Abstract
The polarity of actin is a central determinant of intracellular transport in plant cells. To visualize actin polarity in living plant cells, the tobacco homologue of the actin-related protein 3 (ARP3) was cloned and a fusion with the red fluorescent protein (RFP) was generated. Upon transient expression of these fusions in the tobacco cell line BY-2 (Nicotiana tabacum L. cv. Bright Yellow 2), punctate structures were observed near the nuclear envelope and in the cortical plasma. These dots could be shown to decorate actin filaments by expressing RFP–ARP3 in a marker line, where actin was tagged by GFP (green fluorescent protein)–FABD (fimbrin actin-binding domain 2). When actin filaments were disrupted by latrunculin B or by prolonged cold treatment, and subsequently allowed to recover, the actin filaments reformed from the RFP–ARP3 structures, that therefore represented actin nucleation sites. The intracellular distribution of these sites was followed during the formation of pluricellular files, and it was observed that the density of RFP–ARP3 increased in the apex of the polarized, terminal cells of a file, whereas it was equally distributed in the central cells of a file. These findings are interpreted in terms of position-dependent differences of actin organization.
Keywords: Actin, actin-related protein 3 (ARP3), tobacco BY-2
Introduction
Plant cells can increase in size by up to four orders of magnitude by uptake of water into their vacuoles (Cosgrove, 1987). This cell expansion represents the central adaptive response of the sessile plants to environmental challenges and is therefore highly regulated by stimuli, such as light or gravity, and internal factors including developmental signals and plant hormones. During the last decade, actin filaments have emerged as central elements of vacuole-driven cell growth (for recent reviews, see Bannigan and Baskin, 2005; Smith and Oppenheimer, 2005). The organization and cellular function of actin filaments has therefore attracted considerable attention in the context of growth regulation.
Similar to the situation in animal cells, actin organization in plants is controlled by several, partially directly actin-associated, protein complexes including the Rho-related GTPases of plants (ROPs), the WAVE (for Wiskott–Aldrich syndrome protein family verproline homologous) complex, and the actin-related protein (ARP) 2/3 complex. These regulators modulate the actin cytoskeleton through an elaborate signalling network (for a review, see Xu and Scheres, 2005).
The ARP2/3 complex has been conserved throughout eukaryotic evolution as a modulator of the actin cytoskeleton (Vartiainen and Machesky, 2004). It is essential for amoeboid locomotion and subcellular motility of organelles. When activated, it enhances the nucleation and polymerization of filamentous actin such that the ARP2/3 complex caps the pointed end, and the actin filament grows in the direction of the barbed end (Mullins et al., 1998; Svitkina and Borisy, 1999; Blanchoin et al., 2000). Upon binding to existing actin filaments, the ARP2/3 complex can initiate branches and generate dynamic actin arrays. ARP2 and ARP3 are the largest of seven subunits comprising the multiprotein ARP2/3 complex and share significant structural and sequence similarity with actin (Robinson et al., 2001). In animal cells, this complex has been shown to participate in protrusion-mediated motility. Plant cells with their rigid cellulosic walls are not endowed with such motility; here ARP2/3 is essential for developmental changes of cell polarity and shape (for a review, see Mathur, 2005).
Homologues of all seven subunits of the ARP2/3 complex have recently been identified in plants on the basis of homology searches and loss-of-function mutants. Mutations in genes that encode subunits of the ARP2/3 complex lead to increased bundling of filamentous actin and the formation of aberrant actin patches, culminating in misdirected expansion of various cell types including trichomes, pavement cells, hypocotyl epidermis, and root hairs (Le et al., 2003; Li et al., 2003; Mathur et al., 2003a,b; El-Assal et al., 2004; Saedler et al., 2004). The randomly mis-shaped trichomes of this mutant class (classified as distorted mutants) could be phenocopied in wild-type leaves by pharmacological manipulation of actin (Mathur et al., 1999; Szymanski et al., 1999). Thus, these mutant phenotypes reveal an important role for actin microfilaments and the ARP2/3 complex in the control of polar cell expansion. In cells with pronounced tip growth, such as rhizoids of germinated fucoid zygotes (Hable and Kropf, 2005), or extending root hairs (Van Gestel et al., 2003), elements of this complex could be immunolocalized to the leading edge of these cells, i.e. to the site where actin filaments elongate. Independent observations on apical extension and actin dynamics in tip-growing pollen tubes speak for a major role for actin polymerization in polarized cell expansion (Vidali and Hepler, 2001; Vidali et al., 2001). When actin filaments were transiently eliminated in BY-2 cells (Nicotiana tabacum L. cv. Bright Yellow 2) by a cold treatment, the sites from which new actin filaments regenerated upon rewarming could be immunolabelled using heterologous antibodies against mammalian ARP3 (Fišerová et al., 2006). These data indicated that ARP3 is a component of actin nucleation sites. However, as the different ARP proteins share a high degree of sequence similarity, cross-reactivities of the antibodies are very likely, since these antibodies were raised against heterologous proteins. It is therefore even possible that the signals observed in immunofluorescence originate not from ARP3 but from immunologically related proteins in the complex (or, worse, near the complex). A strategy to reach a clearer outcome would require ARP3 to be tagged and its localization in vivo to be followed.
However, so far, the components of the ARP2/3 complex could not been visualized in living plant cells. In a previous work, it was demonstrated that the organization of actin filaments is important for auxin-dependent, patterned cell division in tobacco BY-2 cells (Maisch and Nick, 2007), implicating that, amongst other factors, it is the polarity of microfilaments that is responsible for polar patterning. By simultaneous visualization of actin and ARP3 in living cells, it could be shown that ARP3 decorated actin filaments. When actin filaments were transiently eliminated by cold or latrunculin B (LatB), and then allowed to recover, ARP3 marked the sites from which the new filaments emanated. The density of ARP3 was increased in the apex of terminal cells, but was equally distributed in the central cells of a file. The difference between these two cell types in terms of actin organization and asymmetry are expected to produce corresponding differences in the directionality of auxin transport and thus the generation of a division pattern. It can be demonstrated that red fluorescent protein (RFP)–ARP3 can be used as reliable marker for actin nucleation sites and thus as an indicator for actin directionality.
