Abstract
Fourier transform ion cyclotron resonance mass spectrometry has the ability to achieve unprecedented mass measurement accuracy (MMA); MMA is one of the most significant attributes of mass spectrometric measurements as it affords extraordinary molecular specificity. However, due to space-charge effects, the achievable MMA significantly depends on the total number of ions trapped in the ICR cell for a particular measurement. Even through the use of automatic gain control (AGC), the total ion population is not constant between spectra. Multiple linear regression calibration in conjunction with AGC is utilized in these experiments to formally account for the differences in total ion population in the ICR cell between the external calibration spectra and experimental spectra. This ability allows for the extension of dynamic range of the instrument while allowing mean MMA values to remain less than 1 ppm. In addition, multiple linear regression calibration is used to account for both differences in total ion population in the ICR cell as well as relative ion abundance of a given species, which also affords mean MMA values at the parts-per-billion level.
Introduction
Fourier transform ion cyclotron resonance (FT-ICR), first developed by Comisarow and Marshall, provides the highest mass resolving power of all mass analyzers, which has the potential to translate into the highest mass measurement accuracy (MMA) over a wide m/z range.1–5 High MMA is important for numerous applications of FT-ICR including the analysis of complex mixtures such as proteins6–8, metabolites9–11, and petroleum products.12, 13 In order to realize the highest achievable MMA for a given FT-ICR MS system, frequency shifts due to space-charge-effects must be accounted for using external or internal calibration stratigies.14, 15 While internal calibration16–21 provides the best correction for space-charge effects it requires specialized hardware or software.
External calibration has been used to account for frequency shifts and thereby improve the MMA of measurements made by FT-ICR.22 For example, Amster and co-workers developed a calibration curve to account for the difference between ion populations of the calibration spectrum and subsequent spectra resulting in <10 parts-per-million MMA.23, 24 Based partly on the work of Amster23, 24 and Smith25, Oberg and Muddiman reported a novel external calibration law which afforded <5 ppm MMA.26 Amster and co-workers recently developed stepwise-external calibration, in which calibration spectra are acquired at low trapping voltages, and experimental spectra are acquired at more routine trapping voltages.27 Smith and co-workers have demonstrated an external calibration method for liquid chromatography-mass spectrometry (LC-MS) that they base on mass accuracy histograms that are derived from sets of tentative assignments for species believed to be contained in the samples.28
The external calibration methods described above attempt to quantitatively account for differences in ion population between spectra. Recently, Hunt and co-workers have demonstrated the effectiveness of combining external calibration with automatic gain control (AGC) where the number of ions in the ICR cell are precisely controlled to fall within the external calibration range.29 This approach allowed them to achieve MMA of <2 ppm. This external calibration strategy has been implemented on commercially available FT-ICR MS instruments equipped with AGC.29, 30 Muddiman and co-workers were able to achieve high MMA using this approach.31 The Smith and Gygi research groups commonly report MMA values of ~5 ppm when utilizing AGC for external calibration procedures.32–34
While AGC does control the total ion population (AT) present in the ICR cell, there is still variation in AT between spectra. Gygi and co-workers have used the calibration coefficients given by the instrument to back calculate cyclotron frequency, and use that frequency to fit their data to a multiple linear regression calibration equation based on that previously reported by Muddiman and Oberg26, which accounts for AT.35
Utilizing previously reported calibration laws in conjunction with AGC, we report improved MMA for a linear ion trap-Fourier transform ion cyclotron resonance mass spectrometer.26, 36 These calibration equations formally account for total ion population due to the inability of AGC to precisely meter ions entering the ICR cell. In addition, one of the calibration equations also accounts for relative ion abundance of particular species. Finally, we demonstrate that the calibration strategies presented in this paper enable higher AGC limits (increased dynamic range), while still maintaining high MMA.
Experimental
Materials
Poly(propylene glycol) with an average molecular weight of 1000 Da, ammonium acetate (>99%), and formic acid were purchased from Sigma (St. Louis, MO). HPLC grade acetonitrile and high purity water were purchased from Burdick and Jackson (Muskegon, MI). 2-propanol (HPLC Grade) was purchased from Fischer Scientific (Fair Lawn, NJ). All materials were used as received.
A modified version of an electrospray ionization (ESI) source developed previously in this laboratory37 was coupled to a hybrid Thermo-Electron (San Jose, CA) LTQ-FT Ultra MS equipped with an Oxford Instruments actively-shielded 7T superconducting magnet (Concord, MA). All spectra were acquired with a resolving power at 400 m/z set to 100,000FWHM, along with AGC settings ranging from 5.0 × 105 to 3.0 × 107. Samples were introduced by direct infusion using a 100 µl gas-tight syringe (Hamilton, Las Vegas, NV) and the syringe pump on the LTQ-FT Ultra at a flow rate of 0.5 µl/min. The ESI emitter tips used were 360 µm o.d., 50 µm i.d. and tapered to 30 ± 1.0 µm i.d. (New Objective, Woburn, MA) and held at a constant potential of 2200 V for all experiments. Electrospray solutions were composed of 70:30 2-propanol:water with 0.5 mM ammonium acetate (NH4OAc).
