Abstract
Prolamins, a group of rice (Oryza sativa) seed storage proteins, are synthesized on the rough endoplasmic reticulum (ER) and deposited in ER-derived type I protein bodies (PB-Is) in rice endosperm cells. The accumulation mechanism of prolamins, which do not possess the well-known ER retention signal, remains unclear. In order to elucidate whether the accumulation of prolamin in the ER requires seed-specific factors, the subcellular localization of the constitutively expressed green fluorescent protein fused to prolamin (prolamin–GFP) was examined in seeds, leaves, and roots of transgenic rice plants. The prolamin–GFP fusion proteins accumulated not only in the seeds but also in the leaves and roots. Microscopic observation of GFP fluorescence and immunocytochemical analysis revealed that prolamin–GFP fusion proteins specifically accumulated in PB-Is in the endosperm, whereas they were deposited in the electron-dense structures in the leaves and roots. The ER chaperone BiP was detected in the structures in the leaves and roots. The results show that the aggregation of prolamin–GFP fusion proteins does not depend on the tissues, suggesting that the prolamin–GFP fusion proteins accumulate in the ER by forming into aggregates. The findings bear out the importance of the assembly of prolamin molecules and the interaction of prolamin with BiP in the formation of ER-derived PBs.
Keywords: Endoplasmic reticulum, Oryza sativa, prolamin, protein body, storage protein, transgenic rice
Introduction
Plants generate a specialized compartment derived from the endoplasmic reticulum (ER) to accumulate the enormous quantities of proteins that are actively synthesized on the ER (Chrispeels and Herman, 2000). ER-derived structures are found in both vegetative and storage organs of plants (Hara-Nishimura et al., 2004). ER bodies of Arabidopsis thaliana, KDEL vesicles of black gram (Vigna mungo), and ricinosomes of castor bean (Ricinus communis) are examples of ER-derived structures that are found in vegetative organs (Schmid et al., 1998; Toyooka et al., 2000; Hayashi et al., 2001). All of these structures have been shown to accumulate a hydrolytic enzyme with a well-known ER retention signal (KDEL) at the C-terminus (Schmid et al., 1998; Toyooka et al., 2000; Matsushima et al., 2003). Protein bodies (PBs) of maize (Zea mays) and precursor-accumulating (PAC) vesicles of pumpkin (Cucurbita maxima) are among the ER-derived structures that are found in storage organs (Hara-Nishimura et al., 1998; Herman and Larkins, 1999). These structures are responsible for the accumulation of seed storage proteins, which do not possess the KDEL signal, in the ER (Hara-Nishimura et al., 1998; Herman and Larkins, 1999).
In rice endosperm, there are two types of PBs (Tanaka et al., 1980; Yamagata et al., 1982) and these are responsible for storing prolamins and glutelins, the major seed storage proteins of rice (Bechtel and Juliano, 1980; Oparka and Harris, 1982; Krishnan et al., 1986; Yamagata and Tanaka, 1986). Although both prolamins and glutelins are synthesized on the rough ER, they are transported by different pathways (Tanaka et al., 1980; Yamagata et al., 1982). Because of their hydrophobicity, prolamins are stored in spherical intracisternal inclusion granules, referred to as type I PBs (PB-Is), within the ER lumen (Tanaka et al., 1980). In contrast, glutelins are introduced into the vesicular transporting system, resulting in accumulation in protein storage vacuoles (PSVs) that are electron-dense and irregularly shaped, referred to as type II PBs (PB-IIs) (Tanaka et al., 1980; Krishnan et al., 1986).
The specific subcellular localization of storage proteins in rice endosperm is a unique process involving seed-specific events. First, the rice storage protein mRNAs are localized to distinct subdomains of the ER. Prolamin mRNAs are targeted to ER that surrounds the PB-I (PB-ER), whereas glutelin mRNAs are localized to the cisternal ER (Crofts et al., 2004). The rice storage proteins are synthesized on each ER membrane, and translocated into the ER lumen. Secondly, chaperone proteins in the ER lumen are crucial for the sorting of storage proteins. The rice prolamin does not contain a KDEL/HDEL ER retention sequence, but its deposition has been shown to be assisted by the binding protein (BiP) (Li et al., 1993). BiP is highly enriched on the periphery of PB-Is (Muench et al., 1997) and has been proposed to be necessary to maintain the prolamin in a competent state for subsequent assembly in the ER. Another molecular chaperone, protein disulphide isomerase, is required for the segregation of prolamins and proglutelins in the ER lumen (Takemoto et al., 2002).
Several recent reports have investigated the intracellular localization of recombinant proteins expressed in seeds, revealing that such proteins can be deposited in unexpected places (Wright et al., 2001; Chikwamba et al., 2003; Arcalis et al., 2004; Petruccelli et al., 2006). Drakakaki et al. (2006) reported that the recombinant phytase was secreted from leaf cells of rice, as expected, whereas it was retained in PB-Is and PB-IIs in the endosperm cells. They concluded that the storage function of rice endosperm may determine whether or not the recombinant phytase is secreted.
Recently, the sorting mechanisms of rice storage proteins have been investigated, although the mechanisms by which prolamins assemble into PB-Is are still poorly understood. To investigate whether ER-derived PB-I formation of prolamin is specific to the highly specialized endosperm, the stability and the subcellular deposition site of the constitutively expressed green fluorescent protein fused to prolamin (prolamin–GFP) in the seeds, leaves, and roots of transgenic rice plants were examined. The results demonstrate that prolamin–GFP fusion proteins are stable not only in the seeds but also in the leaves and roots of transgenic rice. Furthermore, it is shown that prolamin–GFP forms the PB-like structures in leaves and roots of transgenic rice
Materials and methods
Plant materials
Rice (Oryza sativa L. cv Nipponbare) was used in all experiments. The transgenic rice plants were grown with soil in a naturally illuminated temperature-controlled (28 °C) greenhouse at the experimental field of the Kyoto Prefectural Institute of Agricultural Biotechnology. The seedlings were grown at 28 °C under a continuous light condition (30 μmol photons m−2 s−1) in an incubation room.
Plasmid constructions and transformation of rice plants
The p35S:GFP plasmid vector (Takahashi et al., 2004) containing the 35S promoter of the cauliflower mosaic virus (CaMV), the first 5′-untranslated region (UTR) intron of the superoxide dismutase (SOD) gene (sodCc2) (Sakamoto et al., 1995), the coding sequence of sGFP(S65T) (Chiu et al., 1996), and the polyadenylation signal of the nopaline synthase (NOS) gene was used.
