Abstract
MADS-box genes have been shown to play a role in the formation of fruits, both in Arabidopsis and in tomato. In peach, two C-class MADS-box genes have been isolated. Both of them are expressed during flower and mesocarp development. Here a detailed analysis of a gene that belongs to the PLENA subfamily of MADS-box genes is shown. The expression of this PLENA-like gene (PpPLENA) increases during fruit ripening, and its ectopic expression in tomato plants causes the transformation of sepals into carpel-like structures that become fleshy and ripen like real fruits. Interestingly, the transgenic berries constitutively expressing the PpPLENA gene show an accelerated ripening, as judged by the expression of genes that are important for tomato fruit ripening. It is suggested that PpPLENA might interfere with the endogenous activity of TAGL1, thereby activating the fruit ripening pathway earlier compared with wild-type tomato plants.
Keywords: C-type MADS-box genes, fruit ripening, gene expression, peach, PpPLENA gene, Prunus persica, TAGL1 gene, transgenic tomato
Introduction
The existence of different Arabidopsis mutants producing fruits with developmental defects has made it possible to gain significant information regarding patterning and post-fertilization development of the fruit (Dinneny and Yanofsky, 2004; Dinneny et al., 2005; Alonso-Cantabrana et al., 2007). In particular, the use of genetic tools has allowed researchers to demonstrate that many transcription factor encoding genes are involved in the development of the pod dehiscence zone (Dinneny et al., 2005; Ostergaard et al., 2006; Alonso-Cantabrana et al., 2007).
Fleshy fruits have been extensively studied due to their intrinsic economical value. However, those studies have mostly dealt with late stages of fruit development and various aspects of the ripening process (i.e. softening, colour, flavour, etc.). Also the genetic and molecular networks involved in the control of ripening have been extensively studied, since the availability of a command to start and regulate ripening would be of immense economic value. To the latter purpose, it has been found that in climacteric fruits (e.g. tomato) the inability to either synthesize or respond to ethylene results in the inability of the fruit to ripen (Giovannoni, 2004), while no such signal has been found to date in the case of non-climacteric fruit.
In general, the number of available mutants for studying fleshy fruits is limited since plants producing defective fruits were thrown away because they had no commercial value. Only recently have such mutants started to be properly evaluated and saved, as exemplified by the fleshless berry (flb) mutant of grape (Fernandez et al., 2006). However, tomato represents an exception, and many different mutants have been deposited in dedicated collections (the University of California at Davis has a very important one: http://tgrc.ucdavis.edu).
The characterization of tomato mutants unable to ripen their fruits has confirmed that ethylene plays an important role, though not an exclusive one. There are mutants [i.e. nor, rin, and cnr (Tigchelaar et al., 1978; Lincoln and Fisher, 1988; Thompson et al., 1999)] whose fruits do not produce ethylene and are unable to ripen following treatment with the hormone, even though they have a normal signal transduction machinery for ethylene. This finding suggested that factors, other than ethylene, may play fundamental roles in the ripening process.
In particular, the nor (non-ripening) tomato has a mutation that affects a gene encoding a NAC-type [NO APICAL MERISTEM (NAM/ATAF1/CUC2)] transcription factor (Giovannoni et al., 2004), while the cnr (colorless non-ripening) mutant phenotype has been shown to be the consequence of a natural epigenetic mutation that causes a dramatic down-regulation of an SBP-box transcription factor-encoding gene (Manning et al., 2006). In the case of the rin (ripening inhibitor) mutant, it has been demonstrated that the wild-type RIN gene codes for a MADS-box protein (Vrebalov et al., 2002). The latter finding is particularly interesting since it shows that MADS-box genes may play a significant role in the development of fleshy fruits, besides that already evidenced for the dry fruit of Arabidopsis (Roeder and Yanofsky, 2005; Balanzá et al., 2006; Seymour et al., 2008). Homologues of the C-type gene have been found to be expressed in fruits of tomato (Pnueli et al., 1994), grape (Boss et al., 2001), strawberry (Rosin et al., 2003), and others, thus suggesting a possible involvement in fruit development.