Experimental Design
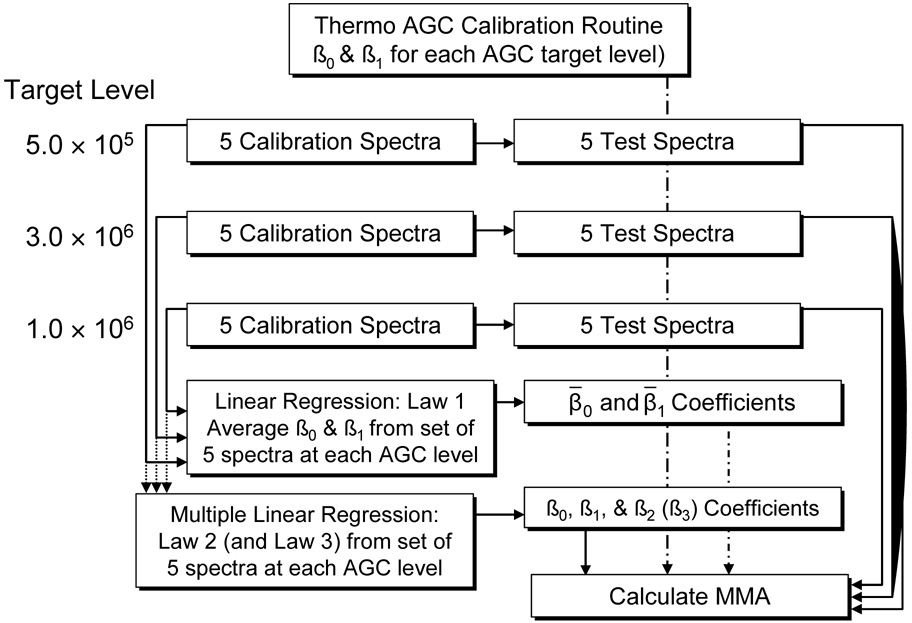
Figure 1 shows the experimental design for the first series of experiments. Calibration of the instrument was carried out using an user-defined list of monoisotoptic m/z values for ammonium-adducted PPG-1000 oligomers, which ranged from m/z = 732 to m/z = 1312. This calibration was conducted using the manufacturer's protocol which generates coefficients for five different AGC target values of which the largest 3 were used: 5.0 × 105, 1.0 × 106, and 3.0 × 106.
Figure 1.
The experimental design is shown, which starts with the calibration of the instrument. This is followed by the collection of 5 calibration and 5 test spectra at each of the specified AGC target levels. Information from the calibration spectra was used in linear regression and multiple linear regression to determine calibration coefficients for Law 1, Law 2 and Law 3. Mass measurement accuracy was calculated using these coefficients and information from the test spectra. MMA was also calculated from the m/z reported by the Thermo software.
To enable the implementation of different calibration methods, the frequencies were obtained utilizing of diagnostic mode in the instrument software. The AGC level was first set to 5.0 × 105 and five calibration spectra were recorded, along with their respective cyclotron frequencies, to generate calibration coefficients (vide infra). This was followed by the collection of five test spectra including their cyclotron frequencies, to determine the achievable mass measurement accuracy (MMA). In an effort to reduce systematic error with respect to AGC levels, this procedure was then repeated for AGC target levels of 3.0 × 106 and 1.0 × 106 in that order. The mass measurement accuracy for the five test spectra at each AGC level were first determined using the manufacturer's software.
The data from each of the five calibration spectra at each AGC target level were fit to Equation 1, developed by Gross and co-workers36, where the response is the product of the observed frequency (fobs) and theoretical m/z values for each oligomer and the predictor is 1/fobs. The y-intercept yields β0 (the scaled magnetic field strength) and the slope yields β1, the contribution for electric fields including trapping voltages (constant during the measurement) and the ions themselves (variable). All five calibration spectra were used to generate average values of β0 and β1. These average calibration coefficients were then used to determine the MMA for the five test spectra at each AGC level using Equation 2. The use of these two equations throughout this paper will be denoted as Law 1.
| Equation 1 |
| Equation 2 |
The data from the calibration spectra at each AGC target level were also fit using a multiple linear regression calibration law previously reported, which is shown in Equation 3.26 β0 accounts for electric field and β1 is the scaled magnetic field strength. This calibration law also formally takes into account the total ion population in the ICR cell (AT), which has the coefficient of β2. These calibration coefficients were then used to determine the MMA for the five test spectra at each AGC level using Equation 4. The use of these two equations will be denoted as Law 2.