To generate a fusion gene, the coding region for rice 13 kDa prolamin (λRM1) (Mitsukawa et al., 1999) was amplified by PCR using the following primer set: forward, 5′-GCGGAGGGATCCATGAAGATCATTTTCGTATTTGCTC-3′ containing a BamHI site; and reverse, 5′-GCGGAGGGATCCGCCGCCGCCGTACCAGACACCACCAACGG-3′ containing a BamHI site. The amplified PCR fragment was digested with BamHI, then inserted into the BamHI site of the N-terminal region of the sGFP(S65T)-coding sequence of the plasmid vector (Takahashi et al., 2004), and the resulting plasmid was named p35S:Pro–GFP.
The expression cassettes of p35S:GFP and p35S:Pro-GFP were transferred into the cloning site of the pIG121Hm plasmid (Ohta et al., 1990). The resulting binary vectors were introduced into rice, using an Agrobacterium-mediated method described previously (Hiei et al., 1994).
RT-PCR analysis
Total RNA was extracted from leaves and roots of 9-day-old seedlings and 12 days after flowering (DAF) developing seeds of wild type (WT) and 35S:Pro-GFP plants with an RNeasy plant mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total RNA (0.5 μg) was converted into cDNA by ReverTra Ace (MMLV Reverse Transcriptase RnaseH-; Toyobo, Osaka, Japan) with oligo(dT) as primer, yielding 20 μl of cDNA solution. PCR was performed in a 10 μl reaction volume containing 1 μl of cDNA solution, 1× PCR buffer, 0.25 μM dNTPs, 1.0 μM of each primer, and 0.5 U of Taq polymerase (Takara, Otsu, Japan). The following primers were used: sGFP (GFP-F, 5′-TATATCATGGCCGACAAGCA-3′; and GFP-R, 5′-GAACTCCAGCAGGACCATGT-3′) and actin (actin-F, 5′-TACAACTCCATCATGAAGTGCG-3′; and actin-R, 5′-AGAAGCATTTCCTGTGCACAAT-3′). The PCR program consisted of 25 repetitive cycles with a denaturation step at 94 °C for 30 s, an annealing step at 57 °C for 30 s, and an elongation step at 72 °C for 1 min. The PCR cycles were preceded by an extra denaturation step at 94 °C for 4 min and ended with an extra elongation step of at 72 °C for 7 min.
Protein extraction and subcellular fractionation
For the extraction of total proteins, the leaves and roots of the 15-day-old seedling and mature seeds were homogenized in SDS sample buffer A [62.5 mM TRIS-HCl (pH 6.8), 4 M urea, 2% (w/v) SDS] supplemented with 5% (v/v) 2-mercaptoethanol (2-ME). The homogenates were centrifuged at 15 000 g for 30 min to obtain the protein extracts as supernatant solutions and then heated (100 °C). An RC DC Protein Assay Kit (Bio-Rad, Hercules, CA, USA) was used for measurement of total proteins according to the manufacturer's instructions. The 10 μg aliquots of proteins were analysed by SDS–PAGE and immunoblot.
For subcellular fractionation, leaf and root cell fractions were prepared according to the method of Tamura et al. (2003). Leaves and roots of the 21-day-old seedlings were chopped with a razor blade on ice in HEPES buffer [50 mM HEPES-KOH (pH 7.5), 0.4 M sucrose, and protease inhibitors (Complete; Roche, Basel, Switzerland)]. The homogenates were filtered through Miracloth (Calbiochem, La Jolla, CA, USA). Filtrates were centrifuged at 15 000 g and 4 °C for 20 min. The pellets were added to HEPES buffer (the P15 fractions), and the supernatants were centrifuged at 100 000 g and 4 °C for 20 min. The pellets were added in HEPES buffer (the P100 fractions), and the supernatants (the S100 fractions) were concentrated by using a Microcon YM-10 centrifugal filter device (Millipore, Billerica, MA, USA). Each of the fractions P15, P100, and S100 was analysed by SDS–PAGE and immunoblot.
For the extraction of proteins under non-reducing conditions, the powder of mature seeds was extracted with the SDS sample buffer A. Leaves were chopped on ice with a razor blade in the HEPES buffer. The homogenates were filtered through Miracloth (Calbiochem). Filtrates were centrifuged at 15 000 g and 4 °C for 20 min. The pellets were added in 1× SDS sample buffer B [62.5 mM TRIS-HCl (pH 7.5), 10% (v/v) glycerol, 2% (w/v) SDS] (the P15 fractions), and the supernatants (the S15 fractions) were concentrated by using a Microcon YM-10 centrifugal filter devices (Millipore). The supernatants were mixed with an equal volume of 2× SDS sample buffer B. For extraction under reducing conditions, the SDS sample buffers A and B were supplemented with 5% (v/v) 2-ME. Each of the fractions P15 and S15 was analysed by SDS–PAGE and immunoblot.
After SDS–PAGE analysis, separated proteins were electrotransferred to an Immun-Blot PVDF Membrane (Bio-Rad), revealed using anti-GFP antibodies (dilution 1:2000; Medical & Biological Laboratories, Nagoya, Japan) and anti-13 kDa prolamin antibodies (1:1000; Furukawa et al., 2003), and detected with the alkaline phosphatase-conjugated goat anti-rabbit IgG secondary antibody (dilution 1:10 000; Promega, Madison, WI, USA). Proteins were stained with 5-bromo-4-chloro-3-indoyl phosphate (BCIP) and nitroblue tetrazolium (NBT) (BCIP/NBT Color Development Substrate; Promega) according to the manufacturer's instructions.
Fluorescence microscopic analysis
Frozen sections of mature rice seeds were prepared by a method described previously (Saito et al., 2008). Mature seeds were vacuum infiltrated overnight with 2% (w/v) carboxymethyl cellulose (CMC) gel. The samples were embedded in 2% (w/v) CMC gel and frozen in cooled hexane (–94 °C). Seed sections were generated using a Cryo-film transfer kit (FINETEC, Tokyo, Japan) and cryostat (Microm Model 500 M; Global Medical Instrumentation, Inc., Ramsey, MN, USA). The 2 μm thick cryostat sections were incubated with 0.1 μM rhodamine B hexyl ester, the ER membrane stainer, in phosphate-buffered saline (PBS)/methanol (50:50) to label the PB-I membrane. After being washed in PBS, the seed sections were inspected with a fluorescence microscope (BX51; Olympus, Tokyo, Japan). Images were analysed with an Aquacosmos system (Hamamatsu Photonics, Hamamatsu, Japan).
The leaves and roots of 18-day-old seedlings of WT, 35S:GFP, and 35S:Pro-GFP were inspected with an E600 microscope and C1si confocal laser scanning microscope system (Nikon, Tokyo, Japan). GFP was excited at a laser wavelength of 488 nm and detected through a filter for a fluorescence wavelength of 500–530 nm.