Materials and methods
Tobacco cell cultures
The tobacco cell line BY-2 (Nagata et al. 1992) was cultivated in liquid medium containing 4.3 g l−1 Murashige and Skoog salts (Duchefa, Haarlem, The Netherlands), 30 g l−1 sucrose, 200 mg l−1 KH2PO4, 100 mg l−1 inositol, 1 mg l−1 thiamine, and 0.2 mg l−1 2,4-dichlorophenoxyacetic acid (2,4-D), pH 5.8. Cells were subcultured weekly, inoculating 1.5–2 ml of stationary cells into 30 ml of fresh medium in 100 ml Erlenmeyer flasks. The cell suspensions were incubated at 25 °C in the dark on an orbital shaker (KS250 basic, IKA Labortechnik, Staufen, Germany) at 150 rpm. Stock BY-2 calli were maintained on medium solidified with 0.8% (w/v) agar and subcultured monthly. The transgenic BY-2 cells and calli were maintained on the same medium supplemented with 25 mg l−1 kanamycin. The BY-2 PIN1-GFP (green fluorescent protein) tobacco cell line was kindly provided by Dr Jan Petrášek (Institute of Experimental Botany, Academy of Sciences of the Czech Republic, Czech Republic), who stably transformed BY-2 cells with a AtPIN1::PIN1:GFP fusion construct (Benková et al., 2003). This cell line is described in more detail in Petrášek and Zažímalová (2006).
Constructs
The region encoding the second fimbrin actin-binding domain (FABD2; amino acids 325–687) was amplified by PCR from pGFPm3abd2 (Voigt et al., 2005) using the following primers: 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TTG GAT CCT CTT G-3′ and 5′-GGGG AC CAC TTT GTA CAA GAA AGC TGG GTT CTA TTC GAT GGA TGC TTC CT-3′. The underlined sites represent the attB sites that have to be integrated into the PCR product for the Gateway® recombination reactions (Hartley et al., 2000). For the generation of stably transformed tobacco cells, the resulting FABD2 region was inserted into the binary vector pK7WGF2 (Karimi et al., 2005) employing the Gateway® technology (Invitrogen Corporation, Paisley, UK) according to the protocol of the manufacturer. The plasmid was sequenced to confirm the accuracy of the sequence. The obtained construct, pK7WGF2-FABD2, was a C-terminal fusion of the second fimbrin actin-binding domain to GFP.
A full-length cDNA of N. tabacum NtARP3 was isolated by reverse transcription-PCR (RT-PCR) using total mRNA from tobacco BY-2 cells as a template. The primers (5′-ATG GAC CCT TCT ACC TCT CG-3′ and 5′-TTGA ATA CAT TCC CTT GAA TAC AGG-3′) were designed based on the alignments of known plant ARP3 sequences. To allow construction of a C-terminal fusion with GFP, the reverse primer was prolonged with an extra T, and a mutation (C→G) was introduced, removing a stop codon (both underlined). The ARP3 PCR fragment was cloned in the pDrive Cloning Vector (Qiagen, Hagen, Germany) and subsequently cleaved with HindIII and BamHI, and transferred to the psmGFP vector (Davis and Vierstra, 1998).
For transient expression of ARP3 in tobacco cells, the coding sequence of N. tabacum NtARP3 was amplified by PCR from psmGFP-ARP3, using the primers 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TAT ATG GAC CCT TCT ACC TCT CG-3′ and 5′-GGGG AC CAC TTT GTA CAA GAA AGC TGG GTT TGA ATA CAT TCC CTT GAA TAC AGG-3′. The resulting ARP3 region was inserted into the transient expression vector p2RGW7 (Dr RY Tsien, University of California, San Diego, CA, USA; Campell et al., 2002) as described above, resulting in p2RGW7-ARP3, a C-terminal fusion of ARP3 to RFP. To confirm the accuracy of the sequence, the plasmid was sequenced. The verified sequence has been deposited at the public database of the European Molecular Biology Laboratory under the accession number AM711117.
Transformation and establishment of a tobacco BY-2 cell line stably expressing the GFP–FABD2 fusion protein
The binary vector construct pK7WGF2-FABD2 was introduced into Agrobacterium tumefaciens (strain LBA4404) by heat shock. A 4 ml−1 aliquot of BY-2 cells that had been cultivated for 3 d was co-incubated for a further 3 d with 100 μl of an overnight culture of the transformed A. tumefaciens at 27 °C as described by An (1985). After incubation, the cells were washed three times in liquid medium containing 100 μg ml−1 cefotaxim and were then plated onto solid medium containing 100 μg ml−1 kanamycin and 100 μg ml−1 cefotaxim. Kanamycin-resistant calli, which appeared after 28 d of incubation in darkness at 25 °C, were transferred onto new plates and cultured separately until they reached ∼1 cm in diameter. Cell suspension cultures established from these calli were maintained as described above, with addition of 25 mg l−1 kanamycin to the cultivation medium. After 6 weeks, a cell line suitable for observing microfilaments was selected by examination of GFP fluorescence by fluorescence microscopy to yield the cell line BY-2 GFP-FABD2.
In silico analysis
The confirmed sequence of the putative tobacco ARP3 (NtARP3) was used for a similarity search on the amino acid level in the Swiss-Prot database (http://www.expasy.org; for Swiss-Prot accession numbers, see Supplementary Table S1 available at JXB online) using a BlastP routine with standard settings. The best hits (E-value ≤e−122) of this search were ARP3 homologues from other organisms. These sequences were aligned using the KALIGN software (http://msa.cgb.ki.se/cgi-bin/msa.cgi; Lassmann and Sonnhammer, 2005) with standard settings. This alignment was imported in FASTA format into the ClustalW software (http://www.ebi.ac.uk/clustalw/index.html) and used to produce a cladogram based on the Neighbor–Joining method (Saitou and Nei, 1987) that could be viewed using the TreeView software (http://taxonomy.zoology.gla.ac.uk). The domains in the sequence were defined using the ProDom database (http://prodom.prabi.fr).