| Equation 3 |
| Equation 4 |
The data from the calibration spectra at each AGC target level, where the instrument performs its calibration routine, were fit using a multiple linear regression calibration law previously reported, which is shown in Equation 5.26 The coefficients for this calibration law are the same as listed for Law 2, with the addition of β3 which formally accounts for the relative ion abundance of a given species (AS). These calibration coefficients were then used to determine the MMA for the five test spectra at each AGC level using Equation 6. The use of these two equations will be denoted as Law 3.
| Equation 5 |
| Equation 6 |
The second set of experiments followed the same scheme except these were performed at AGC levels that were above the range at which the instrument performs mass calibration: 5.0 × 106, 1.0 × 107, and 3.0 × 107; the latter is the highest AGC target level allowed by the software.
All statistical analysis was performed using Analyse-it® for Microsoft Excel®. It is important to note that the numerical values reported herein should be reproducible (but not identical) in other laboratories.
Results and Discussion
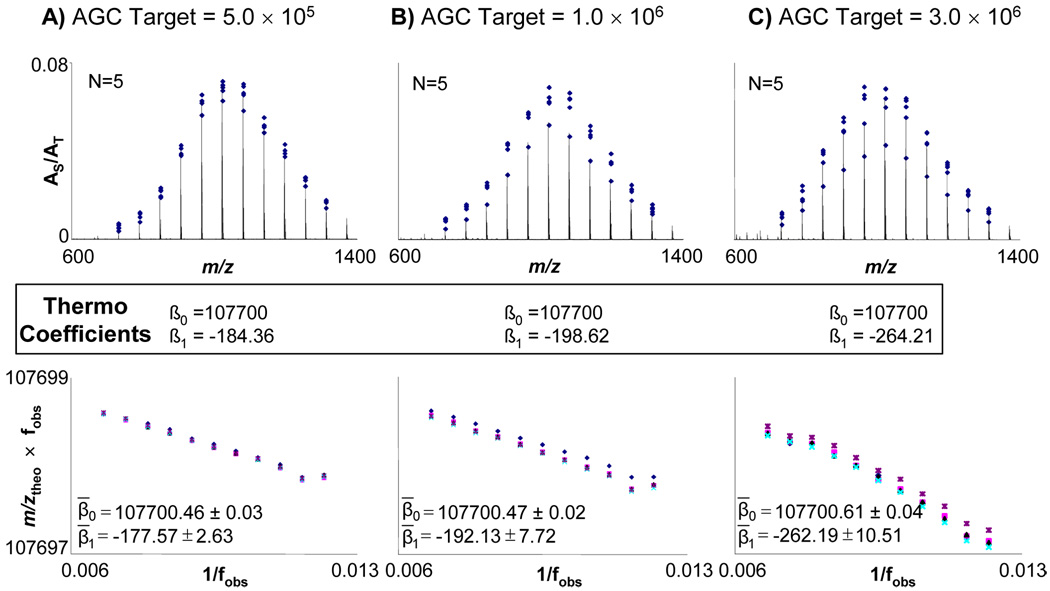
The relative proportion of each monoisotopic m/z for each oligomer to AT as a function of m/z for the three different AGC target values equal to the three highest settings at which the instrument’s calibration routine runs are shown in Figure 2. The overall shape of the oligomer distribution did not change with respect to changes in AGC target value. The coefficients from the instrument’s calibration are shown for comparison to the calculated coefficients from Law 1. As expected, the β0 coefficient remains essentially constant since magnetic field does not significantly decrease throughout the experiment. As the AGC increases, the magnitude of β1 also increases, which accounts for electric field effects in the ICR cell. The variability of β1 also increases which demonstrates that AGC has difficulty precisely reaching its target value as the AGC limit is increased. Also, the data also becomes less linear as AGC target value increase which we attribute to the increase in space charge effects at higher ion population.14, 15
Figure 2.
A) At the top, the range of AS/AT for calibrant peaks in each calibration spectrum are shown; overlaid is a representative spectrum from AGC target level of 5.0 × 105. Below, the calibration coefficients from the RAW file header are given. The bottom is a plot which displays β0 as the y-intercept, β1 as the slope which were acquired from applying Law 1 to the data. Mean values of the coefficients with their 95% confidence interval are displayed. The same information is also shown for AGC target levels of B) 1.0 × 106 and C) 3.0 × 106.
