Abstract
Sucrose binding proteins (SBPs) were predicted to be membrane-associated, but have been shown to localize in the lumen of protein storage vacuoles of various seeds. In this study, a new 64 kDa SBP has been identified from developing mung bean (Vigna radiata) seeds (here termed VrSBP1) via MS/MS analysis and N-terminal amino acid sequencing analysis and specific antibodies were generated using purified VrSBP1 proteins. Western blot analysis with the new VrSBP1 antibodies showed that, similar to most seed storage proteins, VrSBP1 proteins accumulated during seed development and were subsequently mobilized once the mung bean seeds germinated. Immunogold electron microscope (EM) studies on ultra-thin sections of high-pressure freezing/frozen substituted developing mung bean cotyledons demonstrated that VrSBP1 was localized specifically to the tonoplast of the protein storage vacuole and to the limiting membrane of a novel putative prevacuolar compartment. Biochemical and subcellular fractionation studies further demonstrated that VrSBP1 proteins were membrane-associated in developing mung beans, consistent with their tonoplast localization. This study thus shows convincing evidence of tonoplast-localization of a plant SBP for its future functional characterization and provides a model of studying non-integral membrane proteins associated with the tonoplasts in plant cells.
Keywords: High-pressure freezing, immunogold EM, membrane-associated, mung bean, sucrose binding proteins, tonoplast
Introduction
GmSBP1, the first sucrose binding protein (SBP) from soybean (Glycine max) to be studied, was originally identified in membrane preparations from developing soybean cotyledons based on its binding ability to a sucrose derivative which was a competitive inhibitor of sucrose influx into cotyledon protoplasts (Ripp et al., 1988). GmSBP1 was not an integral membrane protein because it did not contain any predicted transmembrane domains (Grimes et al., 1992). However, it was shown to be membrane-associated because Na2CO3 and urea effectively disassociated GmSBP1 from the membrane (Overvoorde and Grimes, 1994). An immunocytochemical study with GmSBP1 antibodies has demonstrated that GmSBP1 located to the plasma membrane (PM) of several cell types engaged in sucrose transport, including the mesophyll cells of young sink leaves, the companion cells of mature phloem, and the cells of the developing cotyledons (Grimes et al., 1992). GmSBP1 may also be involved in sucrose transport because its accumulation pattern correlates with the onset of active sucrose influx, whereas the transcript level of the GmSBP1 gene correlates with the rate of sucrose uptake (Ripp et al., 1988; Grimes et al., 1992); and because GmSBP1 expressed in yeast mediated sucrose uptake in an H+-independent and non-saturable manner (Overvoorde et al., 1996). It is therefore possible that GmSBPs mediate sucrose uptake across the plasma membrane in plants. However, several other studies have indicated that SBPs are members of the seed storage protein super-family where the SBP gene is structurally similar to the 7S vicilin genes (Braun et al., 1996; Overvoorde et al., 1997; Castillo et al., 2000; Heim et al., 2001; Contim et al., 2003).
Sequence alignment analysis has shown that GmSBP does not share any amino acid similarity with known transport proteins. Instead, SBPs show significant sequence and structural homology with the vicilin-like seed storage proteins (Braun et al., 1996; Overvoorde et al., 1997). For example, the soybean GmSBP1 shares 44–61% similarity at the amino acid level with the vicilin-like seed storage proteins. Similarly, the pea (Pisum sativum) SBP (PsSBP) and the faba bean (Vicia faba) SBP (VfSBPL) have also been found to be related to the 7S vicilin-like storage proteins (Castillo et al., 2000; Heim et al., 2001; Wenzel et al., 2005). Furthermore, the structure of the VfSBPL gene has typical characteristics of legume vicilin storage protein genes with six exons and five introns (Heim et al., 2001).
It has been shown that, similar to seed storage proteins, the SBPs of soybean (GmSBPs), faba bean (VfSBPL), and pea (PsSBP) all gradually accumulate during seed development and are subsequently mobilized upon seed germination (Heim et al., 2001; Elmer et al., 2003; Wenzel et al., 2005). Furthermore, even though GmSBP1 was originally found to be localized to plasma membrane in soybean (Grimes et al., 1992), re-examination of the subcellular localization of GmSBPs with newly generated antibodies showed that GmSBPs were localized to the seed protein storage vacuoles (PSVs) in soybean (Elmer et al., 2003). Similar PSV localization for other SBPs including VfSBPL and PsSBP has also been demonstrated (Heim et al., 2001; Wenzel et al., 2005). In each of these immunogold EM studies SBP labelling was found to be evenly distributed across the lumen of seed PSVs, as in the case of the labelling patterns of soluble storage proteins. The results indicate that these SBPs are membrane-associated proteins that are evenly distributed across the lumen of PSVs in various seeds. This is curious, because the membrane association nature of SBPs does not seem to match their lumenal localization inside PSVs.
In this study, a 64 kDa SBP1 has been identified in developing mung bean (Vigna radiata) cotyledons (termed VrSBP1) and specific VrSBP1 antibodies have been raised. It is shown that VrSBP1 proteins are accumulated in developing seeds and mobilized when the mung bean seeds germinate. It has also been demonstrated that VrSBP1 is a membrane-associated protein that is specifically localized to PSV tonoplasts in developing mung bean cotyledons. These observations provide convincing evidence for the tonoplast-localization and membrane-association of the VrSBP1 in developing mung beans. This study will also serve as the first step in molecular and functional characterization of these tonoplast-localized SBPs in plants.
Materials and methods
Plant materials
Mung bean seeds were planted in pots in the greenhouse of The Chinese University of Hong Kong (CUHK). Cotyledons of developing mung bean seeds were used for protein extraction, microsome isolation, and sample preparation for transmission electron microscopy.