Rhodamine–phalloidin staining
For simultaneous observations of GFP–FABD2 and rhodamine–phalloidin, the staining method of Kakimoto and Shibaoka (1987) was modified according to Olyslaegers and Verbelen (1998). Suspended cells were fixed for 10 min in 1.8% (w/v) paraformaldehyde in standard buffer [0.1 M PIPES (pH 7.0), supplemented with 5 mM MgCl2 and 10 mM EGTA]. After a subsequent 10 min fixation in standard buffer containing 1% (v/v) glycerol, cells were rinsed twice for 10 min with standard buffer. Then, 0.5 ml of the resuspended cells were incubated for 35 min with 0.5 ml of 0.66 μM tetramethylrhodamine isothiocyanate (TRITC)–phalloidin (Sigma-Aldrich) freshly prepared from a 6.6 μM stock solution in 96% (w/v) ethanol by dilution [1:10 (v/v)) with phosphate-buffered saline (PBS; 0.15 M NaCl, 2.7 mM KCl, 1.2 mM KH2PO4, and 6.5 mM Na2HPO4 (pH 7.2)]. Cells were then washed three times for 10 min in PBS and observed immediately.
Biolistic, transient expression of RFP–ARP3
For biolistic transformation, gold particles (1.5–3.0 μm; Sigma-Aldrich, Steinheim, Germany) were coated with the RFP–ARP3 vector construct (p2GWR7-ARP3) slightly modified from a manual of Bio-Rad (PDS-1000/He Particle Delivery System Manual). A 1 μg aliquot of DNA was used for transfection. DNA-coated gold particles were placed on macrocarriers (Bio-Rad, Hercules, CA, USA). A cell suspension of 3-day-old transgenic BY-2 GFP-FABD2 cells was transferred into PetriSlides™ (Millipore, Billerica, MA, USA) containing solid medium. The transgenic BY-2 cells were placed in a particle gun, that was constructed according to Finer et al. (1992), and bombarded by three shots at a pressure of 1.5 bar in the vacuum chamber at –0.8 bar. Following bombardment, cells were kept in the dark at 25 °C for 18–60 h before observation.
Latrunculin B treatment, cold treatment, and recovery experiments
Following particle bombardment with the RFP–ARP3 construct, BY-2 GFP-FABD2 cells were diluted in liquid medium containing 500 nM LatB (Sigma-Aldrich) that was added from a stock solution of 2.5 mM LatB in 96% (v/v) ethanol. To eliminate actin filaments for subsequent recovery experiments, the cell suspension was kept in the dark for 10 h at 25 °C. Thereafter, the cells were washed four times in fresh medium to remove LatB and observed immediately over a period of 1 h.
For the cold treatment, the cells were kept in the dark for 18 h at 25 °C after transient transformation in order for the transgene to be adequately expressed. Afterwards, they were stored in a refrigerator at 2 °C for 10 h. Subsequently, they were re-suspended in liquid medium at the control temperature of 25 °C and observed immediately over a period of 1 h.
Quantification of ARP3 expression patterns
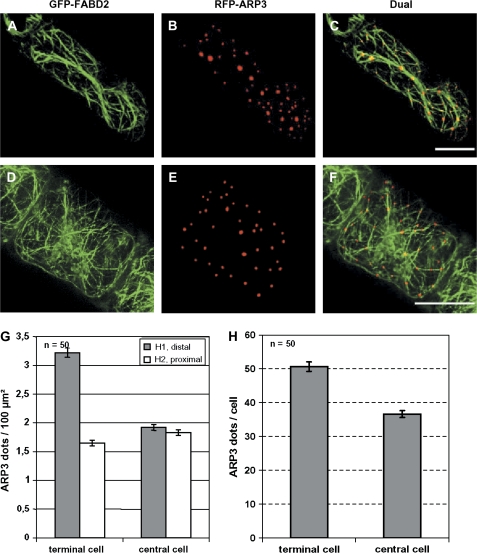
For the quantification of the ARP3 distribution within a given cell file, projections of serial sections of transiently transformed tobacco cells were constructed using the maximum projection function of the AxioVision software (Zeiss, Jena, Germany). The cells were categorized into two classes (terminal cells of a file or cells of the central region of the file). Each cell was divided into two halves and the number of ARP3 dots over the cross-section in the central plane of the cell (subtracted by the cross-sectional area occupied by the nucleus) was determined using the outline function of the AxioVision software. For each value, 50 terminal cells and 50 central cells were used. The results were tested for significance by a t-test at the 95% confidence level.
Microscopy and image analysis
BY-2 cells were examined under an AxioImager Z.1 microscope (Zeiss) equipped with an ApoTome microscope slider for optical sectioning, and a cooled digital CCD camera (AxioCam MRm). For the co-localization of microfilaments and ARP3, GFP and RFP fluorescence were recorded through the filter sets 38 HE (excitation at 470 nm, beamsplitter at 495 nm, and emission at 525 nm) and 43 HE (excitation at 550 nm, beamsplitter at 570 nm, and emission at 605 nm), respectively (both from Zeiss) using either a 63× plan apochromat oil-immersion objective or a 40× objective. Stacks of optical sections were acquired at different step sizes between 0.5 μm and 0.8 μm. Images were processed and analysed using the AxioVision software AxioVision software (Rel. 4.5; Zeiss) as described above. For publication, images were processed with respect to contrast and brightness using the Photoshop software (Adobe Systems, San Jose, CA, USA).