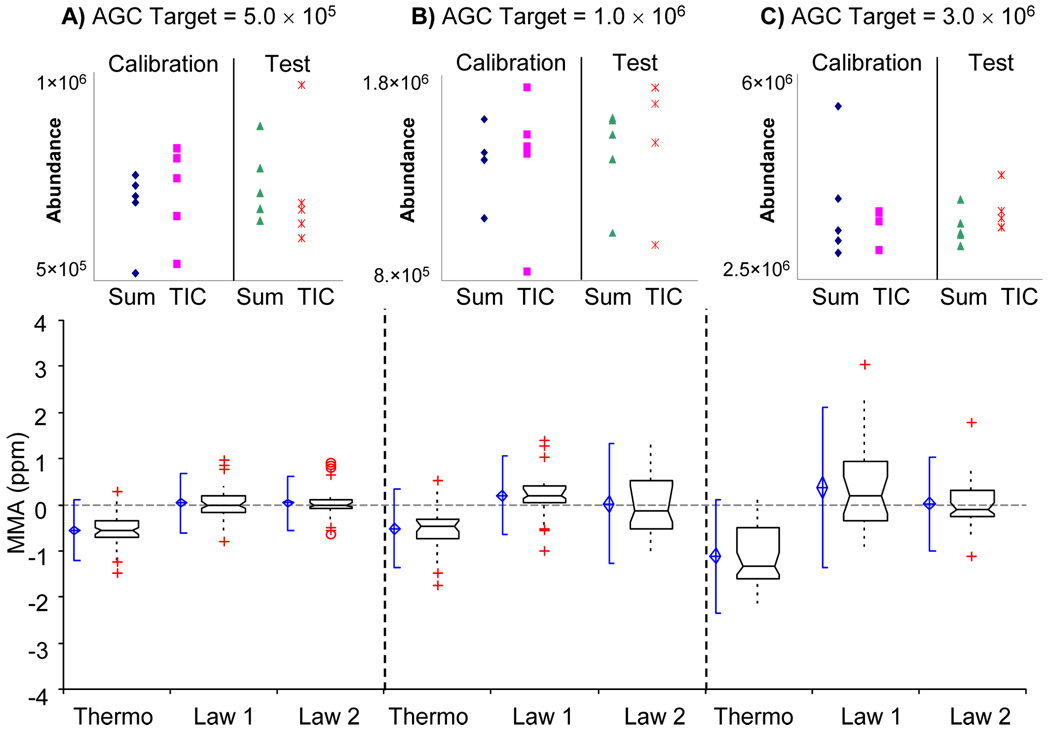
The calculation of the total ion abundance for each spectrum, AT, was carried out using two different methods. Method 1 summed the abundances of all peaks listed in the m/z list (SUM) and method 2 used the "total ion current" (TIC) from the RAW file header. The resultant from both the SUM and TIC were then multiplied by the “ion injection time” listed in the RAW file header to calculate the value of AT. The top of Figure 3 shows the individual AT values for both methods 1 and 2 for the calibrant and test spectra at each AGC target. While the calculated AT’s were different, both methods provided statistical similar results when using the multiple linear regression strategy (data not shown). Method 2 is preferred method, owing to its simplicity, and thus, this method was used herein.
Figure 3.
A) For an AGC target value of 5.0 × 105 the result from calculating AT by methods 1 and 2 for both calibration and test spectra are shown at the top. Box and whisker plots of MMA for Thermo, Law 1, and Law 2 are shown on the bottom. The blue diamond shows the mean and the 95% confidence interval of the mean, while the blue notched lines show the 95% parametric percentile range (2.5–97.5%). The notched box shows the median, lower and upper quartiles. The dotted-line connects the nearest observations within 1.5 IQRs (inter-quartile ranges) of the lower and upper quartiles. Red crosses (+) and circles (o) show observations more than 1.5 IQRs (near outliers) and 3.0 IQRs (far outliers) from the quartiles, respectivly. The same information is also shown for AGC target levels of B) 1.0 × 106 and C) 3.0 × 106.
Figure 3 shows a box and whisker plot of the resulting MMA data for the three highest AGC target values for which the instrument performs its calibration routine. The decreasing MMA using the manufacture’s calibration, as demonstrated by the decrease in MMA with increasing AGC target level, shows systematic error as the AGC target value increases. MMA from the Thermo m/z values was calculated to have a mean of −0.543, −0.513, and −1.117 ppm for the AGC target values of 5.0 × 105, 1.0 × 106, and 3.0 × 106, respectively. It is also important to note that the 95% confidence interval of the mean for these MMA’s does not include zero for any of the specified AGC target levels. The negative values are a result of the ICR cell having fewer ions in the cell, at the time of analysis, than the calibration spectrum.
