Abstract
RNA silencing is a conserved mechanism in eukaryotes that plays an important role in various biological processes including regulation of gene expression. RNA silencing also plays a role in genome stability and protects plants against invading nucleic acids such as transgenes and viruses. Recently, RNA silencing has been found to play a role in defence against bacterial plant pathogens in Arabidopsis through modulating host defence responses. In this study, it is shown that gene silencing plays a role in plant defence against multicellular microbial pathogens; vascular fungi belonging to the Verticillium genus. Several components of RNA silencing pathways were tested, of which many were found to affect Verticillium defence. Remarkably, no altered defence towards other fungal pathogens that include Alternaria brassicicola, Botrytis cinerea, and Plectosphaerella cucumerina, but also the vascular pathogen Fusarium oxysporum, was recorded. Since the observed differences in Verticillium susceptibility cannot be explained by notable differences in root architecture, it is speculated that the gene silencing mechanisms affect regulation of Verticillium-specific defence responses.
Keywords: Abiotic stress, post-transcriptional gene silencing (PTGS), suppressor of gene silencing (SGS), Verticillium dahliae, V. albo-atrum, V. longisporum
Introduction
Plant defence against pathogens is activated through specific host signalling mechanisms (Chisholm et al., 2006; Jones and Dangl, 2006). Microbial intruders can be recognized by extracellular receptor molecules that detect the presence of pathogen-associated molecular patterns (PAMPs) and subsequently activate PAMP-triggered immunity (PTI) as a basal defence response. Virulent pathogen strains are able to interfere with, or suppress, PTI by utilizing effector molecules (Bolton et al., 2008; van Esse et al., 2007, 2008). In turn, some plant genotypes have developed specific receptor molecules, the resistance proteins, to detect the presence of the pathogen effector molecules and activate effector-triggered immunity (ETI; Chisholm et al., 2006; Jones and Dangl, 2006). Only in a few cases has direct interaction of the host resistance protein with the pathogen effector molecule been observed (Scofield et al., 1996; Tang et al., 1996; Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006; Burch-Smith et al., 2007). More often, however, the resistance protein monitors the status of a host target of the pathogen effector molecule in compliance with the guard hypothesis (Dangl and Jones, 2001; Mackey et al., 2002; Rooney et al., 2005; Shao et al., 2003).
Nearly 20 years ago, the phenomenon of RNA silencing was discovered in experiments with transgenic plants that showed silencing of a transgene and also, in a number of cases, of homologous endogenous genes (Napoli et al., 1990; van der Krol et al., 1990). The gene silencing was found to result from the inhibition of gene transcription (transcriptional gene silencing, TGS) or from post-transcriptional degradation of RNA (post-transcriptional gene silencing, PTGS), and correlated with the accumulation of small double-stranded RNA segments of 20–27 nucleotides, so-called small RNAs (sRNAs). These corresponded to the promoter of the silenced gene, or to the degraded RNA in TGS and PTGS, respectively (Hamilton and Baulcombe, 1999; Mette et al., 2000).
RNA silencing is now known as a conserved regulatory mechanism in most eukaryotic organisms that plays a determinant role in various biological processes, including the regulation of endogenous gene expression, genome stability, the taming of transposons, heterochromatin formation, and defence against viruses (Baulcombe, 2004; Brodersen and Voinnet, 2006; Vaucheret, 2006). The key characteristic of RNA silencing is the formation of the sRNAs that are produced by RNaseIII-like Dicer enzymes (Bernstein et al., 2001). These sRNAs can be divided into two major types, the small interfering RNAs (siRNAs) and the micro RNAs (miRNAs), based on their origin and formation. Subsequently, a selected sRNA strand is incorporated into an effector complex that is targeted towards partially or fully complementary RNA or DNA stretches. This so-called RNA-induced silencing complex (RISC) contains an Argonaute (Ago) protein that has an sRNA-binding domain and an endonucleolytic activity to cleave target RNAs (Martinez et al., 2002).
Several studies have shown that PTGS mechanisms are an RNA-based host defence system to control nucleic acid invaders of various natures through the action of cis-acting siRNAs that derive from, and target, the invaders (Vance and Vaucheret, 2001; Bartel, 2004; Baulcombe, 2004; Dunoyer and Voinnet, 2005). These invaders may be endogenous, such as transposons, or exogenous, such as transgenes and viral pathogens. Thus, RNA silencing has been implicated in pathogen defence through its role in viral defence. Upon virus infection, the accumulation of virus-derived sRNAs has been observed (Hamilton and Baulcombe, 1999). Moreover, plant mutants defective in PTGS are often hyper-susceptible to viral infection (Mourrain et al., 2000; Dalmay et al., 2001; Qu et al., 2005; Schwach et al., 2005).
Apart from viral defence, evidence accumulates for RNA silencing to play a role in interactions with other pathogen types, more specifically bacterial defence (Voinnet, 2008). The first example is a miRNA from Arabidopsis that contributes to basal defence against Pseudomonas syringae by regulating auxin signalling (Navarro et al., 2006). The miRNA was induced upon perception of flg-22, a PAMP that is derived from bacterial flagellin, and negatively regulated transcripts of a number of F-box auxin receptors. In turn, repression of auxin signalling was shown to restrict growth of the bacterium P. syringae (Navarro et al., 2006). Another example is an endogenous Arabidopsis siRNA that is specifically induced by avirulent P. syringae carrying AvrRpt2 (Katiyar-Agarwal et al., 2006). This siRNA contributes to RPS2-mediated disease resistance by repressing a putative negative regulator of the RPS2 resistance pathway. Recently, a novel class of small RNAs, long siRNAs (lsiRNAs that are 30–40 nt) that are induced by pathogen infection or under specific growth conditions, was identified. One of the lsiRNAs, AtlsiRNA-1, was specifically induced by avirulent P. syringae carrying AvrRpt2 and induction of AtlsiRNA-1 was found to silence a RAP-domain protein that is involved in disease resistance (Katiyar-Agarwal et al., 2007). Finally, in a forward genetics screen, an Arabidopsis mutant with enhanced disease susceptibility towards a compatible P. syringae strain, an incompatible strain carrying AvrRpm1, and non-adapted P. syringae pv. tabaci was isolated (Agorio and Vera, 2007). Positional cloning revealed a mutation in the Argonaute gene AGO4, that is associated with small interfering RNAs involved in RNA-directed DNA methylation (RdDM), showing that AGO4 plays a role in non-host resistance, basal defence, and effector-triggered immunity against bacterial pathogens (Agorio and Vera, 2007). In addition to P. syringae, it has been shown that RNA silencing mutants are hypersusceptible to the crown gall bacterium Agrobacterium tumefaciens (Dunoyer et al., 2006). Finally, RNA silencing has been shown to be required for the development of nodule differentiation on Medicago truncatula roots in the interaction with the nitrogen fixing Rhizobium bacteria (Combier et al., 2006; Boualem et al., 2008).