Immunoelectron microscopy
Small pieces of 12 DAF developing seeds were vacuum infiltrated for 10 min with a fixative that consisted of 4% (w/v) paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.2) and treated for another 3 h at room temperature with the fixative. After washing with the same buffer, the seeds were dehydrated in a graded ethanol series and then embedded in LR White resin (London Resin Co. Ltd, Hampshire, UK). Blocks were polymerized at 55 °C for 48 h. Ultrathin sections were cut with a diamond knife using a Leica Ultracut UCT (Leica, Wetzlar, Germany) and mounted on nickel grids.
For high pressure freezing and freeze substitution, leaves and roots of 12-day-old seedlings were frozen with a high pressure freezing machine (model HPM010; Bal-Tec, Balzers, Liechtenstein). The frozen samples were treated with acetone for 2 d at –80 °C and warmed at –20 °C for 3 h, 4 °C for 2 h, and room temperature for 2 h. Fixed samples were dehydrated in graded ethanol and embedded in LR White resin.
Ultrathin sections were treated with blocking solution of 1% (w/v) bovine serum albumin in 0.1 M sodium phosphate buffer (pH 7.2) for 30 min at room temperature. The sections were then incubated with anti-GFP antibodies (dilution 1:100; Medical & Biological Laboratories) overnight at 4 °C and with anti-pumpkin BiP antibodies (1:100; Hara-Nishimura et al., 1998) for 1 h at room temperature. After washing with 0.1 M sodium phosphate buffer (pH 7.2), the sections were incubated with a solution of 10 nm gold-labelled goat anti-rabbit IgG antibodies (1:50; GE Healthcare, Buckinghamshire, UK) in the blocking solution for 1 h at room temperature. The sections were washed with distilled water and then stained with 2% (w/v) uranyl acetate. After staining, the sections were examined with a transmission electron microscope (JEM-1220; JEM, Tokyo, Japan) at 100 kV.
Results
Production of transgenic rice constitutively expressing prolamin–GFP
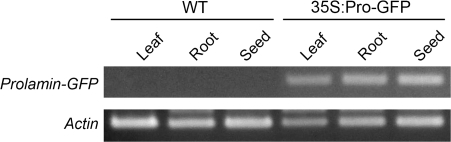
Prolamins are encoded by a multigene family, and separated into three major groups according to their apparent molecular sizes of 10, 13, and 16 kDa (Ogawa et al., 1987). The 13 kDa prolamins are the most abundant group of prolamins in rice (Horikoshi et al., 1991). A chimeric gene encoding a fusion protein of 13 kDa prolamin (λRM1) and GFP was constructed. The prolamin-coding sequence was fused to the region upstream of GFP. GFP is a soluble protein that is secreted efficiently when introduced into plant ER via a signal peptide (Batoko et al., 2000; Frigerio et al., 2001). This chimeric gene was driven by the CaMV 35S promoter (35S:Pro-GFP; Fig. 1). As a control, the GFP gene driven by the CaMV 35S promoter, without the prolamin-coding sequence, was used (35S:GFP; Fig. 1). The first intron of the superoxide dismutase sodCc2 gene (Sakamoto et al., 1995) was inserted between the promoter and the GFP gene to enhance promoter activity. This construct was transferred into rice (O. sativa L. cv. Nipponbare) by an Agrobacterium-mediated transformation method (Hiei et al., 1994). RT-PCR was used to analyse the expression of the prolamin–GFP gene in different tissues of 35S:Pro-GFP plants (Fig. 2). As expected, the transcripts of the prolamin–GFP gene were detected in all tissues analysed, reflecting the activity of the CaMV 35S promoter. The expression of GFP was not detected in tissues of WT plants.
Fig. 1.
Constructs of 35S:GFP and 35S:Pro-GFP that were expressed in transgenic rice plants. 35S-P, the 35S promoter of cauliflower mosaic virus; SOD Intron, the first intron of rice sodCc2; Prolamin, the coding sequence for the 13 kDa prolamin gene (λRM1); GFP, the modified green fluorescent protein gene; nos-T, terminator of the nopaline synthase gene. The arrowheads indicate the RT-PCR primers used for the experiments in Fig. 2.
Fig. 2.
Expression of prolamin–GFP mRNA in different tissues of transgenic plants. RT-PCR analysis of the prolamin–GFP transcript in seeds, leaves, and roots of wild type (WT) and 35S:Pro-GFP plants. The bottom panel shows the expression of the actin genes as a control.
Accumulation of prolamin–GFP fusion proteins in seeds, leaves, and roots of transgenic rice
To investigate the accumulation of the prolamin–GFP fusion proteins in different tissues of the transgenic rice plant, immunoblot analysis of protein extracts from seeds, leaves, and roots was performed with anti-GFP antibodies (Fig. 3). Bands with apparent molecular masses of ∼27 kDa and ∼40 kDa were detected in the 35S:GFP and 35S:Pro-GFP in all tissues, respectively (Fig. 3). The 40 kDa mass is consistent with the predicted molecular size of prolamin–GFP fusion proteins, because the molecular sizes of prolamin and GFP are 13 kDa and 27 kDa, respectively. The ∼40 kDa polypeptides also reacted with anti-13 kDa prolamin antibodies (Fig. 3). These results suggested that the prolamin–GFP fusion proteins accumulated stably not only in the seeds but also in the leaves and roots. The bands of 13, ∼23, and ∼27 kDa were also detected in the seeds of all plants by anti-13 kDa prolamin antibodies. The 13 kDa proteins are endogenous 13 kDa prolamin. The bands of 23 kDa and 27 kDa are non-specific signals. In addition, the processed form of the prolamin–GFP fusion proteins was detected in the leaves and roots of 35S:Pro-GFP plants (Fig. 3).
Fig. 3.
Accumulation of prolamin–GFP fusion proteins in different tissues of transgenic plants. Seeds, leaves, and roots of WT (W), 35S:GFP (G), and 35S:Pro-GFP (P) plants were subjected to SDS–PAGE followed by immunoblot with anti-GFP and anti-13 kDa prolamin antibodies. The upper and lower bands correspond to prolamin–GFP and GFP, respectively. The truncated prolamin–GFP was observed in the leaf and root tissues of 35S:Pro-GFP plants (asterisks). The arrowheads indicate the non-specific signal. The molecular masses are given on the left in kDa.