Using Law 1 to calculate the average calibration coefficients (β0 and β1) for each of the five calibration spectra corrected for this systematic error, which is evident from the mean and median of the MMA data residing very close to zero for all AGC target levels, as shown in Figure 3. Even though the equation for fitting and calculating m/z values for Law 1 and Thermo’s calibration are the same, our data exhibited higher MMA because the calibration coefficients were calculated as the average from 5 different spectra at each AGC level as shown by Figure 1. Using the average calibration coefficients determined from Law 1 we obtained MMA mean values of 0.036, 0.202, and 0.376 ppm at the AGC target levels 5.0 × 105, 1.0 × 106, and 3.0 × 106. The 95% confidence interval of the mean for this data includes zero for 5.0 × 105 (−0.053 to 0.126 ppm); however, not for 1.0 × 106 or 3.0 × 106 data sets.
Law 2 produced MMA mean values of 34, 23, and 27 parts-per-billion (ppb) for the same range of AGC target values as reported above. The 95% confidence interval of the mean includes zero for all AGC target levels in this range: −48 ppb to 116 ppb, −156 ppb to 203 ppb, and −114 ppb to 167 ppb for 5.0 × 105, 1.0 × 106, and 3.0 × 106, respectively. Law 2 enhances MMA because it formally accounts for changes in AT. Law 1 is almost able to perform as well as Law 2, even while not formally accounting for AT, is because Law 1 is an average of 5 spectra instead of a single spectrum, as is the case in Thermo’s calibration procedure. However, since all of the results for Law 2 had 95% confidence intervals that included zero, we can confidently state that Law 2 performed better than alternative calibration laws because it accounts for AT formally.
Gorshkov et. al. has previously shown that the frequency shift due to space-charge effects is dependant on the relative abundance of an ion in the ICR cell.38 This relative ion abundance of a given species (AS) has been previously utilized in Law 3 on a FT-ICR MS without AGC by Muddiman and Oberg.26 Moreover, Muddiman and Oberg demonstrated that both AS and AT were statistically significant in the calculation of MMA for data acquired without AGC.26
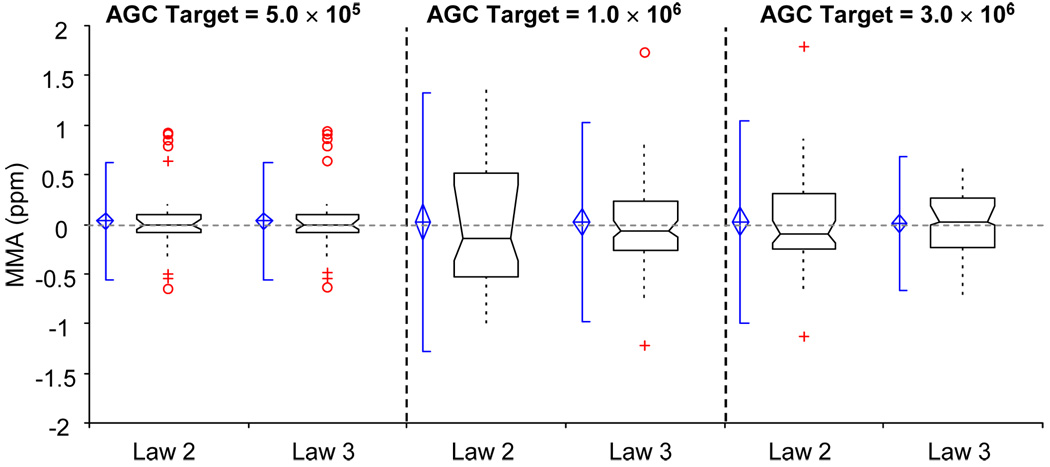
Law 3 formally accounts for both AT and AS, which narrows the range over which the MMA values are spread, especially for high AGC target values, when compared to Law 2, as shown in Figure 4. For this set of experiments the calibration spectra and test spectra had similar AS values, since both were ammonium-adducted PPG-1000 oligomers. The mean MMA values produced by the application of Law 3 are 34, 24, and 9 ppb for AGC target levels of 5.0 × 105, 1.0 × 106, and 3.0 × 106, respectively. The 95% confidence interval of the mean includes zero for all AGC target levels in this range: −48 ppb to 116 ppb, −115 ppb to 163 ppb, and −84 ppb to 102 ppb for the same AGC target values as reported above.
Figure 4.
Box and whisker plots of MMA for Law 2 and Law 3 at each of the specified AGC target levels. The notation for these box and whisker plots is identical to that used in Figure 3.
