predicted by a initio/classical free energy perturbation. J. Phys. Chem. B. 2003;107(35):9505–9513. [Google Scholar]
- 22.Iftimie RS,D, Schofield J. An efficient Monte Carlo method for calculating ab initio transition state theory reaction rates in solution. J. Chem. Phys. 2003;119(21):11285–11297. [Google Scholar]
- 23.Crespo AM,MA, Estrin DA, et al. Multiple-steering QM-MM calculation of the free energy profile in chorismate mutase. J. Am Chem. Soc. 2005;127:6940–6941. doi: 10.1021/ja0452830. [DOI] [PubMed] [Google Scholar]
- 24.Sakane SY,EM, Liu WB, et al. Exploring the ab initio/classical free energy perturbation method: The hydration free energy of water. J. Chem. Phys. 2000;113(7):2583–2593. [Google Scholar]
- 25.Pradipta B. Accelerating quantum mechanical/molecular mechanical sampling using pure molecular mechanical potential as an importance function: The case of effective fragment potential. The Journal of Chemical Physics. 2005;122(9):091102. doi: 10.1063/1.1861890. [DOI] [PubMed] [Google Scholar]
- 26.Hu H, Lu ZY, Yang WT. QM/MM minimum free-energy path: Methodology and application to triosephosphate isomerase. Journal Of Chemical Theory And Computation. 2007;3(2):390–406. doi: 10.1021/ct600240y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishida T, Kato S. Theoretical Perspectives on the Reaction Mechanism of Serine Proteases: The Reaction Free Energy Profiles of the Acylation Process. J. Am. Chem. Soc. 2003;125(39):12035–12048. doi: 10.1021/ja021369m. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Pernia JJ, Silla E, Tunon I, Marti S, Moliner V. Hybrid QM/MM potentials of mean force with interpolated corrections. Journal Of Physical Chemistry B. 2004;108(24):8427–8433. [Google Scholar]
- 29.Warshel A. Computer Modeling of Chemical Reactions in Enzymes and Solutions. John Wiley & Sons; New York: 1991. [Google Scholar]
- 30.Luzhkov V, Warshel A. Microscopic Models for Quantum Mechanical Calculations of Chemical Processes in Solutions: LD/AMPAC and SCAAS/AMPAC Calculations of Solvation Energies. J. Comp. Chem. 1992;13:199–213. [Google Scholar]
- 31.Florián J, Warshel A. Langevin Dipoles Model for Ab Initio Calculations of Chemical Processes in Solution: Parametrization and Application to Hydration Free Energies of Neutral and Ionic Solutes, and Conformational Analysis in Aqueous Solution. J. Phys. Chem. B. 1997;101:5583–5595. [Google Scholar]
- 32.Tapia O, Goscinski O. Self-consistent reaction field theory of solvent effects. Mol. Phys. 1975;29(6):1653–1661. [Google Scholar]
- 33.Cramer CJ, Truhlar DG. Continuum Solvation Models: Classical and Quantum Mechanical Implementations. In: Lipkowitz KB, Boyd DB, editors. Reviews in Computational Chemistry. Vol. 6. VCH; New York: 1995. pp. 1–72. [Google Scholar]
- 34.Tomasi J, Bonaccorsi R, Cammi R, Delvalle FJO. Theoretical Chemistry In Solution - Some Results And Perspectives Of The Continuum Methods And In Particular Of The Polarizable Continuum Model. Theochem-Journal Of Molecular Structure. 1991;80:401–424. [Google Scholar]
- 35.Rivail JL, Rinaldi D. Computational Chemistry: Review of Current Trends. World Scientific Publishing; Singapore: 1995. [Google Scholar]
- 36.Sanchez ML, Martin ME, Galvan IF, del Valle FJO, Aguilar MA. Theoretical calculation of the Stark component of the solute-solvent interaction energy. Validity of the mean field approximation in the study of liquids and solutions. Journal Of Physical Chemistry B. 2002;106(18):4813–4817. [Google Scholar]
- 37.Sanchez ML, Martin ME, Aguilar MA, Del Valle FJO. Solvent effects by means of averaged solvent electrostatic potentials: Coupled method. Journal Of Computational Chemistry. 2000;21(9):705–715. [Google Scholar]