Localization of prolamin–GFP fusion proteins in the PB-Is in starchy endosperm cells
The subcellular localization of prolamin–GFP fusion proteins in the endosperm cells was next investigated. Fluorescence of prolamin–GFP was detected in the starchy endosperm cells of 35S:Pro-GFP plants, and fluorescence signals were also detected in aleurone cells (Fig. 4A, B). However, the particles emitting fluorescence signals in the aleurone layer were smaller than those in the starchy endosperm cells. In the starchy endosperm cells, prolamin–GFP was primarily found in spherical organelles, but it was not observed in starch granules or the intracellular space (Fig. 4B). The GFP fluorescence was not detected in the endosperm cells of the WT (Fig. 4C, D). The co-localization of PB-I and GFP fluorescence was analysed in the endosperm cells of WT, 35S:GFP, and 35S:Pro-GFP plants. When the seed sections were stained with the rhodamine B hexyl ester, which preferentially labels PB-Is (Choi et al., 2000), the fluorescence of prolamin–GFP was localized mainly to the PB-Is (Fig. 4G). In contrast, GFP fluorescence was not detected in the WT endosperm (Fig. 4E), and GFP fluorescence was detected in the cytosol in the 35S:GFP endosperm (Fig. 4F). Immunogold labelling of prolamin–GFP fusion proteins in the starchy endosperm cells of a 35S:Pro-GFP plant showed that the gold particles were distributed in the PB-Is, but not in PB-IIs (Fig. 4H). The gold particles were frequently detected in the core of mature PB-Is (Fig. 4I). Some of the PB-Is in 35S:Pro-GFP endosperm cells had a cracked structure and were larger than in WT and 35S:GFP endosperm cells. It is possible that the structure of PB-Is is influenced by the accumulation of prolamin–GFP fusion proteins. In the endosperm cells of the WT, the gold particles were not detected (data not shown). Without an additional sequence, GFP proteins accumulate diffusely in the cytoplasm and nucleus (Chiu et al., 1996). No significant signals of the gold particles were detected in the endosperm cells of 35S:GFP plants (data not shown); it is more difficult to detect diffuse proteins than aggregated proteins by immunocytochemistry. Immunogold labelling of prolamin–GFP in the aleurone cells of a 35S:Pro-GFP plant showed that the gold particles were detected in novel electron-dense structures with a diameter of ∼300 nm (Fig. 4J, K). This result showed that prolamin–GFP fusion proteins are capable of forming the protein aggregates in the alurone cells.
Fig. 4.
Localization of prolamin–GFP fusion proteins in PB-Is in the endosperm cells. Light transmission (A and C) and GFP fluorescence (B and D) images of the mature seed section of a 35S:Pro-GFP plant (A and B) and the WT (C and D) are shown. PE, pericarp; AL, aleurone layer; SE, starchy endosperm. Bars in A and A and C = 50 μm. (E–G) Mature seed sections of WT (E), 35S:GFP (F), and 35S:Pro-GFP (G) were stained with rhodamine B hexyl ester, which specifically binds to the ER membrane and ER-derived PB-I membrane. Green and magenta fluorescence indicate GFP and PB-I, respectively. Bars in E–G=5 μm. (H–K) Immunogold labelling of prolamin–GFP with anti-GFP antibodies in the developing starchy endosperm cells (H and I) and aleurone cells (J and K) of 35S:Pro-GFP plants. (G) Magnified image of the PB-I. Gold particles are detected in the electron-dense structures in the aleurone cells (J and K, arrows). PB-I, type I protein body; PB-II, type II protein body; SG, starch granule; ER, endoplasmic reticulum; DV, dense vesicle; OB, oil body; AG, aleurone grain; CW, cell wall; Mt, mitochondrion. bar in H and J=1 μm; bar in I and K=250 nm.
Formation of aggregates of prolamin–GFP fusion proteins in the leaves and roots
To investigate the subcellular location of prolamin–GFP in leaves and roots, subcellular fractionation was performed according to the method of Tamura et al. (2003). The homogenates from leaves or roots from the seedlings were separated into three subcellular fractions: a 15 000 g pellet (P15), a 100 000 g pellet (P100), and a 100 000 g supernatant (S100). P100 and S100 contain the microsomal proteins and the mixed cytosolic and vacuolar soluble proteins, respectively. The immunoblots of each fraction with anti-GFP antibodies showed that most of the prolamin–GFP was detected in the P15 fractions of leaves and the P15 and P100 fractions of roots (Fig. 5A, B). In the 35S:GFP plants, the GFP proteins were detected specifically in the S100 fractions. The high concentration of prolamin–GFP in the pellet fractions (P15 and P100) suggested that the aggregated prolamin–GFP fusion proteins are retained in the ER lumen.
Fig. 5.
Subcellular distribution of prolamin–GFP fusion proteins in leaves and roots of transgenic plants. Homogenates from leaves (A) and roots (B) of WT, 35S:GFP, and 35S:Pro-GFP plants were subjected to differential centrifugation to obtain the 15 000 g pellet (P15), the 100 000 g pellet (P100), and the 100 000 g supernatant (S100). Each fraction was subjected to immunoblotting with anti-GFP antibodies. Bands of ∼40 kDa (open arrowheads) correspond to prolamin–GFP, while bands of ∼27 kDa (filled arrowheads) correspond to the truncated form. An asterisk indicates the non-specific signal. Molecular masses are given on the left in kDa.
The subcellular distribution of prolamin–GFP in leaf and root cells was analysed by fluorescence microscopy (Fig. 6). In the leaf and root cells of the 35S:GFP plants, the fluorescence of GFP was detected in the nucleus and cytosol (Fig. 6B, E). The fluorescence of prolamin–GFP showed a bright punctate pattern in both the leaf and root cells of the 35S:Pro-GFP plants, and did not show the network pattern of the ER (Fig. 6C, F). The fluorescence was not detected within the vacuole in these cells. In addition, the fluorescence signals of prolamin–GFP in root cells appeared to be slightly weaker than those in leaf cells (Fig. 6C, F).
Fig. 6.
Fluorescence images of prolamin–GFP in the leaf and root cells. Fluorescence images of the leaves (A–C) and roots (D–F) of WT (A and D), 35S:GFP (B and E), and 35S:Pro-GFP (C and F) plants taken using a confocal laser scanning microscope are shown. Dot-like structures, presumed to be prolamin–GFP proteins, are visible in the leaf cells (C) and the root cells (F) of 35S:Pro-GFP plants. Bars = 20 μm.