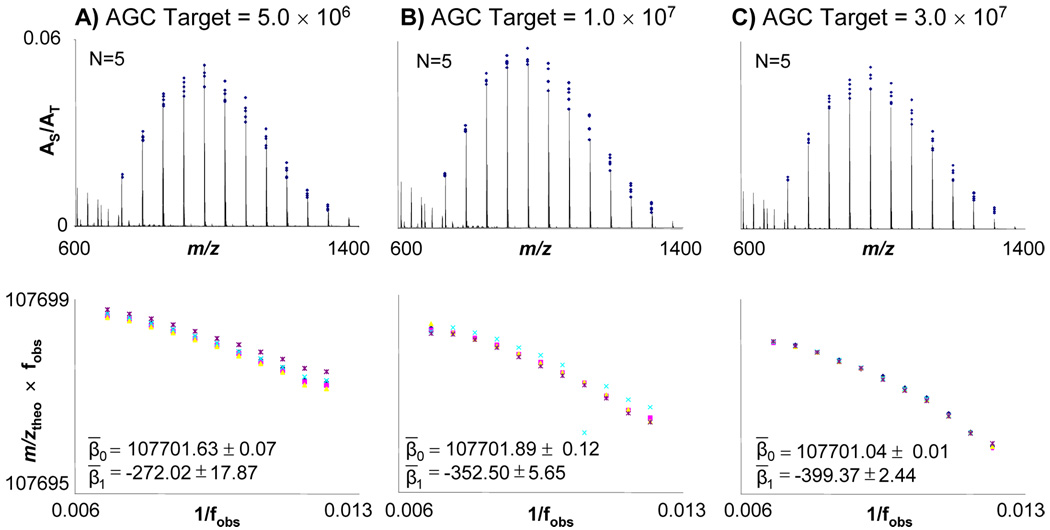
Figure 5 displays the relative proportion of the monoisotopic m/z value for each oligomer to AT as a function of m/z for the range of AGC target values outside of the calibration range of the instrument. The overall distribution does not change over the range of AGC target values. As previously observed, β0 remains similar throughout these experiments and the magnitude of β1 does change systematically as AT is increased.
Figure 5.
A) The top shows the range of AS/AT for calibrant peaks in each calibration spectrum; overlaid is a representative spectrum from AGC target level of 5.0 × 106. Below, the calibration coefficients from the RAW file header are given. The bottom is a plot which displays β0 as the y-intercept, β1 as the slope which were acquired from applying Law 1 to the data. Mean values of these coefficients with their 95% confidence interval are displayed. The same information is also shown for AGC target levels of B) 1.0 × 107 and C) 3.0 × 107.
There are no comparisons to be made to Thermo calibration coefficients for this data set because at AGC target levels above 3.0 × 106 the Thermo coefficients remain the same because 3.0 × 106 is the highest AGC level at which the calibration routine operates. As the AGC target value increases for this data set the departure from linearity of the data becomes greater than that of the data in Figure 2; however, the variation of the β1 coefficient surprisingly decreased, which is under further investigation in our laboratory.
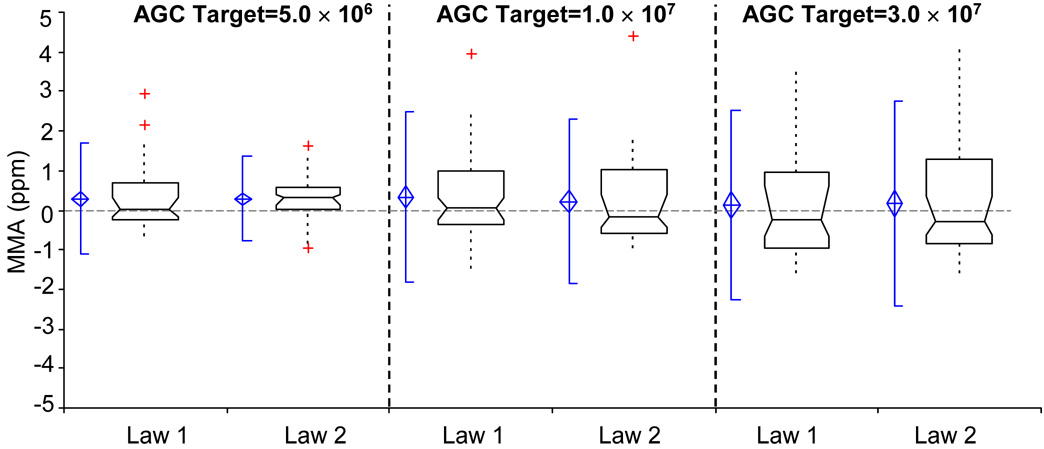
MMA data from experiments with an AGC target value exceeding those at which the Thermo software calibrates the instrument are presented in Figure 6. The resulting MMA from applying the average calibration coefficients obtained using Law 1 for the five calibration spectra to the data at the higher AGC target levels is still able to maintain means close to the sub-ppm level. Law 1 resulted in mean MMA values of 287, 336, and 137 ppb for the AGC target values of 5.0 × 106, 1.0 × 107, and 3.0 × 107, respectively. For the AGC target level of 3.0 × 107, the 95% confidence interval of the mean for Law 1 was −185 ppb to 523 ppb; however, the for other two AGC target levels in this set of experiments the 95% confidence interval did not include zero.