Antibodies
Polyclonal VSRat-1 antibodies and α-TIP antibodies were used in this study, and their characterization has been described in detail in two previous studies (Jauh et al., 1999; Tse et al., 2004). VrSBP1a and VrSBP1b antibodies were generated in this study in the following manner. Developing mung bean cotyledons were ground in sucrose-containing buffer to protect organelle integrity, and then subjected to continuous sucrose gradient ultracentrifugation. Organelle fraction collected from 45% sucrose was collected for protein extraction, followed by protein separation by SDS-PAGE (Wang et al., 2007). VrSBP1 protein bands were cut from the stained gels and VrSBP1 proteins were further recovered using a protein elutor (Bio-Rad). Gel-purified proteins were used as antigens to immunize two rabbits at the Animal House of CUHK. Antibodies were further affinity-purified by using a Protein A column (Sigma) or a column conjugated with the purified VrSBP1 proteins. The mung bean storage protein 8S globulin antibodies were also prepared as follows: 8S globulin proteins were purified from total protein extracts of developing mung bean cotyledons after SDS-PAGE and gel-purification as above for use in the immunization of rabbits. Antibodies were affinity-purified with a CNBr-activated Sepharose (Sigma) conjugated with gel-purified 8S globulin as previously described (Rogers et al., 1997; Jiang and Rogers, 1998).
MS/MS analysis for protein identification
The protein bands of interest were cut out from the Coomassie Brilliant Blue (CBB)-stained protein gel, and stepwise procedures for gel destaining, in-gel digestion, desalting, and spotting on plates were carried out in accordance with the manufacturer's protocols. Prepared samples were then dotted on plates and subjected to analysis by the ABI 4700 proteomics analyser (Applied Biosystems) for subsequent protein identification via database searching. The relative amount of proteins in a CBB-stained protein gel was quantified using the software Quantity One (Bio-Rad).
N-terminal protein sequencing
Purified proteins from the fractions were first separated by SDS-PAGE and then transferred via electroblot onto PVDF membrane. Protein bands on the PVDF membrane were visualized by staining with G-250 Coomassie stain solution whereas the protein bands of interest with the correct molecular weight were cleaved out and subjected to N-terminal protein sequencing according to the manufacturer's protocols.
Protein extraction, fractionation and analysis
Total protein extraction from developing mung bean cotyledons and Western blot analysis has been described previously (Wang et al., 2007). For supernatant and membrane fraction preparations, developing mung bean cotyledons were first ground to a fine powder in liquid nitrogen, suspended in the extraction solution (TRIS-HCl 50 mM, pH 7.4 containing 10 mM KCl, 1 mM EDTA, 0.1 mM phenylmethylsulphonyl fluoride, and 5 mg ml−1 leupeptin), and then centrifuged at 10 000 g for 20 min to produce supernatant (S10) and pellet (P10) fractions. The S10 fraction was further subjected to ultracentrifugation at 100 000 g for 1 h, and the resulting supernatant and pellets were assigned as S100 and P100 fractions, respectively. The P100 pellets were then resuspended in protein extraction solution with the same volume as the supernatant fraction. Proteins of equal volume of S100, P100, and P10 fractions were separated by SDS-PAGE and subjected to Western blot analysis using various antibodies.
For chemical treatment of membrane fraction, the P100 pellet was resuspended in the same extraction solution but containing either 0.1 M Na2CO3, 1% Triton X-100 or 1% SDS, or without chemical addition as control. The suspension was incubated at 4 °C in a shaker set at 100 rpm for 60 min and recentrifuged for 1 h at 100 000 g, and the resulting pellet (P) was resuspended in extraction solution containing 1.5% SDS and 150 mM NaCl with the same volume as the original extraction solution. Equal volumes of the pellet (P) and supernatant (S) protein samples were separated by SDS-PAGE, and Western blot detection was then conducted with various antibodies.
Immunogold electron microscopy (EM)
Immunogold EM on ultrathin sections prepared from high-pressure frozen/freeze substitution of developing mung bean cotyledons was performed essentially as described previously (Tse et al., 2004; Lam et al., 2007; Wang et al., 2007). Briefly, small blocks of cotyledons were frozen in a high-pressure freezing apparatus (EMP2, Leica, Bensheim, Germany). Substitution was performed in dry acetone containing 0.1% uranyl acetate at –85 oC for 5 d in an AFS freeze substitution unit (Leica, Wetzlar, Germany). Samples were stepwise infiltrated with HM20, embedded, and UV polymerized at –35 oC. Immunogold labelling on ultrasections was performed using standard procedures previously described by Tse et al. (2004). The ultrasections post-stained by aqueous uranyl acetate/lead citrate were examined using a JEOL JEM-1200 EXII transmission electron microscope (JEOL) operating at 100 kV or a Hitachi H-7650 transmission electron microscope with CCD camera (Hitachi High-Technologies Corporation, Japan) operating at 80 kV as previously described (Tse et al., 2004).
Results
Identification and antibody generation of mung bean sucrose binding proteins
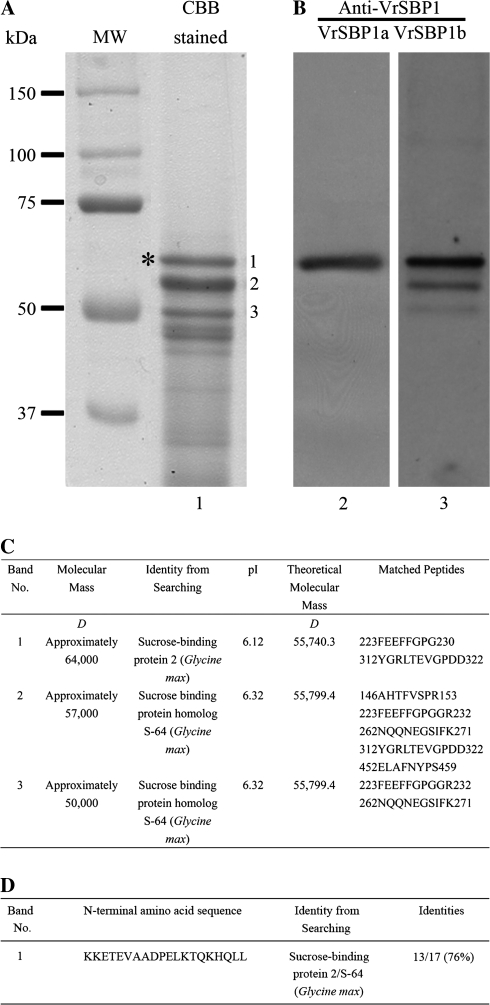
We were interested in the identification of various proteins from various organelle fractions of developing mung bean seeds. As shown in Fig. 1, protein samples from a fraction representing the 45% sucrose gradient contained three major protein bands (Fig. 1A, as indicated by 1–3 in lane 1). Subsequent MS/MS analysis identified all these three protein bands as the soybean GmSBP2/S64 homologues with multiple peptides matching (Pedra et al., 2000; Elmer et al., 2003) sucrose binding proteins (SBPs) (Fig. 1C). To confirm the identity of these proteins as identified by MS/MS analysis further, the 64 kDa protein band was also cut out and used for N-terminal amino acid sequencing analysis by the Edman degradation method. The obtained amino acid sequences, KKETEVAADPELKTQKHQLL, match largely with the GmSBP2/SBP S64 of soybean (Fig. 1D; see Supplementary Fig. S1 at JXB online). Therefore, both MS/MS analysis with multiple peptides matching and the N-terminal amino acid analysis of the 64 kDa protein band of mung bean strongly suggest that it is a homologue of the soybean GmSBP2/S64. These three newly identified SBPs were therefore provisionally named VrSBP1, VrSBP2, and VrSBP3, respectively, as the possibility cannot be ruled out that VrSBP2 and VrSBP3 are the degraded products of VrSBP1. However, any uncertainty in this respect does not affect the findings of this study, which deliberately focused on an investigation of VrSBP1.