Recently it has been demonstrated that miRNAs are key components of plant basal defence as miRNA-deficient Arabidopsis mutants sustained growth of a non-pathogenic, type III secretion-defective P. syringae mutant, non-pathogenic P. fluorescens, and Escherichia coli strains (Navarro et al., 2008). Interestingly, P. syringae effectors were identified that suppressed the transcriptional activation or activity of several PAMP-responsive miRNAs, demonstrating that these bacteria suppress RNA silencing to cause disease (Navarro et al., 2008).
In our research, Arabidopsis thaliana has been used as a host to investigate the biology of the vascular wilt pathogen Verticillium dahliae (Fradin and Thomma, 2006). To investigate the role of putative defence genes against Verticillium infection, transgenic over-expression in wild-type (Col-0) Arabidopsis, but also in the PTGS mutant sgs2 (Butaye et al., 2004), was used. Previously, it has been shown that the inter-transformant variability of transgene expression is reduced in sgs mutants, as the incidence of highly expressing transformants increased from 20% in Col-0 to 100% in sgs mutants (Butaye et al., 2004). Intriguingly, it was observed in several of our experiments that non-transformed sgs2 plants displayed significantly enhanced susceptibility towards V. dahliae when compared with the parental line Col-0. In this paper, the role of RNA silencing in Arabidopsis defence against a number of fungal pathogens, including V. dahliae, was investigated.
Materials and methods
Plant growth conditions
Soil-grown Arabidopsis plants were cultivated in a growth chamber at 22 °C, 72% relative humidity, and a 16 h photoperiod, or in a greenhouse at 21 °C for the 16 h day period and 19 °C for the 8 h night period at 72% relative humidity. In the greenhouse, supplemental light (100 W m−2) was used when the sunlight influx intensity was below 150 Wm−2.
For in vitro growth of Arabidopsis, seeds were surface-sterilized and sown on MS medium (Duchefa, Haarlem, NL) solidified with 1.5% plant agar (Duchefa, Haarlem, NL). For phenotypic evaluations of root growth and development, Arabidopsis plants were grown on vertically oriented half-strength MS plates, supplemented with 1% sucrose and 0.5 g l−1 MES (2-(N-morpholino) ethane-sulphonic acid) (pH 5.8). After sowing, the plates were incubated at 4 °C in the dark for 3 d and subsequently transferred to the growth chamber.
Conditional phenotype assays
To assess susceptibility toward abiotic stress and responsiveness to hormones, in vitro assays were performed (Wang et al., 2008; see Supplementary Table S1 at JXB online). For abiotic stress assays, seeds were sown on MS agar amended with 100 or 150 mM NaCl, 20 or 30 mM LiCl, 150 or 200 mM mannitol, and 3.3 or 6.7 mM H2O2 (see Supplementary Table S1 at JXB online) and evaluated for aberrant growth. To assay heavy metal resistance, plants were grown on vertically oriented half-strength MS plates amended with 2% (w/v) sucrose and 85 μM CdCl2. To assay hormone responsiveness, the sterilized seeds were grown on vertically oriented half-strength MS plates containing different hormones (see Supplementary Table S1 at JXB online). All plates were incubated in the growth chamber. For hypocotyl length assays, plates were incubated in the dark.
Pathogen cultivation
Verticillium dahliae strains JR2 and ST12.01, Verticillium longisporum strain 43, Verticillium albo-atrum strains VA1 and CBS451.88, Fusarium oxysporum f.sp. raphani strain 815 (Diener and Ausubel, 2005), Alternaria brassicicola strain MUCL20297 (Mycotheque Université Catholique de Louvain, Louvain-la-Neuve, Belgium), and Plectosphaerella cucumerina were maintained on potato dextrose agar (PDA; Oxoid, Hampshire, UK). Botrytis cinerea (Brouwer et al., 2003) was grown on half-strength PDA amended with 5 g l−1 agar and 150 g l−1 blended tomato leaves. All fungal cultures were grown at 22 °C. The bacterial strains of Pseudomonas syringae pv. tomato (Pst) DC3000 with or without avrRpt2, avrRpm1, or avrRps4, was grown on King's B agar (King et al., 1954) supplemented with the appropriate antibiotics (25 μg ml−1 rifampicin and 100 μg ml−1 kanamycin). All bacterial strains were grown overnight at 28 °C.
Pathogen inoculations
Inoculum of all fungi (except F. oxysporum f. sp. raphani) was prepared as previously described by Broekaert et al. (1990) and prepared as a suspension of 106 conidia ml−1 in water. For Verticillium inoculations, a minimum of eight 2-week-old Arabidopsis plants were up-rooted and the roots were incubated in the conidial suspension for 3 min. Subsequently, the plants were replanted into fresh soil. Inoculations with F. oxysporum f. sp. raphani were performed in a similar way to the Verticillium inoculations, except that the budcell-inoculum was prepared as described by Diener and Ausubel (2005). All other pathogens were inoculated onto a minimum of four approximately 4-week-old soil-grown plants with fully expanded rosette leaves. Inoculations with A. brassicicola, B. cinerea, and P. cucumerina were performed by placing 6 μl drops of the conidial suspensions on each expanded leaf (Thomma et al., 1998, 2000; Brouwer et al., 2003; O'Connell et al., 2004).