Figure 6.
Box and whisker plots of MMA for Law 1 and Law 2 at each of the specified AGC target levels. The notation for these box and whisker plots is identical to that used in Figure 3.
Law 2 resulted in mean MMA values of 293, 219, and 169 ppb for the same range of AGC target values reported above. For AGC target levels 1.0 × 107 and 3.0 × 107 the 95% confidence interval of the mean included zero (−68 ppb to 506 ppb and −185 ppb to 523 ppb, respectively). Through the use of these calibration procedures we are able to extend the dynamic range of the LTQ-FT, while still maintaining high levels of mass measurement accuracy.
Conclusions
Data was acquired utilizing a LTQ-FT in diagnostic mode, which allowed for the recording of the cyclotron frequency of the ions in the ICR cell. Frequency data allowed for the calibration of spectra of ammonium-adducted PPG-1000 oligomers utilizing two different calibration laws: Law 1 was employed with calibration coefficients determined for each of 5 spectra at a specific AGC target level and then averaged; and Law 2, a multiple linear regression law which formally accounts for the AT at a specific AGC target level. These laws enabled parts-per-billion MMA over a wide range of AGC target levels even in excess of the AGC target level at which the instrument calibration routine permits, thus extending the dynamic range of the instrument. Between the two main laws investigated, Law 2 allowed for the best MMA because of it can correct for the inability of AGC to precisely aliquot a specified number of ions to the ICR cell, especially at higher AGC target levels. Law 3, applied only to the three lower AGC target levels examined in these experiments, was able to attain very high MMA, and fundamentally will likely prove to be the best since it formally accounts for both AT and AS. Coupled with stronger magnetic fields39, this method of calibrating FT-ICR data has the potential to improve MMA even further. These calibration laws should also translate to improving MMA on the LTQ-Orbitrap.40 We are currently working on extending these findings to intact proteins, and protein fragments in MSn data, which should both benefit from the extended dynamic range which these calibration laws and procedures provide.
Acknowledgments
The authors gratefully acknowledge financial support received from the National Cancer Institute, National Institutes of Health (R33 CA105295), the W.M. Keck Foundation, and North Carolina State University.
References
- 1.Comisarow MB, Marshall AG. Chem. Phys. Lett. 1974;25:282–283. [Google Scholar]
- 2.Comisarow MB, Marshall AG. Chem. Phys. Lett. 1974;26:489–490. [Google Scholar]
- 3.Gorshkov MV, Guan SH, Marshall AG. Int. J. Mass Spectrom. Ion Processes. 1993;128:47–60. [Google Scholar]
- 4.Rodgers RP, White FM, Hendrickson CL, Marshall AG, Andersen KV. Anal. Chem. 1998;70:4743–4750. [Google Scholar]
- 5.He F, Hendrickson CL, Marshall AG. Anal. Chem. 2001;73:647–650. doi: 10.1021/ac000973h. [DOI] [PubMed] [Google Scholar]
- 6.Emmett MR, White FM, Hendrickson CL, Shi SDH, Marshall AG. J. Am. Soc. Mass Spectrom. 1998;9:333–340. doi: 10.1016/S1044-0305(97)00287-0. [DOI] [PubMed] [Google Scholar]
- 7.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen YF, Conrads TP, Veenstra TD, Udseth HR. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 8.Kujawinski EB, Hatcher PG, Freitas MA. Anal. Chem. 2002;74:413–419. doi: 10.1021/ac0108313. [DOI] [PubMed] [Google Scholar]
- 9.Brown SC, Kruppa G, Dasseux JL. Mass Spectrom. Rev. 2005;24:223–231. doi: 10.1002/mas.20011. [DOI] [PubMed] [Google Scholar]
- 10.Breitling R, Pitt AR, Barrett MP. Trends Biotechnol. 2006;24:543–548. doi: 10.1016/j.tibtech.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Sanders M, Shipkova PA, Zhang HY, Warrack BM. Curr. Drug Metab. 2006;7:547–555. doi: 10.2174/138920006777697972. [DOI] [PubMed] [Google Scholar]
- 12.Qian KN, Robbins WK, Hughey CA, Cooper HJ, Rodgers RP, Marshall AG. Energy Fuels. 2001;15:1505–1511. [Google Scholar]