Fig. 1.
Identification and antibody generation of sucrose binding protein1 in developing mung bean cotyledons. (A) Developing mung bean seeds were ground in grinding solution containing sucrose for organelle protection and then subjected to continuous sucrose gradient centrifugation. Proteins from fraction 8 (about 45% sucrose) were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (lane 1). Individual protein bands were then isolated for trypsin digestion and subjected to MS/MS analysis for protein identification. The identified 64 kDa sucrose binding protein1 (VrSBP1, as indicated by a single asterisk) proteins were further purified and used as antigens to raise antibodies. CBB, Coomassie Brilliant Blue. MW, molecular weight marker in kDa. (B) Western blot analysis. Lanes 2 and 3 showed Western blot analysis of total proteins from developing mung bean seeds using the two newly generated anti-SBP1a or anti-SBP1b antibodies as indicated. VrSBP1, sucrose binding protein1 of mung bean. (C) MS/MS analysis and protein identification for protein bands 1–3. (D) N-terminal amino acid sequences and protein identification for protein band 1.
VrSBP1 proteins from developing mung bean cotyledons were purified next and used as antigens to raise antibodies. As shown in Fig. 1B, Western blot analysis on total protein extracts from the developing mung bean cotyledons with these two newly generated antibodies (here termed VrSBP1a and VrSBP1b antibodies) detected a major single protein band at about 64 kDa (Fig. 1B, lanes 2 and 3 as indicated by asterisks), which was identical to its corresponding VrSBP1 protein band in the CBB-stained gel. VrSBP1b antibodies also detected two weak bands that corresponded to the VrSBP2 and VrSBP3 protein bands in CBB-stained gel, indicating that VrSBP1b antibodies had a weak cross-reactivity toward both VrSBP2 and VrSBP3 proteins in mung beans.
Expression of SBPs during seed development and germination of mung bean
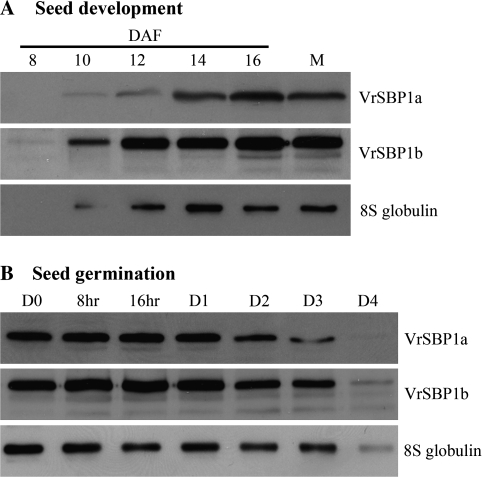
To determine whether the VrSBP1 proteins would display characteristics of storage proteins during seed development and germination, Western blot analysis was performed next with VrSBP1 antibodies on total proteins extracted at various stages from developing mung bean seeds or at various times from germinating seeds. As a control, antibodies for the mung bean major storage protein 8S globulin were also used. As shown in Fig. 2A, little VrSBP1 protein was detected in very young seeds at 8 d after flowering, but the amounts of VrSBP1 proteins as detected by either VrSBP1a or VrSBP1b antibodies gradually increased between 10–16 d after flowering and levelled off thereafter at amounts comparable to those found in mature seeds. Similar patterns of protein accumulation were also observed for the 8S globulin storage proteins during mung bean seed development (Fig. 2A). By contrast, the levels of both VrSBP1 and 8S globulin proteins gradually decreased upon seed germination from day 0 to day 3 after seed imbibitions (Fig. 2B). At day 4 after seed germination, the detectable levels of VrSBP1 or 8S globulin proteins had fallen dramatically (Fig. 2B).
Fig. 2.
Protein profiles of sucrose binding protein1 during mung bean seed development and germination. Total proteins were isolated from various stages of developing (A) or germinating (B) mung bean seeds as indicated, followed by protein separation via SDS-PAGE and Western blot analysis with anti-VrSBP1a, anti-VrSBP1b, and anti-8S globulin antibodies, as indicated. DAF, days after flowering; M, mature dry seed; D, days after imbibitions.