Results
Isolation of tobacco ARP3
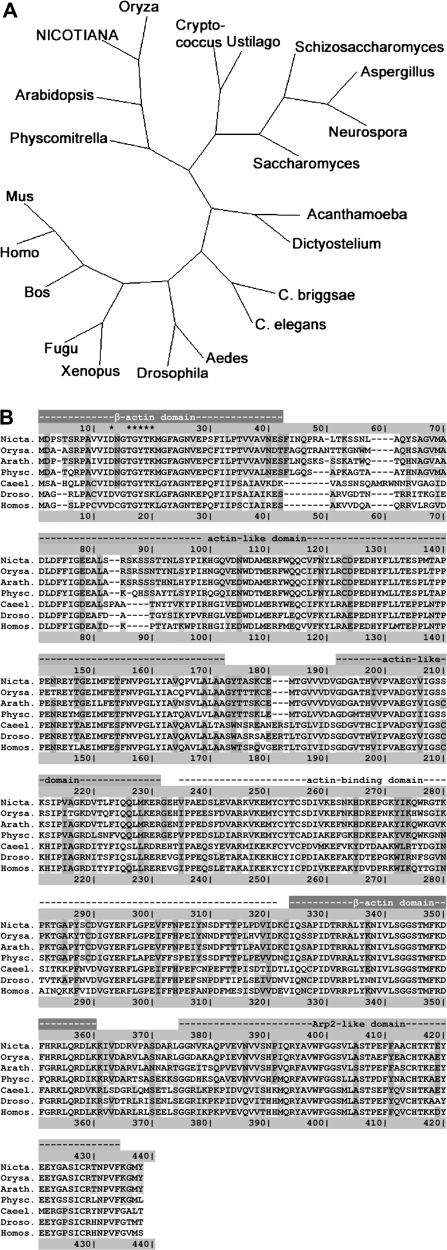
A full-length cDNA for tobacco ARP3 was isolated by RT-PCR using total mRNA from tobacco BY-2 cells as a template. The primers were designed based on alignments of known plant ARP3 sequences. The obtained sequence coded for a protein with 428 amino acid residues (EMBL accession no. AM711117) that was highly homologous to ARP3 from Arabidopsis thaliana (EMBL accession no. AY093149), Oryza sativa (EMBL accession no. AP004092, 428 amino acids), or Physcomitrella patens (EMBL accession no. AM287016, 424 amino acids). A phylogenetic tree was constructed for the most closely related homologues (identified by a BLAST search using the peptide sequence as query, see Supplementary Table S1 at JXB online for a list of the sequences utilized) based on the Neighbor–Joining method (Saitou and Nei, 1987). This tree showed that the bona fide tobacco ARP3 sequence clustered with the other plant ARP3 sequences to a separate branch of the tree (Fig. 1A). The alignment of the corresponding peptide sequences (Fig. 1B) confirmed the high degree of conservation between the tobacco sequence and known ARP3 sequences from other organisms. The similarity is not confined to a high conservation of all the domains that could be identified, but extends even to sequence details. For instance, the sequence motif DxGTGYTK (Fig. 1B, asterisks) that has been associated with nucleotide binding (Beltzner and Pollard, 2004) is conserved in the tobacco sequence. The high conservation with regard to known ARP3 sequences and the presence of the relevant domains and motifs strongly suggests that the cloned sequence represents the tobacco homologue of ARP3.
Fig. 1.
Phylogenetic analysis and alignment of the protein sequences of Nicotiana tabacum ARP3 and homologues from other organisms. (A) A phylogenetic tree of N. tabacum ARP3 and variant ARP3 from diverse organisms based on the Neighbor–Joining method (Saitou and Nei, 1987). GenBank accession numbers (http://www.ncbi.nlm.nih.gov/): Acanthamoeba castellanii, U29610; Aedes aegypti, CH477862; Arabidopsis thaliana, AF507911; Aspergillus terreus, CH476596; Bos taurus, D12816; Caenorhabditis briggsae, CAAC02000457; Caenorhabditis elegans, AC024200; Cryptococcus neoformans, AE017344; Dictyostelium discoideum, Z46418; Drosophila melanogaster, X71789; Fugu rubripes, AF034581; Homo sapiens, AF023453; Mus musculus; BC053106; Neurospora crassa, U79737; Oryza sativa, AP004092; Physcomitrella patens, AM287016; Saccharomyces cerevisiae, L37111; Schizosaccharomyces pombe, M81068; Ustilago maydis, AACP01000036; Xenopus laevis, BC047983. (B) Alignment of the deduced amino acid sequences of ARP3 from N. tabacum, O. sativa, A. thaliana, P. patens, C. elegans, D. melanogaster, and H. sapiens. The alignment was created using the ClustalW program (http://www.ebi.ac.uk/clustalw/index.html) and coloured by residue conservation according to Lassmann and Sonnhammer (2005). The lines above the alignment indicate the conserved regions of ARP3. The asterisks label the putative nucleotide-binding sequence motif (Beltzner and Pollard, 2004).
Visualization of actin filaments by stable GFP–fimbrin expression
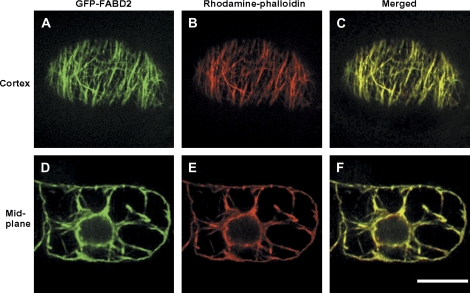
The ABD2 of the A. thaliana fimbrin 1 protein (AtFim1) was fused to the C-terminus of GFP and stably expressed in tobacco BY-2 cells to generate the BY-2 GFP-FABD2 cell line. Projections of optical sections revealed GFP fluorescence of filamentous structures near the cell cortex and along cytoplasmic strands at the mid plane. To ensure that these structures did represent actin filaments, rhodamine–phalloidin staining was performed after mild permeabilization of the cells with glycerol. The filamentous structures labelled by GFP and the rhodamine fluorescence were completely congruent (Fig. 2), demonstrating that the GFP–FABD2 fusion protein labelled actin filaments in tobacco BY-2 GFP-FABD2 cells.
Fig. 2.
Visualization of actin filaments by stably expressed GFP–FABD2 and by rhodamine–phalloidin in tobacco BY-2 cells. GFP–FABD2 fluorescence (A, D), rhodamine–phalloidin fluorescence (B, E), and merged signal (C, F) in focal sections of the cortical region (A–C) or the mid-plane (D–F) of a BY-2 GFP-FABD2 cell. Yellow colour in the merged images (C, F) indicates areas where the two markers co-localize. Bar=20μm.
Localization of actin nucleation sites upon transient expression of RFP–ARP3 in BY-2 GFP-FABD2 cells
For transient expression of ARP3, the coding sequence of full length N. tabacum NtArp3 was inserted into a transient expression vector, resulting in an N-terminal fusion of ARP3 to RFP (RFP–ARP3).
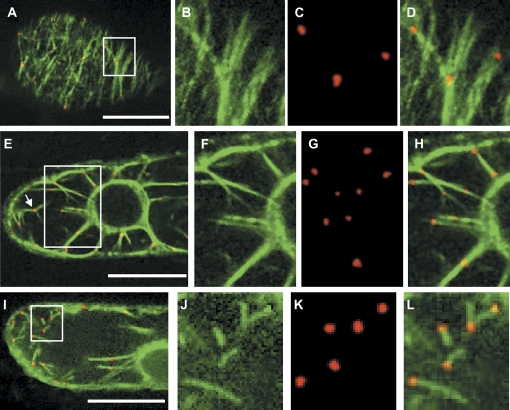
The localization of RFP-labelled ARP3 proteins was analysed upon particle bombardment with this construct into BY-2 GFP-FABD2 cells using projections of serial optical sections. The ARP3 signals appeared as distinct dots and clearly decorated all arrays of the actin cytoskeleton, namely cortical microfilaments, the perinuclear network, and the transvacuolar strands (Fig. 3). The dots were also frequently found at points of actin filament branching (for instance Fig. 3E, white arrow).