- 13.Marshall AG, Rodgers RP. Acc. Chem. Res. 2004;37:53–59. doi: 10.1021/ar020177t. [DOI] [PubMed] [Google Scholar]
- 14.Jeffries JB, Barlow SE, Dunn GH. Int. J. Mass Spectrom. Ion Processes. 1983;54:169–187. [Google Scholar]
- 15.Francl TJ, Sherman MG, Hunter RL, Locke MJ, Bowers WD, Mciver RT. Int. J. Mass Spectrom. Ion Processes. 1983;54:189–199. [Google Scholar]
- 16.Kaiser NK, Anderson GA, Bruce JE. J. Am. Soc. Mass Spectrom. 2005;16:463–470. doi: 10.1016/j.jasms.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Hannis JC, Muddiman DC. J. Am. Soc. Mass Spectrom. 2000;11:876–883. doi: 10.1016/S1044-0305(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 18.Flora JW, Hannis JC, Muddiman DC. Anal. Chem. 2001;73:1247–1251. doi: 10.1021/ac0011282. [DOI] [PubMed] [Google Scholar]
- 19.Nepomuceno AI, Mason CJ, Muddiman DC, Bergen HR, Zeldenrust SR. Clin. Chem. 2004;50:1535–1543. doi: 10.1373/clinchem.2004.033274. [DOI] [PubMed] [Google Scholar]
- 20.Bruce JE, Anderson GA, Brands MD, Pasa-Tolic L, Smith RD. J. Am. Soc. Mass Spectrom. 2000;11:416–421. doi: 10.1016/S1044-0305(00)00096-9. [DOI] [PubMed] [Google Scholar]
- 21.Williams DK, Jr, Hawkridge AM, Muddiman DC. J. Am. Soc. Mass Spectrom. 2007;18:1–7. doi: 10.1016/j.jasms.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Zhang LK, Rempel D, Pramanik BN, Gross ML. Mass Spectrom. Rev. 2005;24:286–309. [Google Scholar]
- 23.Easterling ML, Mize TH, Amster IJ. Anal. Chem. 1999;71:624–632. doi: 10.1021/ac980690d. [DOI] [PubMed] [Google Scholar]
- 24.Taylor PK, Amster IJ. Int. J. Mass Spectrom. 2003;222:351–361. [Google Scholar]
- 25.Masselon C, Tolmachev AV, Anderson GA, Harkewicz R, Smith RD. J. Am. Soc. Mass Spectrom. 2002;13:99–106. doi: 10.1016/S1044-0305(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 26.Muddiman DC, Oberg AL. Anal. Chem. 2005;77:2406–2414. doi: 10.1021/ac048258l. [DOI] [PubMed] [Google Scholar]
- 27.Wong RL, Amster IJ. J. Am. Soc. Mass Spectrom. 2006;17:1681–1691. doi: 10.1016/j.jasms.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 28.Tolmachev AV, Monroe ME, Jaitly N, Petyuk VA, Adkins JN, Smith RD. Anal. Chem. 2006;78:8374–8385. doi: 10.1021/ac0606251. [DOI] [PubMed] [Google Scholar]
- 29.Syka JEP, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. J. Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 30.Peterman SM, Mulholland JJ. J. Am. Soc. Mass Spectrom. 2006;17:168–179. doi: 10.1016/j.jasms.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Johnson KL, Ovsyannikova IG, Madden BJ, Poland GA, Muddiman DC. J. Am. Soc. Mass Spectrom. 2005;16:1812–1817. doi: 10.1016/j.jasms.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Dieguez-Acuna FJ, Gerber SA, Kodama S, Elias JE, Beausoleil SA, Faustman D, Gygi SP. Mol. Cell. Proteomics. 2005;4:1459–1470. doi: 10.1074/mcp.M500137-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Belov ME, Zhang R, Strittmatter EF, Prior DC, Tang K, Smith RD. Anal. Chem. 2003;75:4195–4205. doi: 10.1021/ac0206770. [DOI] [PubMed] [Google Scholar]
- 34.Page JS, Bogdanov B, Vilkov AN, Prior DC, Buschbach MA, Tang K, Smith RD. J. Am. Soc. Mass Spectrom. 2005;16:244–253. doi: 10.1016/j.jasms.2004.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Mol. Cell. Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Ledford EB, Rempel DL, Gross ML. Anal. Chem. 1984;56:2744–2748. doi: 10.1021/ac00278a027. [DOI] [PubMed] [Google Scholar]
- 37.Nepomuceno AI, Muddiman DC, Bergen HR, Craighead JR, Burke MJ, Caskey PE, Allan JA. Anal. Chem. 2003;75:3411–3418. doi: 10.1021/ac0342471. [DOI] [PubMed] [Google Scholar]
- 38.Gorshkov MV, Marshall AG, Nikolaev EN. J. Am. Soc. Mass Spectrom. 1993;4:855–868. doi: 10.1016/1044-0305(93)87003-U. [DOI] [PubMed] [Google Scholar]
- 39.Marshall AG, Guan SH. Rapid Commun. Mass Spectrom. 1996;10:1819–1823. doi: 10.1002/(SICI)1097-0231(199611)10:14<1855::AID-RCM764>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 40.Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. J. Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]