For inoculations with P. syringae, bacteria were grown overnight at 28 °C in liquid King's B medium supplemented with the appropriate antibiotics. Arabidopsis plants were spray-inoculated with a bacterial suspension of OD600 0.3 supplemented with 0.05% (v/v) Silwet L-77 (van Meeuwen Chemicals BV, Weesp, NL).
For all inoculations, except those with F. oxysporum f. sp. raphani and Verticillium spp., plants were kept in boxes with transparent lids at high relative humidity for the remainder of the experiment. All inoculations were performed a minimum of three times with similar results.
V. dahliae biomass quantification in planta
Two-week-old Arabidopsis plants were inoculated with V. dahliae strain JR2 as described above. After visible symptom development at 19–29 d post-inoculation, for each experiment and for each Arabidopsis genotype all above-ground tissues were harvested per plant and flash-frozen in liquid nitrogen. The samples were ground to a powder, of which an aliquot of approximately 100 mg was used for DNA isolation (Fulton et al., 1995). Quantitative real-time PCR was conducted using an ABI7300 PCR machine (Applied Biosystems, Foster City, USA) with the qPCR Core kit for SYBR Green I (Eurogentec Nederland BV, Maastricht, NL). To measure V. dahliae biomass, the internal transcribed spacer region of the ribosomal DNA was targeted using the fungus-specific ITS1-F primer (AAAGTTTTAATGGTTCGCTAAGA; Gardes and Bruns, 1993) in combination with the V. dahliae-specific reverse primer ST-VE1-R (CTTGGTCATTTAGAGGAAGTAA; Lievens et al., 2006), generating a 200 bp amplicon. For sample equilibration, the Arabidopsis large subunit of the RuBisCo gene was targeted using the primer set At-RuBisCo-F3 and -R3 (GCAAGTGTTGGGTTCAAAGCTGGTG and CCAGGTTGAGGAGTTACTCGGAATGCTG, respectively), generating a 120 bp amplicon. Real-time PCR conditions consisted of an initial 95 °C denaturation step for 4 min, followed by 30 cycles of denaturation for 15 s at 95 °C, annealing for 30 s at 60 °C, and extension for 30 s at 72 °C. The average fungal biomass was determined using at least four Verticillium-inoculated plants for each genotype.
Reverse transcription PCR
Total RNA was extracted from plant tissue frozen in liquid nitrogen using the RNeasy Plant Mini kit (Qiagen, Venlo, the Netherlands). On-column DNaseI treatment was performed as described by the manufacturer using the RNase-free DNase Set (Qiagen, Venlo, the Netherlands). Approximately 1.5 μg of total RNA was used for cDNA synthesis using SuperScriptTM III Reverse Transcriptase and Oligo(dT)12–18 primers according to the manufacturer's protocol (Invitrogen, Breda, the Netherlands). PCR amplification of actin (with primer pair Actin2-F2 TAACTCTCCCGCTATGTATGTCGC, and Actin2-R2 GAGAGAAACCCTCGTAGATTGGC) and of PR-1 (with primer pair PR1-F1 AGGCTAACTACAACTACGCTGCG, and PR1-R1 GCTTCTCGTTCACATAATTCCCAC) consisted of an initial denaturing step at 94 °C for 5 min, followed by 30–35 cycles of 20 s at 94 °C, 20 s at 56 °C and 20 s at 72 °C, followed by a final extension step for 5 min at 72 °C. PCR products were visualized on ethidium bromide-stained 1% agarose gels.
Results
sgs mutants display enhanced susceptibility towards V. dahliae
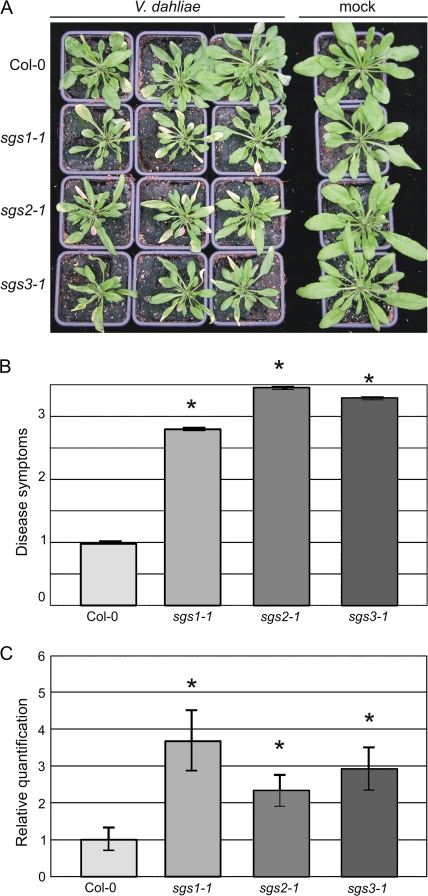
Transgenic expression in the post-transcriptional gene silencing (PTGS) mutant suppressor of gene silencing 2 (sgs2; Elmayan et al., 1998; Mourrain et al., 2000) reduces the inter-transformant variability of transgene expression (Butaye et al., 2004). In several experiments to investigate putative defence genes against V. dahliae in Arabidopsis, transgenic overexpression in Col-0 as well as sgs2-1 was performed. Remarkably, in subsequent disease susceptibility assays with V. dahliae strain JR2 it appeared that untransformed sgs2-1 plants displayed more severe disease symptoms than Col-0 plants (Fig. 1A, B). While Col-0 plants displayed only mild disease symptoms upon V. dahliae inoculation as visualized by rather slight stunting resulting in a reduced rosette diameter at 3 weeks post-inoculation, inoculated sgs2-1 plants showed severe stunting, wilting, anthocyanin accumulation, and tissue necrosis (Fig. 1A, B). The ratio of leaves displaying symptoms of disease was also significantly more for sgs2-1 plants than for Col-0 plants (Fig. 1A, B)
Fig. 1.