Fig. 3.
Transiently expressed RFP–ARP3 decorates actin filaments in tobacco BY-2 cells that stably express GFP–FABD2. (A–D) Co-localization of RFP–ARP3 and GFP–FABD2 in a focal section of the cortical region. (E–H) Co-localization of RFP–ARP3 and GFP–FABD2 in a focal section of the perinuclear region with cytopasmic strands. (I–L) Localization of RFP–ARP3 during the recovery of actin filaments after latrunculin B treatment (500 nM, 10 h). Details boxed in A, E, and I are shown in B, F, and J for the GFP–FABD2 fluorescence, in C, G, and K for the RFP–ARP3 fluorescence, and in D, H, and L for dual fluorescence, respectively. The arrow in E highlights, as an example, a site of actin filament branching. Bars=20 μm.
To test whether the ARP3 dots correspond to sites of actin nucleation, LatB was used as a tool specifically to eliminate actin filaments. A treatment with 500 nM LatB for 10 h removed the actin cytoskeleton completely, as far as could be judged from epifluorescence microscopy. However, when the inhibitor was washed out, new actin structures (short filaments, Y-shaped junctions) reappeared within 15 min emerging from the labelled ARP3 dots (Figs. 3I–L).
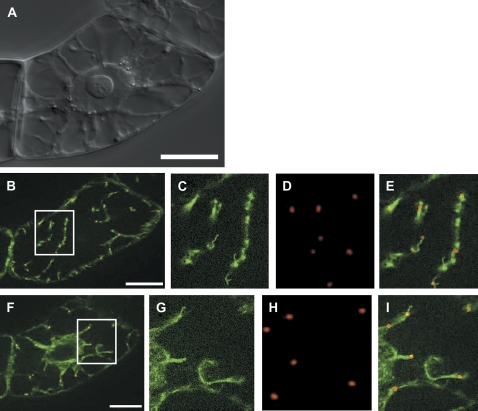
In a previous immunolocalization study, bona fide actin nucleation sites had been identified by cold-induced disintegration of actin filaments (Fišerová et al., 2006). Tests were therefore carried out to determine whether the ARP3 dots could be observed in association with actin in cells that recovered from cold treatment after cold-induced, reversible degradation of actin filaments for 10 h at 2 °C. In fact, it was possible to visualize the RFP–ARP3 signal in close association with short elongating actin rods or short dotted filaments that are formed under these conditions (Fig. 4). This was valid both for the cortical actin filaments (Fig. 4B–E) and for the perinuclear actin network (Fig. 4F–I). It should be noted that the subcellular distribution of the ARP3 dots persisted throughout the treatments and was not affected either by LatB or by cold (data not shown).
Fig. 4.
Transiently expressed RFP–ARP3 decorates sites of actin nucleation during recovery from cold-induced elimination of actin filaments in tobacco BY-2 cells that stably express GFP–FABD2. (A) Differential interference contrast of the target cell. (B–E) Co-localization of RFP–ARP3 and GFP–FABD2 in a focal section of the cell cortex. (F–I) Co-localization of RFP–ARP3 and GFP–FABD2 in a focal section of the cell centre. Details boxed in B and F are shown in C and G for the GFP–FABD2 fluorescence, in D and H for the RFP–ARP3 fluorescence, and in E and I for dual fluorescence, respectively. Bars=20 μm.
Thus, actin-nucleating sites were demonstrated by two independent agents (LatB and cold), and in both cases RFP–ARP3 was observed to localize to these sites.
ARP3 is enriched in the distal halves of terminal cells
In a previous study, it had been demonstrated that the organization of actin filaments is important for patterned cell division in BY-2 cells, implicating that, amongst other factors, it is the polarity of microfilaments that is responsible for patterning (Maisch and Nick, 2007). To gain insight into the role of microfilament polarity in the context of cell file polarity, the RFP–ARP3 construct was now used as a marker for bona fide sites of actin nucleation.
For this purpose, the distribution of ARP3 dots was analyzed either in terminal or in central cells of a given cell file (Fig. 5). To determine the areal densities, the maximum projections from serial stacks of terminal or central cells were subdivided into distal and proximal halves (Fig. 5G, H1 and H2, respectively), and the ARP3 dots scored over the cross-sectional area in the central plane of the cells. In this way, the cross-sectional area occupied by the nucleus was subtracted, because in this region the cytoplasm was almost completely filled such that the density of ARP3 dots was very low. Since the nucleus is relatively large, this inhomogeneity might distort the quantification and even produce (artefactual) gradients if compared with the nuclear-free regions.
Fig. 5.
Distribution of RFP–ARP3 sites in terminal (A–C) versus central (D–F) cells in the GFP-FABD2 cell line. The GFP–FABD2 signals are shown in A and D, the RFP–ARP3 signals in B and E, and the dual fluorescence in C and F, respectively. (G) Areal densities of ARP3 dots in distal (H1) or proximal (H2) halves of terminal versus central cells. Bars=20 μm. (H) Total number of ARP3 dots per cell in terminal versus central cells. The values in G and H represent in each case averages from 50 individuals collected from 32 independent transient transformations.
The distribution of ARP3 was dependent on the cell type: whereas this density ranged around 1.8 ARP3 dots per 100 μm2 in both halves of the central cells or in the proximal halves of terminal cells, it was strongly increased (to 3.2 ARP3 dots per 100 μm2) in the distal halves of terminal cells. As a consequence, the total number of dots per cell was increased by almost 40% in terminal compared with central cells (Fig. 5H). This means that additional ARP3 dots are formed in the distal half of terminal cells.
Behaviour of ARP3 and PIN1 during divisional patterning
To test for possible correlations between ARP3 (as a marker for actin nucleation) and PIN1 (as a marker for cell polarity) during divisional patterning (Fig. 6), the localization of both markers was followed through the formation of the axial, pluricellular files in BY-2 cells. ARP3 was again visualized by an RFP fusion transiently expressed in a GFP–FABD2 background. PIN1 (from A. thaliana) was visualized as a fusion with GFP under control of the homologous PIN1 promoter.