Immunogold EM localization of VrSBP1 proteins
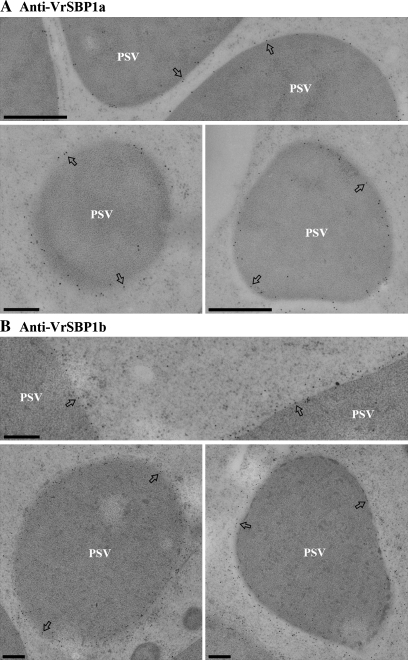
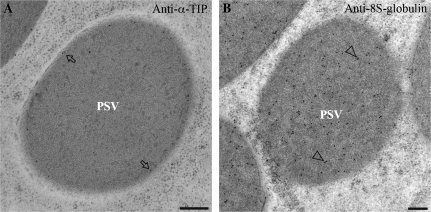
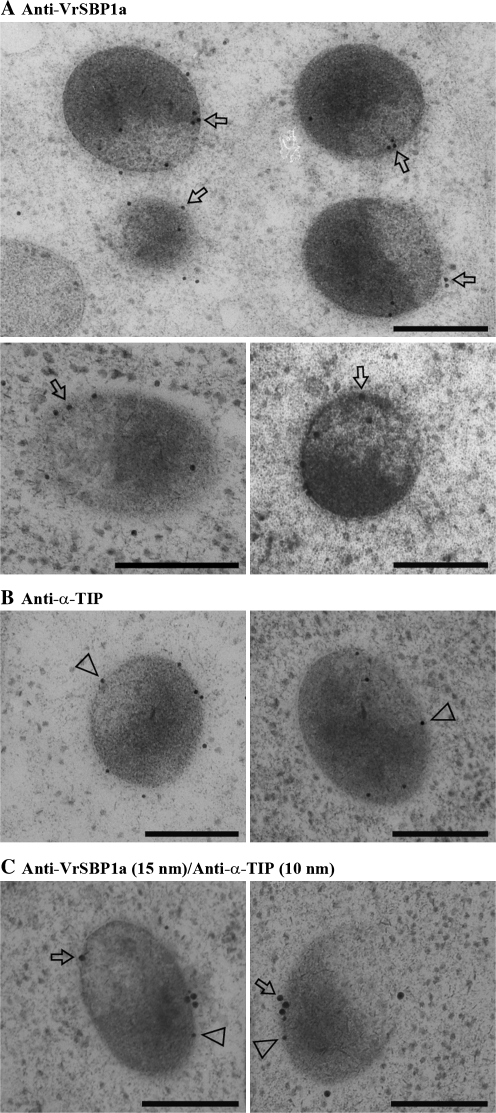
Immunogold EM studies were carried out next to determine the subcellular localization of VrSBP1 in developing mung bean seeds. As shown in Fig. 3, the gold particle labelling with either VrSBP1a or VrSBP1b antibodies was mainly found on the limiting membrane (tonoplast) of the PSVs (Fig. 3A, B, indicated by arrows as examples), and there was very little background labelling. Similarly, as a positive control, α-TIP antibodies also mainly labelled PSV tonoplasts in similar ultra-thin sections (Fig. 4A, examples indicated by arrows). By contrast, gold particle labelling for the 8S globulin storage proteins was found to be evenly distributed inside the PSVs (Fig. 4B, examples indicated by arrowheads).
Fig. 3.
Sucrose binding proteins localized to the PSV tonoplasts of developing mung bean. Ultrathin sections prepared from high-pressure frozen/freeze-substituted developing mung bean seeds were immunogold-labelled with either SBP1a antibodies (A) or SBP1b antibodies (B), as indicated. Arrows indicate examples of gold particles on the tonoplast of protein storage vacuole (PSV). Scale bar=200 nm.
Fig. 4.
Immunogold EM labelling of α-TIP and 8S globulin in developing mung bean. Ultrathin sections prepared from high-pressure frozen/freeze-substituted developing mung bean seeds were immunogold-labelled with either α-TIP antibodies (A) or 8S globulin antibodies (B), as indicated. Arrows and arrowheads indicate examples of gold particles on the tonopalst and lumen of protein storage vacuole (PSV) respectively. α-TIP, alpha-tonoplast intrinsic protein; PSV, protein storage vacuole. Scale bar=200 nm.
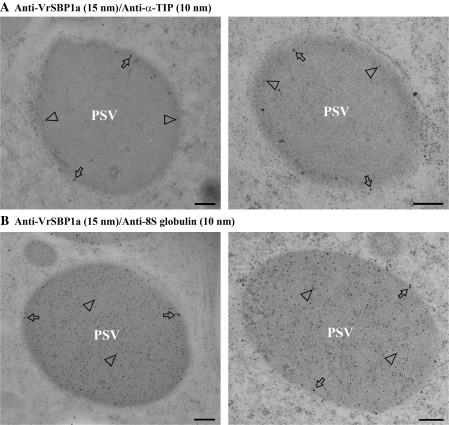
When double-labelling with antibody pairs of either VrSBP1a and α-TIP or VrSBP1a and 8S globulin was performed on similar ultra-thin sections, consistent results were obtained: VrSBP1a and α-TIP were mainly colocalized in the same PSV tonoplasts (Fig. 5A), whereas VrSBP1a and 8S globulin were found to be presented in the tonoplast and lumen of the same PSVs, respectively (Fig. 5B). When multiple VrSBP1 antibody-labelled sections from four independent labelling experiments were counted to estimate the distribution of gold particles across PSVs, the great majority of the gold particles (89%) were found on the PSV tonoplasts, with less than 11% of the gold particles being observed over the PSV lumen (Table 1). This differing distribution of gold particles between the tonoplast and the lumen of PSVs was statistically significant (Table 1). Taken together, these immunogold EM studies demonstrated that both VrSBP1 and α-TIP were mainly localized in PSV tonoplasts while the 8S globulin storage proteins were evenly distributed in the PSV lumen.
Fig. 5.
Immunogold EM double-labelling study. Ultrathin sections prepared from high-pressure frozen/freeze-substituted developing mung bean seeds were immunogold double-labelled with pairs of antibodies of either SBP1a (15 nm) and α-TIP (10 nm) (A) or SBP1a (15 nm) and 8S globulin (B). Arrows and arrowheads indicate examples of gold particles on the tonoplast and lumen of the protein storage vacuole (PSV), respectively. Scale bar=200 nm.
Table 1.
Distribution of gold particles (GPs) across the protein storage vacuoles (PSVs)
| Total no. of GP | No. of PSV | Average GP per PSV | Percentage (%) | |
| Tonoplast | 1297 | 20 | 64.85 | 89.08** |
| Lumen | 159 | 20 | 7.95 | 10.92** |
Significant difference between tonoplast and lumen of PSV was analysed using two-sided paired t test (**P <0.01). Data were collected and analysed from four independent labelling experiments. GP, gold particles.