In core eudicots, two lineages of C-type MADS-box genes can be found: the AGAMOUS lineage, for which the Arabidopsis AGAMOUS and the snapdragon FARINELLI genes are the best known representatives; and the PLENA lineage which also includes, besides the snapdragon PLENA gene, the intensively studied Arabidopsis SHATTERPROOF (1 and 2) genes (Davies et al., 1999; Liljegren et al., 2000; Favaro et al., 2003; Kramer et al., 2004). In particular, PLENA has been shown to specify the C-function in snapdragon, as AGAMOUS does in Arabidopsis, while the FARINELLI gene appears to be mostly involved in stamen development and pollen fertility (Davies et al., 1999). Recently, it has been shown that both AGAMOUS/FARINELLI (Martin et al., 2006) and PLENA/SHATTERPROOF (Tani et al., 2007) orthologues are expressed in peach flower and fruit. In this work, the possible role played by a peach PLENA-like gene named PpPLENA in carpel specification and fruit development has been studied.
Materials and methods
Plant material
Plants of Prunus persica (L.) Batsch cv. Redhaven were grown in a field near Padua. Fruits at various stages of development [S1, S2, S3I, S3II, S4I, and S4II; see Zanchin et al. (1994)], corresponding to 40, 65, 85, 95, 115, and 120–125 d after full bloom, respectively) were collected, frozen in liquid nitrogen, and stored at –80 °C for subsequent use.
Tomato plants belonging to the cultivar ‘Florida Petite’ were used in this work. Seeds were obtained from the Tomato Growers Supply Company, Fort Myers, FL, USA, and plants were grown under standard greenhouse conditions. Fruits at various stages of development (mature green, breaker, and red ripe; see Alba et al., 2005) were collected, frozen in liquid nitrogen, and stored at –80 °C for subsequent use.
RNA extraction and expression analysis
Total RNA was extracted from tissue samples according to Chang et al. (1993). RNA yield and purity were checked by means of UV absorption spectra, whereas RNA integrity was ascertained by electrophoresis in agarose gel.
A 10 μg aliquot of total RNA was pre-treated with 1.5 U of DNase I (Amplification Grade, Invitrogen, Carlsbad, CA, USA). The first-strand cDNA was synthesized from 3 μg of the DNase I-treated RNA by means of the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA), using random hexamers as primers.
Primer sequences for the selected genes are listed in Supplementary Table S1 available at JXB online. The oligonucleotides DZ79 (5′-TGACCTGGGGTCGCGTTGAA-3′, sense) and DZ81 (5′-TGAATTGCAGAATCCCGTGA-3′, antisense), annealing to the internal transcribed spacer of the rRNA, were used to amplify the internal standard with peach samples, whereas oligonucleotides ACT-FOR (AGGCACCCCTTAATCCCAAG) and ACT_REV (AAGCACAGCCTGGATAGCAAC), annealing to actin accession no. U60480, were used with tomato tissues. Reactions were carried out in a final volume of 25 μl containing 5 ng of cDNA, 5 pmol of each primer, and 12.5 μl of the 2× SYBR® Green PCR master mix (Applied Biosystems), according to the manufacturer's instructions. PCR was carried out with the Gene Amp® 7500 Sequence Detection System (Applied Biosystems) for 10 min at 95 °C and then for 40 cycles as follows: 95 °C for 15 s, 60 °C for 15 s, and 65 °C for 34 s. The obtained CT values were analysed by means of the Q-gene software by averaging three independently calculated normalized expression values for each sample. Expression values are given as the mean of the normalized expression values of the triplicates, calculated according to equation 2 of the Q-gene software (Muller et al., 2002). The numerical values obtained from these calculations were transformed into graphics by means of the GraphPad Prism software (GraphPad Software, San Diego, CA, USA).
Cloning of peach MADS cDNAs and isolation of genomic clones
Several MADS cDNAs had been isolated from a peach expressed sequence tag (EST) collection prepared in the authors' laboratory and used for a study of peach softening (Trainotti et al., 2003), but also to carry out preliminary expression studies of a number of transcription factor-encoding genes. Based on those analyses, cognate longer cDNAs were purchased from the GDR (Genome Database for Rosaceae, http://www.bioinfo.wsu.edu/gdr/, now at Washingston State University but then at Clemson University) and fully sequenced on both strands. The sequencing results revealed that both clone PP_Lea0011L16f (accession no. BU042190, unigene Ppe.1710) and PP_Lea0020K12 (accession no. BU044838.1, unigene Ppe.2689) were full-length cDNAs, and corresponded to snapdragon FARINELLI and PLENA, respectively.