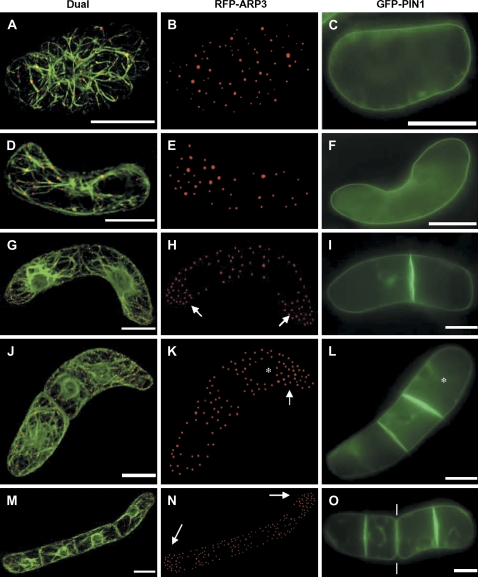
Fig. 6.
Dynamic redistribution of ARP3 and PIN1 during axial division of BY-2 cells. Dual visualization of actin microfilaments by GFP–FABD2 and RFP–ARP3 (A, D, G, J, M), and the distribution of ARP3 (B, E, H, K, N) and PIN1 (C, F, I, L, O) is shown for the symmetric unicellular (A–C), the asymmetric unicellular (D–F), the bicellular (G–I), the tricellular (J–L), and the quadricellular (M–O) state. Bars=20 μm. The arrows point to the increased density of the ARP3 signal in the distal halves of terminal cells (H, K, N). The asterisks label the ‘younger’ terminal cell of tricellular files (K, L). The white lines (O) point at the sites of beginning disintegration of the file of beginning disintegration of the file in smaller subsets (bicellular files).
In the unicellular state, generated by fragmentation of multicellular files, isodiametric single cells (deriving from the central part of a file) could be distinguished from polarized single cells (representing the terminal cells of the disintegrating file). Whereas RFP–ARP3 was distributed homogenously in the symmetric cells (Fig. 6A, B), the density of ARP3 dots was clearly increased in one cell half of unicellular asymmetric files (Fig. 6D, E). The PIN1 signal was localized at the periphery of the cells; plasmolysis experiments (data not shown) suggest that the signal was confined to the plasma membrane. In contrast to the ARP3 signal, PIN1 was found to be uniformly distributed during the unicellular state, irrespective of whether the cells were symmetric (Fig. 6C) or asymmetric (Fig. 6F).
In bicellular files, the density of the ARP3 signal was increased in the distal halves of both cells compared with their proximal halves (Fig. 6G, H). The PIN1 signal was clearly concentrated at the (newly deposited) cross-wall (Fig. 6I); plasmolysis experiments (data not shown) suggested that PIN1 was localized to the plasma membranes of both cells.
Tricellular files occur less frequently than bi- or quadricellular files (Campanoni et al., 2003; Maisch and Nick, 2007). These tricellular files result from an asymmetric first division, followed by a second division of the larger daughter cell that thus is ‘younger’ as compared with the opposite terminal cell (data not shown). In these tricellular files, ARP3 was enriched exclusively in one half of the ‘younger’ terminal cell (Fig. 6J, K). Again, PIN1 was concentrated in the cross-walls. However, the signal was strongest in the most recent cross-wall belonging to the ‘younger’ terminal cell (Fig. 6L, asterisk).
In quadricellular files, the ARP3 signal was enriched in the distal halves of both terminal cells, but equally distributed within the central cells (Fig. 6M, N). The PIN1 signal was still concentrated in the cross-walls, but the signal was most intense in the cross-walls belonging to the two terminal cells. In contrast, the signal was comparably faint in the cross-walls located more centrally in the file. In many quadricellular files, disintegration of the file into smaller subsets became detectable (Fig. 6O, white lines).
Discussion
In addition to their well-known role in cytoplasmic streaming, actin filaments have shifted into the focus of interest because they are central players in signal-regulated cell expansion (Bannigan and Baskin, 2005; Smith and Oppenheimer, 2005). In addition to Rho-related GTPases and WAVE proteins, it is the ARP2/3 complex that mediates the effect of signals upon actin organization (Xu and Scheres, 2005). Despite this central role of the ARP2/3 complex for the signal response of actin filaments, the components of this complex have not been visualized in living plant cells so far. In previous work, it has been demonstrated that the organization of actin filaments is regulated by auxin (Maisch and Nick, 2007), and that the actin response controls development (patterned cell division) in tobacco BY-2 cells. An approach to visualize the ARP2/3 complex in this context in vivo was therefore sought. Tobacco ARP3 was cloned and an RFP–ARP3 fusion construct was generated. It was possible to visualize ARP3 and actin filaments simultaneously in living cells upon biolistic, transient transformation of cells expressing the actin marker GFP–FABD2 with this RFP–ARP3 fusion construct. The RFP-labelled ARP3 was found specifically to decorate all arrays of the actin cytoskeleton.
After recovery from transient elimination of actin filaments (by LatB or cold), the regenerated filaments emerged from sites that were marked by ARP3 proteins (Fig. 3I). This recovery experiment clearly showed that ARP3 marked the sites of actin filament nucleation, consistent with published immunofluorescence studies using antibodies against ARP3 and actin (Fišerová et al., 2006). However, this immunofluorescence study had been performed using heterologous antibody. Since the different ARP proteins share high sequence similarity, cross-reactivities are very likely, such that one cannot be sure about the molecular identity of the visualized nucleation sites. In the present study, it was possible to circumvent this problem by using an RFP fusion with the homologous ARP3 protein and it could be shown that ARP3 is present in actin-nucleating sites. Whether it is directly bound to actin and what other proteins constitute this complex will be the subject of future work, where it is planned to purify those complexes biochemically.
The abundance of the ARP3 signal was significantly increased in the apical half of the terminal cells of a given file (Fig. 5G). This observation is in line with immunolocalization studies in tip-growing plant cells, where elements of the ARP2/3 complex were enriched at the sites of polar growth (Van Gestel et al., 2003; Hable and Kropf, 2005), and indicates a role for ARP3 in the establishment of cell polarity.