Formation of PB-like structures containing prolamin–GFP fusion proteins in leaf and root cells
To characterize these punctate structures of prolamin–GFP in the leaf and root cells, an ultrastructural analysis was performed with an electron microscope using a high pressure frozen/freeze substitution technique for the preparation of the thin sections. Immunogold labelling of prolamin–GFP on these thin-sectioned samples showed that the gold particles were localized in electron-dense structures in the leaf and root cells of 35S:Pro-GFP plants (Fig. 7). Most of the structures containing prolamin–GFP were spherical, with diameters of 50–500 nm in the leaf cells, but some of them were irregular structures (Fig. 7B) or spherical structures with a diameter of 2 μm (Fig. 7C). In addition, these structures in leaf cells were often surrounded by the membrane (Fig. 7D, arrowheads). Some of these structures were also associated with the membrane (Fig. 7E). These structures labelled with anti-GFP antibodies were found in all types of leaf cells, including mesophyll and vascular cells. The novel structures were also found in the root cells (Fig. 7F, G). The characteristics of these structures in the leaves and roots looked similar to those of PB in the endosperms. Examination of several leaf and root sections of WT, 35S:GFP, and 35S:Pro-GFP plants showed that these PB-like structures labelled with anti-GFP antibodies were found only in 35S:Pro-GFP plants and were not present in WT and 35S:GFP plants (data not shown).
Fig. 7.
PB-like structures containing prolamin–GFP fusion proteins in the vegetative tissues. (A–E) Immunogold labelling of prolamin–GFP with anti-GFP antibodies in the leaf cells of 35S:Pro-GFP plants as determined using a high pressure frozen/freeze substitution technique. Numerous electron-dense structures labelled with anti-GFP antibodies were visible in the leaf cells (A, arrows). These structures had different shapes and sizes (A–E), and some of them were in close proximity to the rough ER (D and E, arrowheads). (F and G) Immunogold labelling of prolamin–GFP in the root cells of 35S:Pro-GFP plants. The PB-like structures were also visible in the root cells (F and G). Cp, chloroplast; LV, lytic vacuole; CW, cell wall. Bars in A–C, E, and F = 500 nm; bar in D = 200 nm.
Association of the ER-resident molecular chaperone, BiP, with PB-like structures in the leaf and root cells
The PB-like structures in the leaf cells were often observed in close proximity to the membrane (Fig. 7D, E). To determine whether the PB-like structures were derived from the ER, an immunoelectron microscopy analysis was performed with antibodies raised against pumpkin BiP, an ER-resident molecular chaperone. The anti-BiP antibodies were localized in the peripheral region of PB-Is in WT rice endosperm (Takahashi et al., 2005). Immunogold labelling of BiP revealed the presence of gold particles within PB-Is in the endosperm of the 35S:Pro-GFP plants (Fig. 8A). In the leaf and root cells, the gold particles were detected in electron-dense structures (Fig. 8B, C), and the size and shape of these structures were highly similar to those of the PB-like structures observed in Fig. 7. Figure 8B shows the presence of electron-dense structures on the membrane, causing the area to appear swollen. Gold particles were also detected within the continuous ER membrane region (Fig. 8B). The electron-dense structures labelled with anti-BiP antibodies were not present in WT and 35S:GFP plants (data not shown). These results showed that the PB-like structures are derived from rough ER.
Fig. 8.
Association of an ER-resident molecular chaperone, BiP, with PB-I and PB-like structures of 35S:Pro-GFP plants. Immunoelectron micrographs of PB-Is in starchy endosperm cells of seeds (A) and PB-like structures in leaf cells (B) and root cells (C) of 35S:Pro-GFP plants obtained using anti-pumpkin BiP antibodies. Bar in A = 500 nm; bars in B and C = 200 nm.
2-ME increases the solubility of prolamin–GFP fusion proteins
The rice 13 kDa prolamin encoded by λRM1 contains four cysteine residues. Because little is known about which cysteine residues in rice prolamins engage in intermolecular and/or intramolecular disulphide bonds, an investigation was carried out to determine whether cysteine residues of prolamin–GFP fusion proteins could form intermolecular disulphide bonds. In the seeds of 35S:Pro-GFP plants, the addition of 4% 2-ME to the extraction buffer reduced the number of disulphide bonds, leading to prolamin–GFP solubilization (Fig. 9A, lane 3, open arrowhead). When proteins were extracted from seeds of 35S:Pro-GFP plants in the absence of 2-ME, prolamin–GFP was not solubilized (Fig. 9A, lane 6). The presence or absence of 2-ME did not affect the GFP solubilization in protein extracts from the seeds of 35S:GFP plants (Fig. 9A, lanes 2 and 5, closed arrowhead). These results suggest that the cysteine residues of prolamin–GFP formed intermolecular disulphide bonds with prolamin–GFP in the endosperm cells. To investigate whether the cysteine residues of prolamin–GFP expressed in the leaves could form intermolecular disulphide bonds with prolamin–GFP molecules, the influence of 2-ME on the solubility of prolamin–GFP fusion proteins in the leaves was examined. The homogenates from leaves were separated into two subcellular fractions: a 15 000 g pellet (P15; the PB fractions) and a 15 000 g supernatant (S15). When the proteins in the P15 fractions from 35S:Pro-GFP plants were extracted in the presence or absence of 2-ME, prolamin–GFP was solubilized under reducing conditions, whereas the prolamin–GFP was only partially solubilized under non-reducing conditions (Fig. 9B, lanes 3 and 6, open arrowhead). In contrast, GFP was not affected by the absence of 2-ME in the S15 fractions from 35S:GFP plants (Fig. 9B, lanes 2 and 4, closed arrowhead). These results indicate that the prolamin–GFP fusion proteins in the PB-like structures form the intermolecular disulphide bonds.
Fig. 9.
Influence of 2-ME on prolamin–GFP fusion protein solubilization. In the experiment shown in A, proteins from seeds of WT (lanes 1 and 4), 35S:GFP (lanes 2 and 5), and 35S:Pro-GFP (lanes 3 and 6) plants were extracted with 5% (v/v) 2-ME (lanes 1–3) or without 2-ME (lanes 4–6). In B, homogenates from leaves of WT (lanes 1 and 4), 35S:GFP (lanes 2 and 5), and 35S:Pro-GFP (lanes 3 and 6) plants were subjected to centrifugation to obtain the 15 000 g pellet (P15) and the 15 000 g supernatant (S15). Proteins in each fraction were extracted with 5% (v/v) 2-ME (lanes 1–3) or without 2-ME (lanes 4–6). The proteins were separated by SDS–PAGE and immunoblotted with anti-GFP antibodies. The upper bands (open arrowheads) and lower bands (filled arrowheads) correspond to prolamin–GFP and GFP, respectively.