Arabidopsis sgs mutants display enhanced susceptibility towards Verticillium dahliae. (A) Typical symptoms of V. dahliae on Arabidopsis sgs mutants. The mutants sgs1-1, sgs2-1, sgs3-1, and the corresponding wild type Col-0 were inoculated with V. dahliae strain JR2 or mock-inoculated. V. dahliae-inoculated sgs mutants show enhanced symptom development, including more severe stunting, wilting, anthocyanin accumulation, and tissue necrosis, when compared with Col-0 plants at 19 d post-inoculation. (B) Quantification of symptom development at 19 d post-inoculation shown as a ratio of diseased rosette leaves with standard deviation. The ratio of diseased rosette leaves for Col-0 is set to one. Asterisks indicate significant differences when compared with the wild type Col-0 (P <0.05). (C) Quantitative real-time PCR of fungal colonization by comparing V. dahliae internal transcribed spacer (ITS) transcript levels (as a measure for fungal biomass) relative to Arabidopsis Rubisco transcript levels (for equilibration) at 19 d post-inoculation. The mutants sgs1-1, sgs2-1, sgs3-1, and the corresponding wild type Col-0 were inoculated with V. dahliae strain JR2 and the relative average fungal biomass is shown with standard errors. Asterisks indicate significant differences when compared with colonization of the wild type Col-0.
In addition to V. dahliae strain JR2, our analysis was extended to include other Verticillium pathogens of Arabidopsis (Fradin and Thomma, 2006). These included V. dahliae strain ST12.01, the V. albo-atrum strains VA1 and CBS451.88, and V. longisporum strain Vl43. All these Verticillium strains caused more disease symptoms on sgs2-1 plants when compared with Col-0 plants (see Supplementary Fig. S1 at JXB online), confirming that the enhanced susceptibility of the sgs2-1 mutant broadly concerns plant pathogenic Verticillium species.
In addition to sgs2-1, reduced inter-transformant variability in transgene expression was similarly demonstrated in the non-allelic sgs3-1 mutant (Butaye et al., 2004). To investigate the role of PTGS in Arabidopsis defence against Verticillium further, the two additional non-allelic PTGS mutants; sgs1-1 and sgs3-1 (Elmayan et al., 1998; Mourrain et al., 2000), were tested for their susceptibility towards V. dahliae strain JR2. Similar to the sgs2-1 plants, sgs1-1 and sgs3-1 plants also consistently displayed enhanced disease development upon V. dahliae inoculation (Fig. 1A, B).
To quantify V. dahliae colonization in the different Arabidopsis genotypes, the fungal biomass was measured with real-time PCR. Determination of the average fungal biomass revealed significantly enhanced fungal colonization in V. dahliae-inoculated sgs1-1, sgs2-1, and sgs3-1 plants when compared with the inoculated Col-0 plants (Fig. 1C), since at least double the amount of fungal biomass was detected in these mutants at 3 weeks post-inoculation.
sgs mutants do not display enhanced susceptibility towards other pathogens
To investigate whether the enhanced pathogen susceptibility phenotype of the sgs mutants extended to other pathogens in addition to Verticillium species, the susceptibility of the sgs1-1, sgs2-1, and sgs3-1 mutants towards the vascular fungus F. oxysporum f.sp. raphani (Diener and Ausubel, 2005) was tested. However, disease development on the three sgs mutants did not differ from disease development on Col-0 plants upon inoculation with this pathogen (Fig. 2). Furthermore, a number of additional fungal and bacterial pathogens was tested on the sgs mutants (see Supplementary Table S1 at JXB online; Wang et al., 2008). These comprised the foliar fungal pathogens Botrytis cinerea, Alternaria brassicicola, and Plectosphaerella cucumerina, and virulent and avirulent strains of the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000. However, for none of these pathogens was altered susceptibility observed in the sgs mutants when compared with Col-0 (data not shown). Thus, the enhanced susceptibility of the sgs mutants is specific for Verticillium pathogens and does not extend to other pathogens.
Fig. 2.
Typical symptoms caused by F. oxysporum on Arabidopsis sgs mutants. The mutants sgs1-1, sgs2-1, sgs3-1, and the corresponding wild type Col-0 were inoculated with F. oxysporum f.sp. raphani, or mock-inoculated. The picture was taken at 12 d post-inoculation.
sgs mutants do not display altered sensitivity towards abiotic stress
RNA silencing has also been implicated in abiotic stress resistance (Borsani et al., 2005; Sunkar et al., 2007). Therefore, the sgs mutants were screened for their responses towards treatment with different hormones (abscisic acid, auxin, brassinolide, cytokinin, ethylene, gibberellic acid, and jasmonate) and sensitivity towards salt, heavy metal, reactive oxygen, and osmotic stress (see Supplementary Table S1 at JXB online; Wang et al., 2008). However, none of the sgs mutants showed significantly altered phenotypes towards these treatments when compared with Col-0 plants (data not shown).