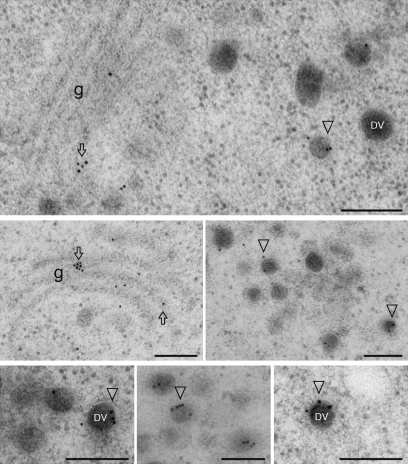
In addition to PSV tonoplast labelling, immunogold EM labelling with VrSBP1a or VrSBP1b antibodies was also found in the Golgi apparatus and dense vesicles (DVs) (Fig. 6). Interestingly, VrSBP1 antibodies also labelled a novel compartment about 200–500 nm in diameter (Fig. 7A, B), which might represent the prevacuolar compartment (PVC) for PSV (see Discussion). Similar to the tonoplast labelling of VrSBP1 in PSVs, the gold particle labelling of VrSBP1 in the novel compartments was also mainly found on their limiting membranes (Fig. 7C, examples indicated by arrows), a result consistent with the membrane association nature of VrSBPs (see below).
Fig. 6.
Immunogold EM localization of VrSBP1 to Golgi and dense vesicles. Ultrathin sections prepared from high-pressure frozen/freeze-substituted developing mung bean seeds were immunogold single-labelled with either VrSBP1a antibodies. Arrows and arrowheads indicate examples of gold particles on the Golgi apparatus and the dense vesicles (DVs). Scale bar=200 nm.
Fig. 7.
Immunogold EM localization of VrSBP1 to a novel compartment. Ultrathin sections prepared from high-pressure frozen/freeze-substituted developing mung bean seeds were immunogold single-labelled with either VrSBP1a (A) or α-TIP antibodies (B), or double-labelled with VrSBP1a and α-TIP antibodies (C). Arrows indicate examples of gold particles on the limiting membrane of the novel organelle. Scale bar=200 nm.
VrSBP1 proteins are membrane-associated
The immunogold EM results so far demonstrated that VrSBP1 localized to the PSV tonoplast and limiting membrane of putative novel PVCs in developing mung bean seeds. To confirm this membrane localization of VrSBP1 proteins further, a biochemical analysis of the membrane association nature of VrSBP1 proteins was carried out. Three known marker proteins were used as controls in this analysis to compare with the VrSBP1: α-TIP, a type IV integral membrane protein with six transmembrane domains localizing on the PSV tonoplasts (Jauh et al., 1999); vacuolar sorting receptor (VSR), a type I integral membrane protein with a single transmembrane domain (Paris et al., 1997; Jiang and Rogers, 1998); and the 8S globulin, a soluble lumenal PSV-localized storage protein.
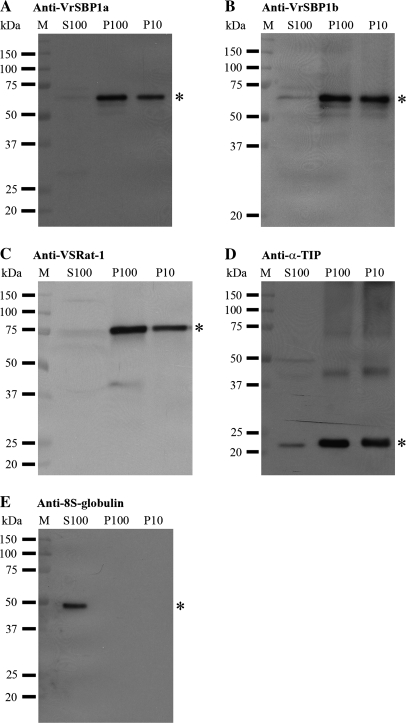
As shown in Fig. 8, VrSBP1 proteins detected by either VrSBP1a or VrSBP1b antibodies were mainly found in both P100 and P10 fractions but were absent in the S100 fraction (Fig. 8A, B), a result similar to the expected distributions of the two tested integral membrane proteins of VSR (Fig. 8C) and α-TIP (Fig. 8D). By contrast, the 8S globulin was found in S100 only as expected for soluble storage proteins (Fig. 8E). Taken together, these results indicated that VrSBP1 proteins were present in the membrane fractions, and were therefore membrane proteins in developing mung bean seeds. These results were also consistent with the PSV tonoplast localization of VrSBP1 in the immunogold EM study.
Fig. 8.
VrSBP1 is present in the membrane fraction. Three fractions were first isolated from developing mung bean cotyledons: S100 representing soluble proteins, P100 representing small vesicles, and P10 representing large membranes. Total proteins from an equal volume of the three individual fractions were next separated by SDS-PAGE, followed by Western blot analysis with various antibodies, as indicated. Asterisks indicate the position of the detected proteins. M, molecular weight marker in kDa.
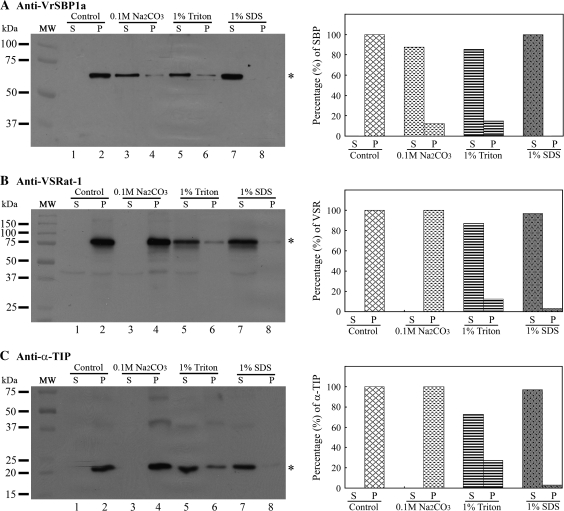
Since SBPs did not contain any predicted transmembrane domain (Overvoorde and Grimes, 1994; Castillo et al., 2000; Heim et al., 2001; Wenzel et al., 2005) but might be membrane-associated (Overvoorde and Grimes, 1994), an attempt was made to determine the possible membrane-association nature of VrSBP1 in mung beans by a further analysis of the property of VrSBP1 within the P100 vesicle fraction. Three chemicals were used to treat the P100 fraction to test for their ability to release VrSBP1 from membrane association into the soluble fraction: 0.1 M Na2CO3, 1% Triton X-100, and 1% SDS.