Discussion
The aim of this study was to determine whether accumulation of prolamin in the ER is specific to endosperm tissue. The principal findings of this study are as follows: (i) the prolamin–GFP fusion proteins accumulated not only in the storage organs but also in the vegetative organs of transgenic rice plants; (ii) the protein aggregates containing the prolamin–GFP were observed in the leaf and the root cells; (iii) some of these structures were surrounded by the membrane; (iv) the ER-resident molecular chaperone BiP proteins accumulated in these structures; and (v) when the leaves were homogenized without 2-ME, the prolamin–GFP fusion proteins were only partially solubilized. The findings indicate that the seed-specific factors are not essential for the aggregation of prolamin–GFP fusion proteins, suggesting that the assembly of prolamin molecules and interaction of prolamin–GFP with BiP results in the formation of PB.
Prolamin–GFP fusion proteins specifically localize in PB-Is in endosperm cells
In rice endosperm cells, prolamins are deposited within the ER and then form PB-Is (Tanaka et al., 1980). In the endosperm cells of 35S:Pro-GFP plants in the present study, the prolamin–GFP fusion proteins localized in PB-Is (Fig. 4G–I). Kawagoe et al. (2005) showed that GFP fused to the α-globulin signal peptide was partitioned primarily into the lumen, rather than into PB-Is, in the ER. They pointed out that GFP itself does not have structural characteristics that promote protein integration into PB-Is. These results indicate that the coding sequence of prolamin contains the determinants required for protein localization in the PB-Is in rice endosperm. Hamada et al. (2003) reported that the prolamin RNA transport pathway to the PB-ER requires two partially redundant cis-elements, one located in the coding sequence and a second residing in the 3′-UTR. The 35S:Pro-GFP construct used in this study contains the coding sequence of prolamin, but lacks the 3′-UTR (Fig. 1). Further studies are needed to determine whether the prolamin–GFP mRNA is targeted to the PB-ER in starchy endosperm cells.
Accumulation of prolamin–GFP fusion proteins in leaves and roots
The immunoblot analysis revealed that the prolamin–GFP fusion proteins accumulated not only in the seeds but also in the leaves and roots of 35S:Pro-GFP plants (Fig. 3). The subcellular fractionation in leaves and roots revealed that prolamin–GFP fusion proteins were present in the 15 000 g pellet (the PB fraction) and the 100 000 g pellet (the microsomal fraction) (Fig. 5). These results suggest that the prolamin–GFP was aggregated in the leaves and roots. On the other hand, the processed forms of prolamin–GFP were also detected in the leaves and roots (Fig. 3). Moreover, a portion of the prolamin–GFP expressed in leaves and roots was detected in the 100 000 g supernatant (mixed cytosolic and vacuolar soluble proteins fraction) (Fig. 5). The endogenous ER residents are transported constitutively to the vacuoles by bypassing the Golgi complex, and are then degraded (Tamura et al., 2004). It is possible that, in these ways, some prolamin–GFP fusion proteins are transported to the vacuoles in the leaves and roots. Tamura et al. (2003) succeeded in stabilizing GFP fluorescence within the acidic vacuoles of Arabidopsis plants by incubating them in darkness. In the 35S:Pro-GFP plants grown in darkness, however, the fluorescence of prolamin–GFP was not detected in the vacuoles (data not shown).
Prolamin–GFP fusion proteins form the PB-like structures in leaves and roots
The fluorescence of prolamin–GFP showed a bright punctate pattern in both the leaf and the root cells of the 35S:Pro-GFP plants, and did not show the network pattern of the ER (Fig. 6C, F). Immunoelectron microscopy analysis showed that the prolamin–GFP accumulates in the electron-dense structures surrounded by the membrane in the aleurone layer (Fig. 4) and the leaves and roots (Fig. 7). The ER-resident protein BiP was observed in the PB-like structures (Fig. 8B, C). These results suggest that the PB-like structures develop inside the rough ER. The ER-targeted GFP fusion protein, which contains an N-terminal signal peptide and the C-terminal amino acid HDEL, has been shown to form a characteristic reticulate network in transgenic Arabidopsis plants (Haseloff et al., 1997). These results show that the addition of a prolamin molecule is responsible for the formation of PB-like structures.
Zeins, the storage proteins of maize seeds, belong to the prolamin class of storage proteins and are deposited in ER-derived PBs (Larkins and Hurkman, 1978). Zeins consist of several types of polypeptides, the α-, β-, γ-, and δ-zeins, which are structurally distinct (Thompson and Larkins, 1989). The γ-zein, β-zein, and δ-zein have been expressed individually in the leaves of transgenic Arabidopsis (γ-zein) and tobacco (β-zein and δ-zein) plants by using the CaMV 35S promoter, and, in all cases, the proteins were retained in the ER-derived PBs (Geli et al., 1994; Bagga et al., 1995, 1997). Furthermore, the N-terminal domains of γ-zein can confer the ability to form ER-derived PBs when fused to the bean vacuolar storage phaseolin in the chimeric protein zeolin (Mainieri et al., 2004). The results agree with the experiments on PB formation of the γ-zein, β-zein, and δ-zein in the leaves of transgenic plants. Meanwhile, the α-zein protein does not accumulate to measurable levels in transgenic plants (Coleman et al., 1996). When co-expressed with β- and γ-zein, the stability of α-zein increased (Coleman et al., 1996, 2004). These results suggest that not only the hydrophobic nature of the prolamins, which is a common characteristic of prolamins, but also additional factors are involved in the ER-derived PB formation.
Retention mechanism of prolamin–GFP fusion proteins in the ER
Although the prolamins of rice and maize are deposited inside ER-derived PBs, they lack the C-terminal ER retention signal KDEL/HDEL. Other than the fact that both are rich in proline residues and overall hydrophobicity, there is a low degree of similarity between the rice prolamins and zeins. The expression of γ-zein in transgenic Arabidopsis and Xenopus oocytes and γ-gliadin in Xenopus oocytes has been employed to determine whether other retention signals exist (Altschuler et al., 1993; Geli et al., 1994; Torrent et al., 1994). These studies indicated that an N-terminal region containing a tandem repeat of PPPVHL (γ-zein) and a tandem repeat of PQQPFPQ (γ-gliadin) is responsible for the localization of these prolamins within the ER. However, rice prolamins lack such tandem repeats.
Prolamin mRNA is transported to the PB-ER in the endosperm cells (Crofts et al., 2004). Hamada et al. (2003) demonstrated that prolamin–GFP hybrid RNAs which lack the 3′-UTR are partially targeted to the PB-ER in the endosperm cells. The prolamin–GFP mRNA expressed in leaf and root cells may be segregated on the ER membrane in a similar manner. There is a possibility that the targeting of prolamin–GFP mRNA is responsible for concentration of prolamin–GFP proteins in the PB-like structures.