From previous work (Maisch and Nick, 2007), it was expected that the dynamic organization of actin relevant for the patterned cell division in BY-2 should be accompanied by dynamic changes in the localization of actin-organizing factors such as the ARP2/3 complex. To test this working hypothesis, the intracellular distribution of RFP–ARP3 was monitored during the patterned cell division that generates axial, pluricellular files. As a reference for the behaviour of cell polarity in this context, the localization of PIN1 was used. Although the role of PIN1 for auxin efflux is not completely understood, it is now generally accepted as a reliable marker for the direction of auxin flux (e.g. during primordia formation in the apical meristem; Reinhard et al., 2003).
The GFP–PIN1 fusion was predominantly localized to the plasma membranes of the cross-walls, resembling the distribution observed in the root elongation zone of A. thaliana (for a review, see Friml, 2003). However, this localization was not uniform over all cells of a file. It was the terminal cells where the PIN1 signal was clearest and most strictly confined to the cross-wall (Fig. 6O). In the more central cells, the PIN1 signal was progressively fading away. Since the polar distribution of PIN1 is most conspicuous in the two terminal cells, it can be inferred that the polarity of auxin flux is most pronounced in those cells and expressed to a lesser extent in the central cells of a file, possibly indicating opposing auxin fluxes passing through these cells.
Cell polarity, as visualized by GFP–PIN1, matched the localization of RFP–ARP3, which formed a clear gradient that was confined to the terminal cells of the files. In contrast, the RFP–ARP3 signal was evenly distributed in the central cells of the files, where cell polarity (as evident from a reduced GFP–PIN1 signal) was much less pronounced. One observation should be emphasized in this context: upon disintegration of a cell file, PIN1 redistributes such that it is found uniformly along all cell walls, the former cross-wall as well as the side walls (Fig. 6C, F). This would indicate that auxin efflux is temporarily delocalized in these single cells. In contrast, the ARP3 gradient found in terminal cells is conserved in the polarized cells that originate from the terminal cells upon disintegration of the file. This suggests that ARP3 is a more persistent marker of cell polarity than PIN1. This would be consistent with a role for ARP3 in the positioning of auxin efflux carriers by nucleating actin filaments, which in turn serve as tracks for the vesicle trafficking machinery. In other words, ARP3 might represent a direct manifestation of cell polarity.
Recent studies have linked the auxin signalling machinery to the organization of actin (for a review, see Xu and Scheres, 2005). However, this presumed link has been questioned recently by experiments where PIN1 and PIN2 maintained their polar localization, although actin filaments had been eliminated by the artificial auxin 2,4-D, or the phytotropin naphthylphthalamic acid (NPA) (Rahman et al. 2007). For the phytotropins 2,3,5-triiodobenzoic acid (TIBA) and 2-(1-pyrenoyl) benzoic acid (PBA), it was shown very recently that they induce actin bundling not only in plants, but also in mammalian and yeast cells, i.e. in cells that are not to be expected to utilize auxin as a signalling compound (Dhonukshe et al., 2008). This has been presented as supportive evidence for a role for actin filaments in polar auxin transport. However, it was mentioned in the same work that NPA did not cause actin bundling in non-plant cells, suggesting that its mode of action is different.
Thus, although actin seems to play a role in the polarity of auxin fluxes, the relationship is, first, not simple, and, secondly, far from being understood. The inconsistencies in the published reports might be caused by a branching of the causal chains: vesicle trafficking mediated by ADP ribosylation factors (ARFs) is required for the polar localization of Rho-related GTPases (ROPs) in plants which control regulators of the ARP2/3 complex (Frank et al., 2004). On the other hand, ARF-mediated vesicle trafficking also controls the localization of PIN proteins which is known to rely on the activity of the serine-threonine kinase PINOID (PID; Friml et al., 2004) and on the function of P-glycoprotein (PGP)/multiple drug resistance (MDR) proteins (Noh et al., 2001). Thus, ARF-dependent vesicle flow controls actin nucleation (activity of the ARP2/3 complex) and the multifaceted membrane localization patterns of PIN proteins in parallel. However, the initial cue that controls the spatial pattern of ARF activity that results in a polar localization of ROP and PIN proteins remains unknown.
However, the finding that ARP3 marks cell polarity more persistently than PIN1, which is redistributed during the single-cell state, suggests that the actin cytoskeleton might act upstream of PIN1 localization. It will also be interesting to follow the localization of ARP3 during the dynamic repolarization of tip-growing cells such as pollen tubes or root hairs. This could be due to actin-dependent movement of the vesicles that deliver the auxin efflux carriers to the plasma membrane or to actin-dependent tethering of these carriers in the plasma membrane.
Future work will be directed to test and understand the possible link between actin organization and auxin flux at both the molecular and the cellular level. On the one hand, it is desirable to use manipulation of ARP3 expression as a tool specifically to modulate the organization of actin filaments, and on the other hand the distribution and density of ARP3 both after inhibition of the polar auxin transport and during mitosis should be studied in stably transformed BY-2 cell lines.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Swiss-Prot accession numbers of ARP3 homologues from diverse organisms used for the construction of the phylogenetic tree in Fig. 1A.
Supplementary Material
Acknowledgments
This work was supported by a fellowship from the Landesgraduierten-Programm of the State of Baden-Württemberg to JM, the Ministry of Education, Youth and Sports of the Czech Republic (project number LC06034) to JF and LF, the Landesschwerpunkt-Programm of the State of Baden-Württemberg (LuNaCell) to PN, and the German–Czech Scientific-Technical Cooperation program (WTZ). The authors thank Dr Boris Voigt and Professor Dr Diedrik Menzel (Department of Plant Cell Biology, Institute of Cellular and Molecular Botany, University of Bonn, Germany) for the pGFPm3abd2 construct, and Professor Dr Zdeněk Opatrný (Department of Botany, Charles University Prague, Czech Republic) for critical reading of the manuscript. The BY-2 PIN1-GFP tobacco cell line was kindly provided by Dr Jan Petrášek (Institute of Experimental Botany, Academy of Sciences of the Czech Republic, Czech Republic).