As shown in Fig. 9, after the P100 fraction was treated with 0.1 M Na2CO3 more than 87% of the VrSBP1 proteins were released from membrane association (Fig. 9A, lane 2) into the soluble fraction (Fig. 9A, lanes 3 versus 4), indicating that Na2CO3 was effective in disassociating VrSBP1 from the membrane. A similar result was obtained when the P100 fraction was treated with 1% Triton X-100 (Fig. 9A, lanes 5 and 6), whereas treatment of 1% SDS caused the nearly complete release of VrSBP1 into the soluble fraction (Fig. 9A, lanes 7 and 8). By contrast, when the same P100 fraction was treated with 0.1 M Na2CO3, followed by detection using either VSR or α-TIP antibodies, neither VSR nor α-TIP protein was released from the membrane because no protein was detected in the soluble fraction after 0.1 M Na2CO3 treatment (Fig. 9B, C, lane 3). However, both 1% Triton X-100 and 1% SDS treatments caused VSR or α-TIP to be released from the membrane (Figs 9B and 9C, lanes 5–8). These results strongly indicate that VrSBP1 proteins in mung bean are membrane-associated and are different from the integral membrane proteins of VSR or α-TIP.
Fig. 9.
VrSBP1 is membrane-associated. The P100 vesicle fraction from developing mung bean cotyledons were first resuspended in solutions containing one of the three chemicals (0.1 M Na2CO3, 1% Triton X-100, and 1% SDS), as indicated, and incubated for 1 h at 4 °C, followed by centrifugation at 100 000 g for 1 h, resulting in pellet (P) and supernatant (S). The pellet (P) was resolubilized in the same volume of extraction solution containing 1.5% SDS and 150 mM NaCl. Equal volumes of protein samples were separated by SDS-PAGE, followed by Western blot analysis using anti-VrSBPa (A), anti-VSRat-1 (B), and anti-α-TIP (C), as indicated. Asterisks indicate the position of the detected proteins. The signals of the detected protein bands were further quantified with the software Quantity One (Bio-Rad) to determine the percentages of protein distribution between the pellet (P) and supernatant (S) in each treatment. MW, molecular weight marker in kDa.
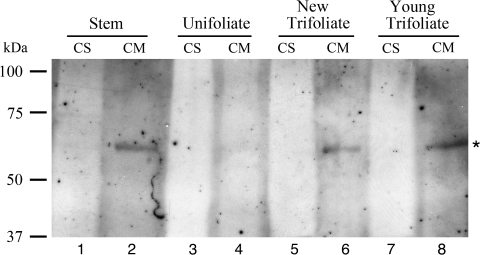
To find out if VrSBP1 proteins were also present in other tissues of mung bean, Western blot analysis was also performed with VrSBP1 antibodies on proteins isolated from various tissues of mung bean seedlings. Both soluble proteins (CS) and membrane proteins (CM) were first isolated from various tissues of mung bean seedlings including stems, unifoliate, new trifoliate, and young trifoliate leaves, followed by protein separation via SDS-PAGE and Western blot analysis using VrSBP1 antibodies. As shown in Fig. 10, similar amounts of VrSBP1 proteins were detected in the CM fractions of stem, new trifoliate, and young trifoliate leaves (Fig. 10, lanes 2, 6, 8), but little VrSBP1 was present in unifoliate leaves (Fig. 10, lane 4). These results indicated that, in addition to developing seeds, the presence of VrSBP1 in mung bean seedlings might also suggest its roles in these tissues. In addition, the presence of VrSBP1 in the CM fraction rather than in the CS fraction in the seedlings is also consistent with its nature of membrane association.
Fig. 10.
VrSBP1 proteins are present in mung bean seedlings. Both soluble proteins (CS) and membrane proteins (CM) were first isolated from various tissues of mung bean seedlings including stems, unifoliate, new trifoliate, and young trifoliate leaves, followed by protein separation via SDS-PAGE and Western blot analysis using VrSBP1 antibodies. An asterisk indicates the position of the VrSBP1.
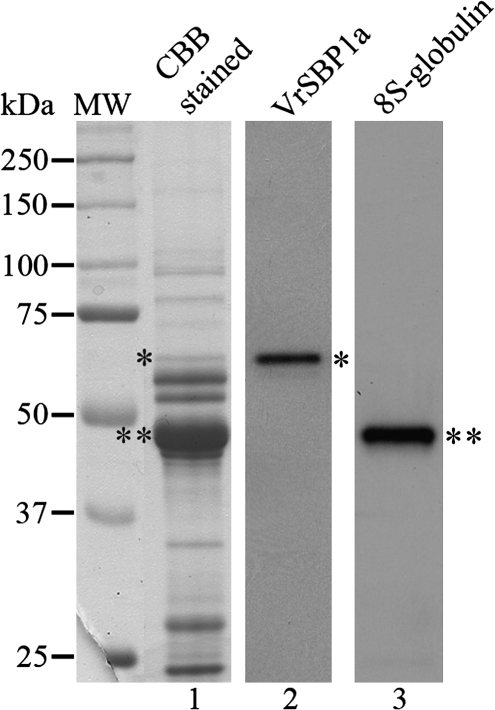
To determine the relative protein abundance of VrSBP1, total proteins were also extracted from developing mung bean cotyledons for analysis. As shown in Fig. 11, the amount of VrSBP1 proteins, as detected and quantified in a CCB-stained protein gel (Fig. 11, lane 1, as indicated by single asterisk), only represents about 1.98% of the total seed protein, while the major storage protein 8S globulin accounts more than 70% of the total proteins in developing mung bean cotyledons (Fig. 11, lane 1, as indicated by double asterisks). Thus, VrSBP1 is unlikely to be a storage protein in developing mung bean.
Fig. 11.
VrSBP1 is not a major protein in developing mung bean cotyledons. Total proteins from developing mungbean cotyledons were extracted and separated by SDS-PAGE, followed by either Coomassie Brilliant Blue staining for protein visulization (lane 1) or Western blot analysis using anti-VrSBP1a (lane 2) and anti-8S-globulin antibodies (lane 3), respectively. A single asterisk and a double asterisk indicate the positions of VrSBP1 and 8S-globulin, respectively.