The role of disulphide bonds in the sorting of storage proteins in rice endosperm cells has been investigated. Glutelins are separated into two groups according to the number of cysteine residues: a high molecular weight and low molecular weight type (Sugimoto et al., 1986). In endosperm of the esp2 rice mutant, which lacks the protein disulphide isomerase, glutelin precursors were deposited with prolamin polypeptides in the ER (Takemoto et al., 2002). α-Globulin is a monomeric storage protein and forms intramolecular disulphide bonds (Kawagoe et al., 2005). The disulphide bonds formed at the dicysteine residues in the CCxQL motif of α-globulin play a critical role in protein sorting in rice endosperm (Kawagoe et al., 2005). These results suggest that it is important to form the appropriate disulphide bonds for the accumulation of each rice storage protein in the appropriate location. In this study, it was shown that the prolamin–GFP fusion proteins expressed in the seeds and leaves required 2-ME to be completely solubilized (Fig. 9). This suggests that prolamin–GFP fusion proteins form the intermolecular and/or intramolecular disulphide bonds in PB-like structures. Mitsukawa et al. (1999) indicated that prolamins are polymerized in PB-Is by intermolecular disulphide bonds. Kawagoe et al. (2005) suggested that Cys135 engages in the polymerization of the epitope-tagged prolamin (λRM1) in vitro. It is possible that rice prolamins form intermolecular bonds and are polymerized in the ER.
BiP is a major chaperone of the ER (Vitale and Denecke, 1999). BiP transiently binds many newly synthesized secretory proteins and probably prevents unspecific aggregation by interacting with regions rich in hydrophobic amino acids. However, the interaction of BiP and prolamins is peculiar. BiP interacts with rice prolamins during their co-translational translocation into the ER lumen and has been detected at the periphery of rice PBs (Li et al., 1993; Muench et al., 1997). In 35S:Pro-GFP leaves and roots in the present study, BiP was localized in PB-like structures (Fig. 8B, C). The results support the hypothesis that BiP retains prolamins in the ER lumen by facilitating their folding and assembly into PB-Is in rice endosperm (Li et al., 1993; Muench et al., 1997).
In conclusion, the results show that the addition of a single prolamin polypeptide to GFP causes the formation of PB-like structures. The findings bear out the importance of the assembly of prolamin molecules and the interaction of prolamin with BiP in the formation of ER-derived PBs.
Acknowledgments
We thank Professor T Shiina (Kyoto Prefectural University, Kyoto) for assistance with the confocal laser scanning microscope; Dr Y Niwa (Shizuoka University, Shizuoka) for providing the 35S-GFP(S65T) plasmid; and Professor Ikuko Hara-Nishimura (Kyoto University, Kyoto) for providing the rabbit polyclonal antibodies against pumpkin BiP. This work was supported in part by Grants-in-Aid for Scientific Research to KT (#12138207) and TM (#18658140) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and in part by a grant for ‘Breeding and Integrated Research Toward Enhancing Consumption of Domestic Farm Products in the Food Service Industry: Rice’ to TM from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
References
- Altschuler Y, Rosenberg N, Harel R, Galili G. The N- and C-terminal regions regulate the transport of wheat γ-gliadin through the endoplasmic reticulum in Xenopus oocytes. The Plant Cell. 1993;5:443–450. doi: 10.1105/tpc.5.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcalis E, Marcel S, Altmann F, Kolarich D, Drakakaki G, Fischer R, Christou P, Stoger E. Unexpected deposition patterns of recombinant proteins in post-endoplasmic reticulum compartments of wheat endosperm. Plant Physiology. 2004;136:3457–3466. doi: 10.1104/pp.104.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Adams H, Kemp JD, Sengupta-Gopalan C. Accumulation of 15-kilodalton zein in novel protein bodies in transgenic tobacco. Plant Physiology. 1995;107:13–23. doi: 10.1104/pp.107.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Adams HP, Rodriguez FD, Kemp JD, Sengupta-Gopalan C. Coexpression of the maize δ-zein and β-zein genes results in stable accumulation of δ-zein in endoplasmic reticulum-derived protein bodies formed by β-zein. The Plant Cell. 1997;9:1683–1696. doi: 10.1105/tpc.9.9.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. The Plant Cell. 2000;12:2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel DB, Juliano BO. Formation of protein bodies in the starchy endosperm of rice (Oryza sativa L.): a re-investigation. Annals of Botany. 1980;45:503–509. [Google Scholar]
- Chikwamba RK, Scott MP, Mejia LB, Mason HS, Wang K. Localization of a bacterial protein in starch granules of transgenic maize kernels. Proceedings of the National Academy of Sciences, USA. 2003;100:11127–11132. doi: 10.1073/pnas.1836901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Current Biology. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Herman EM. Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiology. 2000;123:1227–1234. doi: 10.1104/pp.123.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Herman EM, Takasaki K, Larkins BA. The maize γ-zein sequesters α-zein and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. The Plant Cell. 1996;8:2335–2345. doi: 10.1105/tpc.8.12.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Yoho PR, Escobar S, Ogawa M. The accumulation of α-zein in transgenic tobacco endosperm is stabilized by co-expression of β-zein. Plant and Cell Physiology. 2004;45:864–871. doi: 10.1093/pcp/pch104. [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Ogawa M, Kumamaru T, Satoh H. Targeting of proteins to endoplasmic reticulum-derived compartments in plants. The importance of RNA localization. Plant Physiology. 2004;136:3414–3419. doi: 10.1104/pp.104.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Marcel S, Arcalis E, Altmann F, Gonzalez-Melendi P, Fischer R, Christou P, Stoger E. The intracellular fate of a recombinant protein is tissue dependent. Plant Physiology. 2006;141:578–586. doi: 10.1104/pp.106.076661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, Foresti O, Hernandez Felipe D, Neuhaus JM, Vitale A. The C-terminal tetrapeptide of phaseolin is sufficient to target green fluorescent protein to the vacuole. Journal of Plant Physiology. 2001;158:499–503. [Google Scholar]
- Furukawa S, Mizuma T, Kiyokawa Y, Masumura T, Tanaka K, Wakai Y. Distribution of storage proteins in low-glutelin rice seed determined using a fluorescent antibody. Journal of Bioscience and Bioengineering. 2003;5:467–473. doi: 10.1016/S1389-1723(03)70133-9. [DOI] [PubMed] [Google Scholar]