Assessment of Verticillium susceptibility in additional gene-silencing mutants
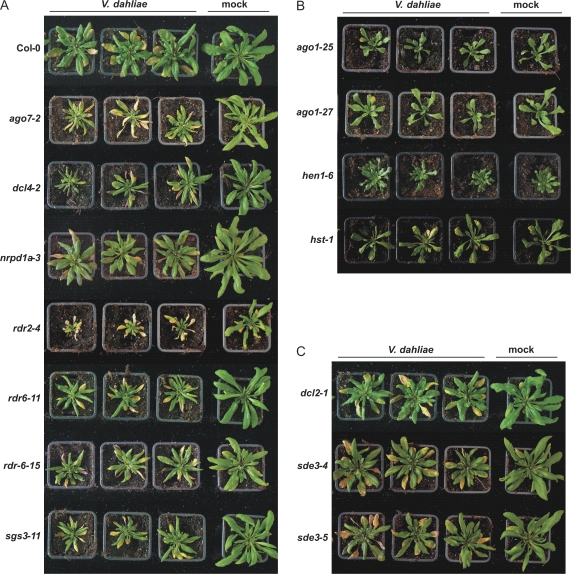
The enhanced susceptibility phenotype of the sgs mutants upon Verticillium inoculation directed us to assess susceptibility towards this pathogen in additional gene-silencing mutants. These comprised additional mutant alleles of SGS2 (also known as RDR6), namely rdr6-11 and rdr6-15, and for SGS3, namely sgs3-11. Furthermore, mutants of other components of RNA-silencing pathways were also included (Table 1). These included mutants of genes that encode different enzyme families, such as the argonautes AGO1 and AGO7, the dicers DCL2 and DCL4, the methyltransferase HEN1, the putative sRNA transporter HST, the DNA-dependent RNA polymerase NRPD1a, the RNA-dependent RNA polymerase RDR2, and the RNA helicase SDE3 that all have been implicated in different RNA-silencing pathways (Table 1; Voinnet, 2008). All mutants, derived from a Col-0 parental line, were challenged with V. dahliae strain JR2. As expected, additional mutant alleles of SGS2 and SGS3 (rdr6-11, rdr6-15, and sgs3-11) were more susceptible than Col-0 plants upon V. dahliae inoculation (Fig. 3A), thus confirming the enhanced susceptibility observed in the sgs2-1 and sgs3-1 mutants. The other PTGS mutants could be divided into three classes, based on the phenotypes after V. dahliae inoculation; those displaying enhanced susceptibility (Fig. 3A), mutants displaying enhanced resistance (Fig. 3B), and mutants displaying similar disease phenotypes as Verticillium-inoculated Col-0 plants (Fig. 3C). The mutants ago7-2, dcl4-2, nrpd1a-3, and rdr2-4 were found to be more susceptible to V. dahliae challenge by showing more severe stunting and necrosis when compared with inoculated Col-0 plants (Fig. 3A; see Supplementary Fig. S2 at JXB online). By contrast, the mutants ago1-25, ago1-27, hen1-6, and hst-1 were found to be more resistant because they displayed less necrosis and no anthocyanin production when compared with Col-0 plants upon V. dahliae inoculation (Fig. 3B; see Supplementary Fig. S2 at JXB online). Finally, the mutants dcl2-1, sde3-4, and sde3-5 showed a disease susceptibility phenotype that was similar to that of Col-0 with respect to severity of stunting, necrosis, and anthocyanin production (Fig. 3C; see Supplementary Fig. S2 at JXB online).
Table 1.
Arabidopsis mutants used in this study
| Gene name | AGI code | Protein function | Mutant allele | Reference |
| AGO1 | At1g48410 | slicer in RISC | ago1-25 | Morel et al., 2002 |
| ago1-27 | Morel et al., 2002 | |||
| AGO7 | At1g69440 | slicer in RISC | ago7-2 | SALK_095997a |
| DCL2 | At3g03300 | dicer | dcl2-1 | Xie et al., 2004 |
| DCL4 | At5g20320 | dicer | dcl4-2 | Yoshikawa et al., 2005 |
| HEN1 | At4g20910 | methyltransferase | hen1-6 | Li et al., 2005 |
| HST | At3g05040 | transporter | hst-1 | Telfer and Poethig, 1998 |
| NRPD1a/SDE4 | At1g63020 | polymerase | nrpd1a-3 | Herr et al., 2005 |
| RDR2 | At4g11130 | RDR | rdr2-4 | Smith et al., 2007 |
| RDR6/SDE1/SGS2 | At3g49500 | RDR | sgs2-1 | Elmayan et al., 1998 |
| rdr6-11 | Peragine et al., 2004 | |||
| rdr6-15 | Allen et al., 2004 | |||
| SDE3 | At1g05460 | RNA helicase | sde3-4 | Vazquez et al., 2004b |
| sde3-5 | SALK_003347a | |||
| SGS1 | Unknown | Unknown | sgs1-1 | Elmayan et al., 1998 |
| SGS3/SDE2 | At5g23570 | CC-domain | sgs3-1 | Mourrain et al., 2000 |
| protein | sgs3-11 | Peragine et al., 2004 |
SALK T-DNA insertion mutant (Alonso et al., 2003).
Fig. 3.
Typical symptoms caused by V. dahliae on various Arabidopsis silencing mutants. Arabidopsis gene-silencing mutants and the corresponding wild type Col-0 were inoculated with V. dahliae strain JR2, or mock-inoculated. (A) V. dahliae-inoculated ago7-2, dcl4-2, rdr6-11, rdr6-15, and sgs3-11 plants show enhanced symptom development, including more severe stunting, wilting, anthocyanin accumulation, and tissue necrosis, compared with inoculated Col-0 plants at 20 d post-inoculation. (B). V. dahliae-inoculated ago1-25, ago1-27, hen1-6, and hst-1 mutants develop fewer symptoms than inoculated Col-0 plants (A) at 25 d post-inoculation. (C) V. dahliae-inoculated dcl2-1, sde3-4, and sde3-5 mutants show similar disease symptoms as inoculated Col-0 plants (A) at 20 d post-inoculation.
Quantification of V. dahliae biomass in planta
To quantify V. dahliae colonization in the different Arabidopsis genotypes, the fungal biomass was measured in individual plants with real-time PCR. For each of the genes tested, the average fungal colonization of at least one mutant allele was quantified with real-time PCR. This analysis demonstrated that the altered susceptibility phenotypes correlated with the degree of fungal colonization when compared with inoculated Col-0 plants (Table 2). The mutants displaying enhanced symptoms upon Verticillium inoculation (sgs1-1, sgs2-1, sgs3-1, ago7-2, dcl4-2, nrpd1a-3, rdr2-4, and rdr6-15) accumulated significantly more fungal biomass when compared with inoculated Col-0 plants, while the mutants that showed reduced symptom development (ago1-27, hen1-6, and hst-1) accumulated significantly less fungal biomass. By contrast, fungal biomass accumulation in Verticillium-inoculated dcl2-1 and sde3-4 plants was not significant different from that of inoculated Col-0 plants (Table 2).