Discussion
In this study, a 64 kDa sucrose binding protein was identified from developing mung bean seeds and termed VrSBP1. Very specific antibodies for VrSBP1 were generated and used in this study. The identity of the 64 kDa mung bean VrSBP1 as the homologue of the soybean GmSBP2/S64 was confirmed by both MS/MS analysis with multiple peptide matching and N-terminal amino acid sequencing analysis. Further immunocytochemical and biochemical analysis demonstrated that VrSBP1 was a membrane-associated protein that was specifically localized to the PSV tonoplasts in developing mung bean seeds. This tonoplast-localized and membrane-associated feature of VrSBP1 is unique, and our findings will lay the foundations for further physiological and functional characterization of SBPs in future studies via molecular cloning of VrSBP1 and genetic knock-out or over-expression approaches in transgenic plants. Furthermore, our findings will also provide a model of studying non-integral membrane proteins associated with the tonoplasts in plant cells.
The accumulation of VrSBP is developmentally regulated and its functional implications in seeds
Several previous studies of SBPs in plants have demonstrated that SBPs show characteristics of storage proteins in terms of PSV localization and accumulation during seed development (Heim et al., 2001; Elmer et al., 2003; Wenzel et al., 2005).
Similarly, it has been shown in this study that the VrSBP1 proteins gradually accumulated during mung bean seed development, but these developmentally-accumulating VrSBP1 proteins were degraded upon seed germination. Such VrSBP1 protein profiles are characteristic of PSV-localized soluble storage proteins in most seeds (Jiang et al., 2000). However, since the VrSBP1 proteins were found to be localized in or associated with the PSV tonoplast of developing mung bean seeds rather than in the PSV lumen, and that the VrSBP1 proteins represented only 1.98% of the total proteins in developing mung bean seeds (Fig. 11), it is thus unlikely that VrSBP1 proteins would have the same function as storage proteins and contribute significantly to provide amino acids for seedling growth upon seed germination.
In addition, SBPs in soybean (GmSBP) were quite similar to 7S vicilin-like seed storage proteins in terms of amino acid sequences and predicted structures, but the biological functions of SBPs seem to be distinct from those of storage proteins (Braun et al., 1996; Overvoorde et al., 1997). These conclusions are also true for the VrSBP1 proteins of mung bean in this study. It is thus likely that there could be an evolutionary link between the 7S or 8S storage proteins and the SBPs in plants as previously suggested by Elmer et al. (2003).
Tonoplast localization of VrSBP1
Several previous studies of the subcellular localization of SBPs in seeds found that SBPs were localized mainly inside (in the lumen of) the protein storage vacuoles (PSVs) in soybean, faba bean, and pea seeds (Heim et al., 2001; Elmer et al., 2003; Wenzel et al., 2005). However, such lumenal localization of SBPs in seed PSVs cannot explain the membrane association nature of SBPs from biochemical analysis (Grimes et al., 1992; Overvoorde et al., 1994). For example, the soybean SBP GmSBPs were showed to be membrane-associated, but immunogold EM localization of GmSBPs did not show any membrane localization in PSVs. More recently, the pea SBPs (PsSBPs) have been shown to be concentrated on the membranes of dense vesicles (DVs), but, like the storage protein legumin, are still found to be evenly distributed in the lumen of PSVs (Wenzel et al., 2005). It is thus likely that the PsSBPs reached PSVs via the pea DVs, although the mechanism by which the PsSBPs were disassociated from membrane localization in DVs into the lumenal localization of PSVs upon reaching the PSVs remains unknown (Wenzel et al., 2005).
In this study, three SBP protein bands were identified from the sucrose gradient sample of developing mung bean cotyledons as the soybean SBP homologues of GmSBP2/S64 via MS/MS analysis (Fig. 1). The two newly generated antibodies for VrSBP1 proteins, VrSBP1a and VrSBP1b, labelled PSVs but concentrated on the tonoplast in developing mung bean cotyledons, while little labelling was found at the plasma membrane or in the nucleus (Fig. 3). Therefore, VrSBP1 is probably a tonoplast-localized or tonoplast-associated protein in developing mung bean cotyledons. In addition, the biochemical data indicate that VrSBP1 proteins in developing mung bean cotyledons are membrane-associated proteins, supporting the tonoplast localization of VrSBP1. The results of immunocytochemical analysis of VrSBP1 are therefore consistent with the biochemical data in this study. This unique tonoplast localization of VrSBP1 in developing mung beans might implicate the functional site of VrSBP1 in developing mung bean cotyledons. It has been known that some vacuoles in plant cells accumulate sugars, therefore, the tonoplast-association of VrSBP1 in the PSV of developing mung bean might implicate the possible functional role of VrSBP1 as a sugar transporter in PSV membranes of mung bean seeds. The speculation needs experimental proof in future study.
PSV transport of VrSBP1 in developing mung bean cotyledons
Vacuolar targeting of soluble proteins to LV and PSV is believed to be a receptor-mediated process involving the interaction between the vacuolar sorting determinant (VSDs) of cargo proteins and VSR proteins (Jiang and Rogers, 2003). So far, no conserved VSDs have been identified in the primary sequence of storage proteins. Although GmSBP2/S64, PsSBP, and VfSBPL contain three hydrophobic amino acid residues similar to VSD of phaseolin, GmSBP1 does not possess a hydrophobic C-tail. Therefore, putative VSDs for SBPs (including the VrSBP1) remain to be investigated.
The pea SBPs, PsSBPs, are believed to reach PSVs via the Golgi-derived DVs in developing peas (Wenzel et al., 2005). Similarly, the VrSBP1 identified in this study probably reached the PSVs via similar Golgi-derived DVs because VrSBP1 antibodies labelled both Golgi apparatus and DVs in developing mung bean cotyledons (Fig. 6). In addition, VrSBP1 antibodies also labelled the limiting membranes of novel organelles of 200–500 nm in size in developing mung bean cotyledons (Fig. 7). Since VrSBP1 proteins were found in Golgi, DVs, PSV tonoplast, and the novel vesicles of 200–500 nm in size, these novel organelles would probably serve as the intermediate PVCs for VrSBP1 proteins on their path from Golgi to PSVs via DVs in developing mung bean cotyledons, but final proof must come from future experimental evidence. Such PVCs for PSVs in developing mung bean cotyledons are morphologically distinct from the multivesicular nature of PVCs/MVBs identified in tobacco BY-2 cells (Tse et al., 2004), developing Arabidopsis seeds (Otegui et al., 2006), and germinating mung bean seeds (Wang et al., 2007). Future studies are needed to address the biochemical nature of these PVCs for PSVs that are defined by the VrSBP1 in developing mung bean cotyledons.