Glossary
Abbreviations
- ARP3
actin-related protein 3
- BY-2
tobacco (Nicotiana tabacum L.) cell line ‘Bright Yellow 2’
- FABD2
fimbrin actin-binding domain 2
- GFP
green fluorescent protein
- RFP
red fluorescent protein
References
- An G. High efficiency transformation of cultured tobacco cells. Plant Physiology. 1985;79:568–570. doi: 10.1104/pp.79.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A, Baskin TI. Directional cell expansion—turning toward actin. Current Opinion in Plant Biology. 2005;8:619–624. doi: 10.1016/j.pbi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. Journal of Molecular Biology. 2004;336:551–565. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Campanoni P, Blasius B, Nick P. Auxin transport synchronizes the pattern of cell division in a tobacco cell line. Plant Physiology. 2003;133:1251–1260. doi: 10.1104/pp.103.027953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proceedings of the National Academy of Sciences, USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. Assembly and enlargement of the primary cell wall in plants. Annual Review of Cell and Developmental Biology. 1987;13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Molecular Biology. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Grigoriev I, Fischer R, et al. Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proceedings of the National Academy of Sciences, USA. 2008;105:4489–4494. doi: 10.1073/pnas.0711414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal SD, Le J, Basu D, Saad ME, Mallery EL, Szymanski DB. DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. The Plant Journal. 2004;38:526–538. doi: 10.1111/j.1365-313X.2004.02065.x. [DOI] [PubMed] [Google Scholar]
- Finer JJ, Vain P, Jones MW, McMullen MD. Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Reports. 1992;11:323–328. doi: 10.1007/BF00233358. [DOI] [PubMed] [Google Scholar]
- Fišerová J, Schwarzerová K, Petrášek J, Opartný Z. ARP2 and ARP3 are localized to sites of actin filament nucleation in tobacco BY-2 cells. Protoplasma. 2006;227:119–128. doi: 10.1007/s00709-006-0146-6. [DOI] [PubMed] [Google Scholar]
- Frank M, Egile C, Dyachok J, Djakovic S, Nolasco M, Li R, Smith LG. Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proceedings of the National Academy of Sciences, USA. 2004;101:16379–16384. doi: 10.1073/pnas.0407392101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. Auxin transport: shaping the plant. Current Opinion in Plant Biology. 2003;6:7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, et al. A PINOID-dependent binary switch in apical–basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Hable WE, Kropf DL. The Arp2/3 complex nucleates actin arrays during zygote polarity and growth. Cell Motility and the Cytoskeleton. 2005;61:9–20. doi: 10.1002/cm.20059. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Research. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T, Shibaoka H. Actin filaments and microtubules in the preprophase band and phragmoplast of tobacco cells. Protoplasma. 1987;140:151–156. [Google Scholar]
- Karimi M, De Mayer B, Hilson P. Modular cloning and expression of tagged fluorescent protein in plant cells. Trends in Plant Science. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, El-Assal SD, Basu D, Saad ME, Szymanski DB. Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Current Biology. 2003;13:1341–1347. doi: 10.1016/s0960-9822(03)00493-7. [DOI] [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM. The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiology. 2003;132:2034–2044. doi: 10.1104/pp.103.028563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch J, Nick P. Actin is involved in auxin-dependent patterning. Plant Physiology. 2007;143:1695–1704. doi: 10.1104/pp.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J. The ARP2/3 complex: giving plant cells a leading edge. BioEssays. 2005;27:377–387. doi: 10.1002/bies.20206. [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hülskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. The Plant Cell. 2003a;15:1632–1645. doi: 10.1105/tpc.011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hülskamp M. Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development. 2003b;130:3137–3146. doi: 10.1242/dev.00549. [DOI] [PubMed] [Google Scholar]
- Mathur J, Spielhofer P, Kost B, Chua NH. The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development. 1999;126:5559–5568. doi: 10.1242/dev.126.24.5559. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollar TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proceedings of the National Academy of Sciences, USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezava S. Tobacco BY-2 cell line as the ‘Hela’ cell in the cell biology of higher plants. International Review of Cytology. 1992;132:1–30. [Google Scholar]
- Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. The Plant Cell. 2001;13:2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olyslaegers G, Verbelen JP. Improved staining of F-actin and colocalization of mitochondria in plant cells. Journal of Microscopy. 1998;192:73–77. [Google Scholar]
- Petrašek J, Zažímalová E. The BY-2 cell line as a tool to study auxin transport. In: Nagata T, Matsuoka K, Inzé D, editors. Tobacco BY-2 cells: from cellular dynamics to omics. Berlin: Springer; 2006. pp. 107–115. [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI. Auxin, actin and growth of the Arabidopsis thaliana primary root. The Plant Journal. 2007;50:514–528. doi: 10.1111/j.1365-313X.2007.03068.x. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Saedler R, Mathur N, Srinivas BP, Kernebeck B, Hülskamp M, Mathur J. Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant and Cell Physiology. 2004;45:813–822. doi: 10.1093/pcp/pch103. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. Spatial control of cell expansion by the plant cytoskeleton. Annual Review of Cell and Developmental Biology. 2005;21:271–295. doi: 10.1146/annurev.cellbio.21.122303.114901. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. Journal of Cell Biology. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM. Organized F-actin is essential for normal trichome cell morphogenesis in Arabidopsis. The Plant Cell. 1999;11:2331–2348. doi: 10.1105/tpc.11.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gestel K, Stegers H, von Witsch M, Šamay J, Baluška F, Verbelen JP. Immunological evidence for the presence of plant homologues of actin-related protein Arp3 in tobacco and maize: subcellular localization to actin-enriched pit fields and emerging root hairs. Protoplasma. 2003;222:45–52. doi: 10.1007/s00709-003-0004-8. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Machesky LM. The WASP–Arp2/3 pathway: genetic insights. Current Opinion in Cell Biology. 2004;16:174–181. doi: 10.1016/j.ceb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Vidali L, Hepler PK. Actin and pollen tube growth. Protoplasma. 2001;215:64–76. doi: 10.1007/BF01280304. [DOI] [PubMed] [Google Scholar]
- Vidali L, McKenna ST, Hepler PK. Actin polymerization is essential for pollen tube growth. Molecular Biology of the Cell. 2001;12:2534–2545. doi: 10.1091/mbc.12.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Šamaj J, Müller J, Baluška F, Menzel D. GFP–FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. European Journal of Cell Biology. 2005;84:595–608. doi: 10.1016/j.ejcb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Xu J, Scheres B. Cell polarity: ROPing the ends together. Current Opinion in Plant Biology. 2005;8:613–618. doi: 10.1016/j.pbi.2005.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.