Table 2.
Quantification of Verticillium dahliae biomass in Arabidopsis gene-silencing mutants by real-time PCR comparison of V. dahliae internal transcribed spacer (ITS) transcript levels (as a measure for fungal biomass) relative to Arabidopsis RuBisCo transcript levels (for equilibration) at 19–29 d post-inoculation with V. dahliae strain JR2
| Gene name | Genotype | Symptom displaya | Biomass fold changeb | Significancec |
| Col-0 | – | 1 | – | |
| AGO1 | ago1-27 | Reduced | 0.007 | P <0.1 |
| AGO7 | ago7-2 | Enhanced | 3.174 | P <0.2 |
| DCL2 | dcl2-1 | Similar | 0.829 | No |
| DCL4 | dcl4-2 | Enhanced | 2.422 | P <0.05 |
| HEN1 | hen1-6 | Reduced | 0.045 | P <0.1 |
| HST | hst1-1 | Reduced | 0.039 | P <0.05 |
| NRPD1a/SDE4 | nrpd1a-3 | Enhanced | 1.816 | P <0.2 |
| RDR2 | rdr2-4 | Enhanced | 2.701 | P <0.05 |
| RDR6/SDE1/SGS2 | sgs2-1 | Enhanced | 2.279 | P <0.05 |
| rdr6-15 | Enhanced | 3.286 | P <0.05 | |
| SDE3 | sde3-4 | Similar | 1.674 | No |
| SGS1 | sgs1-1 | Enhanced | 3.729 | P <0.05 |
| SGS3/SDE2 | sgs3-1 | Enhanced | 2.938 | P <0.05 |
Symptom display upon V. dahliae inoculation when compared with Col-0 (also see Fig. 3).
The relative average fungal biomass is indicated as relative fold-change when compared with fungal biomass in V. dahliae-inoculated Col-0 plants of which the average fungal biomass was set to one.
Statistically significant differences are given as P-values according to a Student's t test with a 95% to an 80% confidence interval (P <0.05–0.2).
Assessment of root development and architecture
Being a root pathogen, differences in Verticillium susceptibility of the different mutants may be explained by differences in root architecture, the tissues that are inoculated. Although no obvious differences in root architecture were observed during uprooting and inoculation of the mutants, except for the ago mutants that had developed shorter roots, root development and architecture was assessed upon in vitro growth on MS medium. However, apart from rather slight growth differences, no notable differences in root development and architecture were observed for the RNA-silencing mutants that correlated with the differences in Verticillium susceptibility (Fig. 4). For all mutants, development of the primary, dominant, root was followed by production of lateral roots in a later stage.
Fig. 4.
Typical root architecture of in vitro-grown Arabidopsis gene silencing mutants. Roots were grown on vertically oriented MS plates and pictures were taken 10 d after sowing. (This figure is available in colour at JXB online.)
Assessment of basal defence responses
To investigate whether the altered Verticillium susceptibility phenotypes of the various PTGS mutants can be explained by defects in basal defence signalling pathways, the expression of molecular markers for salicylic acid- (SA) and jasmonic acid- (JA) mediated defence response pathways was assessed. Expression of the SA marker gene PR-1 (Uknes et al., 1992) was clearly induced in Col-0 plants as well as in all PTGS mutants at 24 h after drop-inoculation with 2 mM SA (see Supplementary Fig. 3 at JXB online). In non-treated plants, little or no PR-1 expression was monitored in these genotypes (data not shown). Thus, the altered susceptibility phenotypes could not be correlated to changes in SA-mediated defence responses. Similarly, the expression patterns of the JA-marker PDF1.2 (Penninckx et al., 1996; Thomma et al., 1998) and the chitin elicitor-responsive marker MPK3 (Wan et al., 2008) could not be correlated to the altered susceptibility phenotypes (data not shown).
Discussion
Recent evidence indicates that, apart from defence against viruses, RNA silencing plays a role in defence against bacterial pathogens (Voinnet, 2008), and that, similar to viruses, bacteria have also developed mechanisms to suppress RNA silencing in order to cause disease (Navarro et al., 2008). It is shown here that RNA silencing is also important for defence against multicellular, eukaryotic, microbial pathogens, namely vascular fungi of the Verticillium genus. These include strains of the species V. dahliae, V. albo-atrum, and V. longisporum that are all pathogenic on Arabidopsis (Fradin and Thomma, 2006). Various components of RNA-silencing pathways were tested and most of them were found to affect Verticillium resistance, some positively and others negatively. Furthermore, our results show that PTGS is truly affecting Verticillium resistance and not merely symptom development or display, since altered symptom development of the Verticillium inoculated RNA-silencing mutants correlated with altered Verticillium colonization in these mutants as shown by real-time PCR-based fungal biomass quantification (Table 2).
The altered susceptibility phenotypes of the RNA-silencing mutants is specific for Verticillium defence as shown for the sgs mutants. Inoculation of the sgs mutants with strains belonging to different pathogenic species of the Verticillium genus all resulted in a similar increased susceptibility phenotype. Inoculations with other pathogens that use different colonization and feeding styles did not show altered susceptibility phenotypes. This suggests that the enhanced susceptibility is not due to defects in any of the well-known basal defence signalling pathways (Thomma et al., 2001a). Indeed, in our analysis it was not possible to correlate altered susceptibility to SA or JA signalling. However, this is not surprising because alterations in these basal defence responses would most likely be reflected in altered susceptibility towards some of the other pathogens that were tested. For instance, altered SA signalling would most likely lead to altered susceptibility towards P. syringae and P. cucumerina, while altered JA-signalling would be reflected in A. brassicicola and B. cinerea resistance (Thomma et al., 1998, 2001a, b). Our assays also included the vascular fungal pathogen F. oxysporum f sp. raphani that displays a similar life-style to Verticillium spp. Both F. oxysporum and Verticillium spp infect plants through the roots and enter the xylem where they release conidia that spread upwards through the vessels with the transpiration stream (Di Pietro et al., 2001; Fradin and Thomma, 2006; Berrocal-Lobo and Molina, 2008). Despite these similarities in host colonization, the susceptibility of the RNA silencing mutants is specific towards Verticillium spp, suggesting that a highly specific disease mechanism is affected in these mutants. Since the different RNA-silencing mutants did not show obvious alterations in root development or architecture that correlated with the altered susceptibility phenotypes, this mechanism could not be linked to root development.