Supplementary data
Supplementary Fig. S1, Alignment of VrSBP1 and GmSBPs amino acid sequences, can be found at JXB online.
Supplementary Material
Acknowledgments
This work was supported by grants from the Research Grants Council of Hong Kong (CUHK4307/03M. CUHK4580/05M, CUHK488707, and CUHK465708), NSF of China (30529001), and 863 Program of China (2007AA02Z102), CUHK Scheme B and C, and UGC-AoE to L Jiang.
Glossary
Abbreviations
- SBP
sucrose binding protein
- TIP
tonoplast intrinsic protein
References
- Braun H, Czihal A, Shutov AD, Baumlein H. A vicilin-like seed protein of cycads: similarity to sucrose-binding proteins. Plant Molecular Biology. 1996;31:35–44. doi: 10.1007/BF00020604. [DOI] [PubMed] [Google Scholar]
- Castillo J, Rodrigo MI, Marquez JA, Zuniga A, Franco L. A pea nuclear protein that is induced by dehydration belongs to the vicilin superfamily. European Journal of Biochemistry. 2000;267:2156–2165. doi: 10.1046/j.1432-1327.2000.01229.x. [DOI] [PubMed] [Google Scholar]
- Contim LA, Waclawovsky AJ, Delu-Filho N, Pirovani CP, Clarindo WR, Loureiro ME, Carvalho CR, Fontes EP. The soybean sucrose binding protein gene family: genomic organization, gene copy number and tissue-specific expression of the SBP2 promoter. Journal of Experimental Botany. 2003;54:2643–2653. doi: 10.1093/jxb/erg301. [DOI] [PubMed] [Google Scholar]
- Elmer A, Chao W, Grimes H. Protein sorting and expression of a unique soybean cotyledon protein, GmSBP, destined for the protein storage vacuole. Plant Molecular Biology. 2003;52:1089–1106. doi: 10.1023/a:1025483809791. [DOI] [PubMed] [Google Scholar]
- Grimes HD, Overvoorde PJ, Ripp K, Franceschi VR, Hitz WD. A 62-kD sucrose binding protein is expressed and localized in tissues actively engaged in sucrose transport. The Plant Cell. 1992;4:1561–1574. doi: 10.1105/tpc.4.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim U, Wang Q, Kurz T, et al. Expression patterns and subcellular localization of a 52 kDa sucrose-binding protein homologue of Vicia faba (VfSBPL) suggest different functions during development. Plant Molecular Biology. 2001;47:461–474. doi: 10.1023/a:1011886908619. [DOI] [PubMed] [Google Scholar]
- Jauh GY, Phillips TE, Rogers JC. Tonoplast intrinsic protein isoforms as markers for vacuolar functions. The Plant Cell. 1999;11:1867–1882. doi: 10.1105/tpc.11.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Phillips TE, Rogers SW, Rogers JC. Biogenesis of the protein storage vacuole crystalloid. Journal of Cell Biology. 2000;150:755–770. doi: 10.1083/jcb.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Rogers JC. Integral membrane protein sorting to vacuoles in plant cells: evidence for two pathways. Journal of Cell Biology. 1998;143:1183–1199. doi: 10.1083/jcb.143.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Rogers JC. Sorting of lytic enzymes in the plant Golgi apparatus. Annual Plant Reviews. 2003;9:114–140. [Google Scholar]
- Lam SK, Siu CL, Hillmer S, Jang S, An G, Robinson DG, Jiang L. Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. The Plant Cell. 2007;19:296–319. doi: 10.1105/tpc.106.045708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA. The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. The Plant Cell. 2006;18:2567–2581. doi: 10.1105/tpc.106.040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Chao WS, Grimes HD. A plasma membrane sucrose-binding protein that mediates sucrose uptake shares structural and sequence similarity with seed storage proteins but remains functionally distinct. Journal of Biological Chemistry. 1997;272:15898–15904. doi: 10.1074/jbc.272.25.15898. [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Frommer WB, Grimes HD. A soybean sucrose binding protein independently mediates nonsaturable sucrose uptake in yeast. The Plant Cell. 1996;8:271–280. doi: 10.1105/tpc.8.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Grimes HD. Topographical analysis of the plasma membrane-associated sucrose binding protein from soybean. Journal of Biological Chemistry. 1994;269:15154–15161. [PubMed] [Google Scholar]
- Paris N, Rogers SW, Jiang L, Kirsch T, Beevers L, Phillips TE, Rogers JC. Molecular cloning and further characterization of a probable plant vacuolar sorting receptor. Plant Physiology. 1997;115:29–39. doi: 10.1104/pp.115.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedra JHF, Delu-Filho N, Pirovani CP, Contim LAS, Dewey RE, Otoni WC, Fontes EPB. Antisense and sense expression of a sucrose binding protein homologue gene from soybean in transgenic tobacco affects plant growth and carbohydrate partitioning in leaves. Plant Science. 2000;152:87–99. [Google Scholar]
- Ripp KG, Viitanen PV, Hitz WD, Franceschi VR. Identification of membrane protein associated with sucrose transport into cells of developing soybean cotyledons. Plant Physiology. 1988;88:1435–1445. doi: 10.1104/pp.88.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SW, Burks M, Rogers JC. Monoclonal antibodies to barley aleurain and homologs from other plants. The Plant Journal. 1997;11:1359–1368. doi: 10.1046/j.1365-313x.1997.11061359.x. [DOI] [PubMed] [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. The Plant Cell. 2004;16:672–693. doi: 10.1105/tpc.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li Y, Lo SW, Hillmer S, Sun SS, Robinson DG, Jiang L. Protein mobilization in germinating mung bean seeds involves vacuolar sorting receptors and multivesicular bodies. Plant Physiology. 2007;143:1628–1639. doi: 10.1104/pp.107.096263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D, Schauermann G, von Lupke A, Hinz G. The cargo in vacuolar storage protein transport vesicles is stratified. Traffic. 2005;6:45–55. doi: 10.1111/j.1600-0854.2004.00243.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.