- Geli MI, Torrent M, Ludevid D. Two structural domains mediate two sequential events in γ-zein targeting: protein endoplasmic reticulum retention and protein body formation. The Plant Cell. 1994;6:1911–1922. doi: 10.1105/tpc.6.12.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW. Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. The Plant Cell. 2003;15:2265–2272. doi: 10.1105/tpc.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R, Shimada T, Nishimura M. Diversity and formation of endoplasmic reticulum-derived compartments in plants. Are these compartments specific to plant cells? Plant Physiology. 2004;136:3435–3439. doi: 10.1104/pp.104.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Simada T, Hatano K, Takeuchi Y, Nishimura M. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. The Plant Cell. 1998;10:825–836. doi: 10.1105/tpc.10.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I. A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant and Cell Physiology. 2001;42:894–899. doi: 10.1093/pcp/pce144. [DOI] [PubMed] [Google Scholar]
- Herman EM, Larkins BA. Protein storage bodies and vacuoles. The Plant Cell. 1999;11:601–614. doi: 10.1105/tpc.11.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Kobayashi H, Yamazoe Y, Mikami B, Morita Y. Purification and complete amino acid sequence of major prolamin of rice endosperm. Journal of Cereal Science. 1991;14:1–14. [Google Scholar]
- Kawagoe Y, Suzuki K, Tasaki M, Yasuda H, Akagi K, Katoh E, Nishizawa NK, Ogawa M, Takaiwa F. The critical role of disulfide bond formation in protein sorting in the endosperm of rice. The Plant Cell. 2005;17:1141–1153. doi: 10.1105/tpc.105.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta. 1986;169:471–480. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- Larkins BA, Hurkman WJ. Synthesis and deposition of zein in protein bodies of maize endosperm. Plant Physiology. 1978;62:256–263. doi: 10.1104/pp.62.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, Okita TW. Rice prolamine protein body biogenesis: a BiP-mediated process. Science. 1993;262:1054–1056. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- Mainieri D, Rossi M, Archinti M, Bellucci M, De Marchis F, Vavassori S, Pompa A, Arcioni S, Vitale A. Zeolin. A new recombinant storage protein constructed using maize γ-zein and bean phaseolin. Plant Physiology. 2004;136:3447–3456. doi: 10.1104/pp.104.046409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nshimura I. A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER-retention signal in Arabidopsis. The Plant Journal. 2003;33:493–502. doi: 10.1046/j.1365-313x.2003.01636.x. [DOI] [PubMed] [Google Scholar]
- Mitsukawa N, Konishi R, Kidzu K, Ohtsuki K, Masumura T, Tanaka K. Amino acid sequencing and cDNA cloning of rice seed storage proteins, the 13kDa prolamins, extracted from type I protein bodies. Plant Biotechnology. 1999;16:103–113. [Google Scholar]
- Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant and Cell Physiology. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, Kasai Z, Tanaka K. Purification of protein body-I of rice seed and its polypeptide composition. Plant and Cell Physiology. 1987;28:1517–1527. [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K. Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant and Cell Physiology. 1990;31:805–813. [Google Scholar]
- Oparka KJ, Harris N. Rice protein-body formation: all types are initiated by dilation of the endoplasmic reticulum. Planta. 1982;154:184–188. doi: 10.1007/BF00387914. [DOI] [PubMed] [Google Scholar]
- Petruccelli S, Otegui MS, Lareu F, et al. A KDEL-tagged monoclonal antibody is efficiently retained in the endoplasmic reticulum in leaves, but is both partially secreted and sorted to protein storage vacuoles in seeds. Plant Biotechnology Journal. 2006;4:511–527. doi: 10.1111/j.1467-7652.2006.00200.x. [DOI] [PubMed] [Google Scholar]
- Saito Y, Nakatsuka N, Shigemitsu T, Tanaka K, Morita S, Satoh S, Masumura T. Thin frozen film method for visualization of storage proteins in mature rice grains. Bioscience, Biotechnology, and Biochemistry. 2008;72:2779–2781. doi: 10.1271/bbb.80337. [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Okumura T, Kaminaka H, Sumi K, Tanaka K. Structure and differential response to abscisic acid of two promoters for the cytosolic copper/zinc-superoxide dismutase genes, SodCc1 and SodCc2, in rice protoplasts. FEBS Letters. 1995;358:62–66. doi: 10.1016/0014-5793(94)01396-i. [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson D, Kalousek F, Gietl C. A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta. 1998;206:466–475. doi: 10.1007/s004250050423. [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Tanaka K, Kasai Z. Molecular species in the protein body type II (PB-II) of developing rice endosperm. Agricultural and Biological Chemistry. 1986;50:3031–3035. [Google Scholar]
- Takahashi H, Morita S, Masumura T, Tanaka K. Preparation of endosperm protoplasts from a dwarf rice variety and transient expression of green-fluorescent protein. Plant Biotechnology. 2004;21:109–112. [Google Scholar]
- Takahashi H, Saito Y, Kitagawa T, Morita S, Masumura T, Tanaka K. A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant and Cell Physiology. 2005;46:245–249. doi: 10.1093/pcp/pci019. [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiology. 2002;128:1212–1222. doi: 10.1104/pp.010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi SI, Watanabe M, Nishimura M, Hara-Nishimura I. Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. The Plant Journal. 2003;35:545–555. doi: 10.1046/j.1365-313x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Yamada K, Shimada T, Hara-Nishimura I. Endoplasmic reticulum-resident proteins are constitutively transported to vacuoles for degradation. The Plant Journal. 2004;39:393–402. doi: 10.1111/j.1365-313X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. Isolation and characterization of two types of protein bodies in the rice endosperm. Agricultural and Biological Chemistry. 1980;44:1633–1639. [Google Scholar]
- Thompson GA, Larkins BA. Structural elements regulating zein gene expression. Bioessays. 1989;10:108–113. doi: 10.1002/bies.950100404. [DOI] [PubMed] [Google Scholar]
- Torrent M, Geli MI, Ruiz-Avila L, Canals JM, Puigdomenech P, Ludevid D. Role of structural domains for maize γ-zein retention in Xenopus oocytes. Planta. 1994;192:512–518. doi: 10.1007/BF00203589. [DOI] [PubMed] [Google Scholar]
- Toyooka K, Okamoto T, Minamikawa T. Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. The Journal of Cell Biology. 2000;148:453–464. doi: 10.1083/jcb.148.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Denecke J. The endoplasmic reticulum—gateway of the secretory pathway. The Plant Cell. 1999;11:615–628. doi: 10.1105/tpc.11.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KE, Prior F, Sardana R, Altosaar I, Dudani AK, Ganz PR, Tackaberry ES. Sorting of glycoprotein B from human cytomegalovirus to protein storage vesicles in seeds of transgenic tobacco. Transgenic Research. 2001;10:177–181. doi: 10.1023/a:1008912305913. [DOI] [PubMed] [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiology. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K. The site of synthesis and accumulation of rice storage proteins. Plant and Cell Physiology. 1986;27:135–145. [Google Scholar]