In contrast to SGS1, both SGS2 (also known as RDR6 and SDE1) and SGS3 were cloned and found to encode an RNA-dependent RNA polymerase (RDR) and a protein of unknown function, respectively. SGS2 and SGS3 are required for the synthesis of dsRNA in different RNA-silencing pathways (Dalmay et al., 2000; Mourrain et al., 2000; Brodersen and Voinnet, 2006; Vaucheret, 2006). Furthermore, our analysis comprised mutants for the argonautes AGO1 and AGO7, the dicers DCL2 and DCL4, the methyltransferase HEN1, the putative sRNA transporter HST, the DNA-dependent RNA polymerase NRPD1a, the RNA-dependent RNA polymerase RDR2, and the RNA helicase SDE3, all of which have been implicated in different RNA-silencing pathways and regulate processes including TGS, PTGS, antiviral defence, plant development, hormone signalling, and abiotic and biotic stress tolerance (Brodersen and Voinnet, 2006; Vaucheret, 2006; Voinnet, 2008). While HEN1 methylates small RNA species and thus protects these sRNAs from degradation and polyuridylation (Chen et al., 2002; Li et al., 2005; Yu et al., 2005), HST possibly mediates the transport of miRNAs from the nucleus to the cytoplasm (Mallory and Vaucheret, 2006; Sunkar et al., 2007). SDE3 acts as an RNA helicase and may facilitate the synthesis of dsRNA by SGS2/RDR6/SDE1 (Dalmay et al., 2001). Although its precise function is unclear, NRPD1a is suggested to be a silencing-specific polymerase (Herr et al., 2005). In this study, as many as ten different RNA-silencing components, namely AGO7, DCL4, NRPD1a, RDR2, SGS1, SGS2/RDR6/SDE1, SGS3, AGO1, HEN1, and HST were all shown to affect Verticillium defence.
The combination of RNA-silencing components that is involved in altered Verticillium susceptibility does not comply with one single RNA-silencing pathway among those that are currently discriminated. However, the identification and full characterization of such pathways is still in its infancy. Defence against Verticillium might trigger a novel RNA-silencing pathway that is similar to the natural cis-antisense transcript-derived siRNA (nat-siRNAs) pathway that is induced upon stresses including bacterial infection (Borsani et al., 2005; Wang et al., 2005; Katiyar-Agarwal et al., 2006). In this case siRNAs might be specifically produced upon induction of NATs by the action of RDR6/SGS2/SDE1, SGS3 NRPD1a3, RDR2, and DCL4 and incorporated in AGO7 to trigger a defence response by repression of AGO1, HEN1, and HST. Alternatively, the observed phenomena may be the result of the cross-interaction of multiple RNA-silencing pathways that influence the defence response. Furthermore, the presence of ten AGOs, four DCLs and six RDRs in Arabidopsis (Morel et al., 2002; Schauer et al., 2002; Yu et al., 2003) may reflect the versatility of these components in RNA-silencing pathways.
Whatever the exact pathway that is involved, it is likely that RNA silencing is involved either in a highly specific defence response against Verticillium pathogens or, alternatively, is involved in a developmental cue that is of particular importance for Verticillium infections. Interestingly, it was recently demonstrated that inoculation of Arabidopsis with non-pathogenic P. syringae that triggers a robust basal defence response in Arabidopsis leads to altered accumulation of several microRNAs, including those targeting multiple components of auxin signalling pathways (Fahlgren et al., 2007). Furthermore, it was recently suggested that the transcriptional regulation of resistance gene loci may be under the control of RNA silencing, as demonstrated for the RPP5-locus for recognition of the oomycete downy mildew pathogen Peronospora parasitica (Yi and Richards, 2007). This demonstrates that RNA silencing may affect diverse pathogens by regulating various modulators of host defence (Voinnet, 2008). Relatively little is known about the biology of vascular wilt diseases, and processes that are involved in defence against these pathogens (Fradin and Thomma, 2006). This makes it difficult to identify the physiological process that is affected in the RNA-silencing mutants and that explains the observed disease phenotypes. It is possible that microarray analyses on inoculated wild-type plants and RNA-silencing mutants will facilitate the identification of this process. However, the main challenge will be to identify the small RNAs that are the basis of the altered Verticillium susceptibility in these mutants.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Conditional phenotype assays for sgs1-1, sgs2-1, and sgs3-1 mutants.
Supplementary Fig. S1. Typical symptoms of Arabidopsis sgs2-1 mutants upon inoculation with plant pathogenic Verticillium species.
Supplementary Fig. S2. Quantification of symptom development at 20 dpi shown as the ratio of diseased rosette leaves with standard deviation.
Supplementary Fig. S3. Salicylic acid-induced PR-1 expression in Arabidopsis gene silencing mutants.
Supplementary Material
Acknowledgments
This work was supported, in part, by the Dutch Graduate School of Experimental Plant Sciences (EPS) and co-financed by the Centre for BioSystems Genomics (CBSG) which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research. BPHJT is supported by a Vidi grant of the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO). The authors thank Dr H Vaucheret and Professor Dr RS Poethig for providing seeds of RNA-silencing mutants, and Drs P Crous, M Höfte, B Lievens, J Robb, and A von Tiedemann for providing Verticillium isolates.