- 38.Mendoza MLS, Aguilar MA, del Valle FJO. A mean field approach that combines quantum mechanics and molecular dynamics simulation: the water molecule in liquid water. Theochem-Journal Of Molecular Structure. 1998;426:181–190. [Google Scholar]
- 39.Sanchez ML, Aguilar MA, delValle FJO. Study of solvent effects by means of averaged solvent electrostatic potentials obtained from molecular dynamics data. Journal Of Computational Chemistry. 1997;18(3):313–322. [Google Scholar]
- 40.Warshel A, Sussman F, King G. Free Energy of Charges in Solvated Proteins: Microscopic Calculations Using a Reversible Charging Process. Biochemistry. 1986;25:8368–8372. doi: 10.1021/bi00374a006. [DOI] [PubMed] [Google Scholar]
- 41.Lee FS, Chu ZT, Bolger MB, Warshel A. Calculations Of Antibody Antigen Interactions - Microscopic And Semimicroscopic Evaluation Of The Free-Energies Of Binding Of Phosphorylcholine Analogs To Mcpc603. Protein Engineering. 1992;5(3):215–228. doi: 10.1093/protein/5.3.215. [DOI] [PubMed] [Google Scholar]
- 42.Lee FS, Chu ZT, Warshel A. Microscopic and Semimicroscopic Calculations of Electrostatic Energies in Proteins by the POLARIS and ENZYMIX Programs. J. Comp. Chem. 1993;14:161–185. [Google Scholar]
- 43.Warshel A, Russel ST. Calculations of electrostatic interactions in biological systems and in solutions. Q. Rev. Biophys. 1984;17:283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- 44.King G, Warshel A. A Surface Constrained All-Atom Solvent Model for Effective Simulations of Polar Solutions. J. Chem. Phys. 1989;91(6):3647–3661. [Google Scholar]
- 45.Lee FS, Warshel A. A Local Reaction Field Method for Fast Evaluation of Long-Range Electrostatic Interactions in Molecular Simulations. J. Chem. Phys. 1992;97:3100–3107. [Google Scholar]
- 46.Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 47.Becke AD. A New Mixing of Hartree-Fock and Local Density-Functional Theories. Journal of Chemical Physics. 1993;98(2):1372–1377. [Google Scholar]
- 48.Kendall RA, Dunning TH, Harrison RJ. Electron-Affinities Of The 1st-Row Atoms Revisited - Systematic Basis-Sets And Wave-Functions. Journal Of Chemical Physics. 1992;96(9):6796–6806. [Google Scholar]
- 49.Besler BH, Merz KM, Kollman PA. Atomic Charges Derived From Semiempirical Methods. Journal Of Computational Chemistry. 1990;11(4):431–439. [Google Scholar]
- 50.Haranczyk M, Gutowski M. Quantum mechanical energy-based screening of combinatorially generated library of tautomers. TauTGen: A tautomer generator program. Journal Of Chemical Information And Modeling. 2007;47(2):686–694. doi: 10.1021/ci6002703. [DOI] [PubMed] [Google Scholar]
- 51.Cramer CJ, Truhlar DG. AM1-SM2 and PM3-SM3 parameterized SCF Solvation Models for Free Energies in Aqueous Solution. J. of Computer-Aided. Molecular Design. 1992 doi: 10.1007/BF00126219. [DOI] [PubMed] [Google Scholar]
- 52.Wei D, Salahub DR. A Combined Density Functional and Molecular Dynamics Simulation of a Quantum Water Molecule in Aqueous Solution. Chem. Phys. Lett. 1994;24:291. [Google Scholar]
- 53.Tunon I, MartinsCosta MTC, Millot C, RuizLopez MF, Rivail JL. A coupled density functional-molecular mechanics Monte Carlo simulation method: The water molecule in liquid water. Journal Of Computational Chemistry. 1996;17(1):19–29. [Google Scholar]
- 54.Laasonen K, Sprik M, Parrinello M, Car R. Ab-Initio Liquid Water. Journal Of Chemical Physics. 1993;99(11):9080–9089. [Google Scholar]
- 55.Klahn M, Braun-Sand S, Rosta E, Warshel A. On Possible Pitfalls in ab initio QM/MM Minimization Approaches for Studies of Enzymatic Reactions. J. Phys. Chem. B. 2005 doi: 10.1021/jp0521757. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warshel A, Kato M, Pisliakov AV. Polarizable Force Fields: History, Test Cases and Prospects. Journal of Chemical Theory and Computation. 2007;3(6):2034–2045. doi: 10.1021/ct700127w. [DOI] [PubMed] [Google Scholar]