References
- Agorio A, Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. The Plant Cell. 2007;19:3778–3790. doi: 10.1105/tpc.107.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature Genetics. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A. Arabidopsis defence response against Fusarium oxysporum. Trends in Plant Science. 2008;13:145–150. doi: 10.1016/j.tplants.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Esse HP, Vossen JH, et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologs in other fungal species. Molecular Microbiology. 2008;69:119–136. doi: 10.1111/j.1365-2958.2008.06270.x. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F. MicroRNA166 controls root and nodule development in Medicago truncatula. The Plant Journal. 2008;54:876–887. doi: 10.1111/j.1365-313X.2008.03448.x. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. The diversity of RNA-silencing pathways in plants. Trends in Genetics. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Vanderleyden J. An automated quantitative assay for fungal growth inhibition. FEMS Microbiology Letters. 1990;69:55–59. [Google Scholar]
- Brouwer M, Lievens B, Van Hemelrijck W, Van den Ackerveken G, Cammue BPA, Thomma BPHJ. Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real-time fluorescence PCR. FEMS Microbiology Letters. 2003;228:241–248. doi: 10.1016/S0378-1097(03)00759-6. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biology. 2007;5:501–514. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye KMJ, Goderis IJWM, Wouters PFJ, Pues JM-TG, Delaure SL, Broekaert WF, Depicker A, Cammue BPA, De Bolle MFC. Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. The Plant Journal. 2004;39:440–449. doi: 10.1111/j.1365-313X.2004.02144.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host–microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes and Development. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for posttranscriptional gene silencing in Arabidopsis. EMBO Journal. 2001;20:2069–2077. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich L, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proceedings of the National Academy of Sciences, USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A, Garcia-Maceira FI, Meglecz E, Roncero MIG. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Molecular Microbiology. 2001;39:1140–1152. [PubMed] [Google Scholar]
- Diener AC, Ausubel FM. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics. 2005;171:305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CIA, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proceedings of the National Academy of Sciences, USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. Induction, suppression and requirement of RNA-silencing pathways in virulent Agrobacterium tumefaciens infections. Nature Genetics. 2006;38:258–263. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Voinnet O. The complex interplay between plant viruses and host RNA-silencing pathways. Current Opinion in Plant Biology. 2005;8:415–423. doi: 10.1016/j.pbi.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Elmayan T, Balzergue S, Beon F, et al. Arabidopsis mutants impaired in cosuppression. The Plant Cell. 1998;10:1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Thomma BPHJ. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Molecular Plant Pathology. 2006;7:71–86. doi: 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- Fulton TM, Chunwongse J, Tanksley SD. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Molecular Biology Reporter. 1995;13:207–209. [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes, application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in post-transcriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO Journal. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes and Development. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu JK, Staskawicz BJ, Jin HL. A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences, USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of phycocyanin and fluorescin. Journal of Clinical and Laboratory Medicine. 1954;44:301–307. [PubMed] [Google Scholar]
- Li JJ, Yang ZY, Yu B, Liu J, Chen XM. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Current Biology. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens B, Brouwer M, Vanachter ACRC, Cammue BPA, Thomma BPHJ. Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Science. 2006;171:155–165. [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nature Genetics. 2006;38:850–850. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJM. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO Journal. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. The Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, et al. Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. The Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerre-Tugaye MT, Dumas B. A novel Arabidopsis–Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Molecular Plant–Microbe Interactions. 2004;17:272–282. doi: 10.1094/MPMI.2004.17.3.272. [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux J-P, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. The Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes and Development. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye XH, Hou GC, Sato S, Clemente TE, Morris TJ. RDR6 has a broad-spectrum but temperature-dependent antiviral defence role in Nicotiana benthamiana. Journal of Virology. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney HC, van 't Klooster JW, van der Hoorn RAL, Joosten MHAJ, Jones JDG, de Wit PJGM. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends in Plant Science. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. The Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu JH, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Tang XY, Frederick RD, Zhou JM, Halterman DA, Jia YL, Martin GB. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Telfer A, Poethig RS. HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development. 1998;125:1889–1898. doi: 10.1242/dev.125.10.1889. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Broekaert WF, Cammue BPA. Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiology and Biochemistry. 2000;38:421–427. [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defence-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences, USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA. The complexity of disease signaling in Arabidopsis. Current Opinion in Immunology. 2001a;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Tierens FM-J, Penninckx IAMA, Mauch-Mani B, Broekaert WF, Cammue BPA. Different micro-organisms differentially induce Arabidopsis disease response pathways. Plant Physiology and Biochemistry. 2001b;39:673–680. [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. The Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol AR, Mur LA, Beld M, Mol JNM, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene-expression. The Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse HP, Bolton MD, Stergiopoulos I, de Wit PJGM, Thomma BPHJ. The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Molecular Plant–Microbe Interactions. 2007;20:1092–1101. doi: 10.1094/MPMI-20-9-1092. [DOI] [PubMed] [Google Scholar]
- van Esse HP, van't Klooster JW, Bolton MD, Yadeta K, van Baarlen P, Boeren S, Vervoort J, de Wit PJGM, Thomma BPHJ. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defence. The Plant Cell. 2008;20:1948–1963. doi: 10.1105/tpc.108.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants: defense and counter defense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes and Development. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Post-transcriptional RNA silencing in plant–microbe interactions: a touch of robustness and versatility. Current Opinion in Plant Biology. 2008;11:464–470. doi: 10.1016/j.pbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Wan J, Patel A, Mathieu M, Kim SY, Xu D, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. The Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ellendorff U, Kemp B, et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiology. 2008;147:503–517. doi: 10.1104/pp.108.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biology. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biology. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. The Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes and Development. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DQ, Fan BF, MacFarlane SA, Chen ZX. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defence. Molecular Plant–Microbe Interactions. 2003;16:206–216. doi: 10.1094/MPMI.2003.16.3.206. [DOI] [PubMed] [Google Scholar]
- Yu B, Yang ZY, Li JJ, Minakhina S, Yang MC, Padgett RW, Steward R, Chen XM. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.