The cognate genomic clones λMADS-462-13 and λMADS-794-4 were isolated from a library constructed by the cloning of peach DNA partially digested with MboI into the BamHI site of the λEMBL3 SP6/T7 vector. The PP_Lea0011L16 and PP_Lea0020K12 cDNA clones were used as probes to screen the library following standard procedures (Sambrook et al., 1989). DNAs from the purified λ clones were extracted with a commercial kit (Qiagen), digested with HindIII and, after electrophoresis and blotting, probed again with the corresponding cDNAs. The hybridizing bands were subcloned in the pGEM 7Zf+ (Promega) plasmid vector and fully sequenced on both strands. The sequences have been deposited in the GenBank database with the following accession numbers: FJ188413 for PpPLENA and FJ184275 for PpFAR.
DNA sequencing and analysis
DNA sequencing was performed at the University of Padua sequencing facility (CRIBI) using a PCR-based dideoxynucleotide terminator protocol and automated sequencers (Applied Biosystems). Sequence manipulations, analyses, and alignments were performed using the ‘Lasergene’ software package (DNASTAR).
Transformation of tomato
The peach MADS 794 cDNA was cloned into the pBin-AR vector (Hoefgen and Willmitzer, 1988). The resulting binary plasmid was inserted in Agrobacterium tumefaciens (strain LBA4404) cells that were then used to transform tomato according to Fillati et al. (1987).
Kanamycin-resistant plants have been confirmed for the presence of the transgene by means of both PCR and Southern analysis.
Microscopy and in situ expression analyses
Tomato flower buds were observed without any treatment under low pressure conditions by means of environmental scanning electron microscopy (ESEM) at the CUGAS facilities (University of Padua).
For the in situ expression analysis, pre-anthesis floral buds, closed flowers, and open flowers (cut in two half) were fixed and embedded in paraffin according to Brambilla et al. (2007). The digoxigenin-labelled antisense mRNA probes, derived from sequences downstream of the MADS box and the K-box of the two peach genes (PpFAR, MADS 462 and PpPLE, MADS 794), were generated using an in vitro transcription kit (Roche Diagnostics GmbH, Mannheim, Germany). The region of PpFAR (MADS 462) cDNA from nucleotides 675 to 1045 was used as a template and was amplified using the following primers: TAATACGACTCACTATAGGGAGAGAGTTGGAGGAACTTG and GAGATCATGCAGTCTCAGCC. Likewise, the region of PpPLE (MADS 794) cDNA from nucleotides 791 to 1076 was used as a template and was amplified using the following primers: TAATACGACTCACTATAGGGTGAGAACATTGAGAAGCTGG and GAGGGCACAACAGCAGCAAAC. When compared with the Wibur–Lipman algorithm, these two DNA regions have a 40.5% similarity, thus the hybridization signals observed in the high stringency in situ experiments can be considered gene specific.
Hybridization and immunological detection were performed as described by Brambilla et al. (2007) with minor modifications. The hybridization was carried out at 45 °C overnight. The detection was performed using the Dig-detection kit (Roche Diagnostics).
Results
Characterization of two peach C-type MADS-box genes
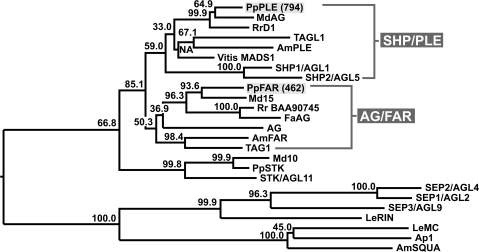
Two peach C-class MADS-box cDNAs were obtained as described in Materials and methods and, in order to characterize them in detail, the predicted protein sequences have been compared with C-class proteins identified in other species (Fig. 1). This analysis showed that one gene belongs to the AGAMOUS/FARINELLI lineage while the other is part of the SHATTERPROOF/PLENA clade.
Fig. 1.
Phylogenetic tree of several plant MADS-box proteins. The peach proteins have been highlighted by a grey background. A peach MADS-box protein highly similar to Arabidopsis seedstick has also been included (PpSTK: ABQ85556). Protein sequences from other plants have been retrieved from public databases. Their GenBank accession numbers are as follows: Malus×domestica (apple), ‘MdAG’ AF401637, ‘Md10’ CAA04324, and ‘Md15’ CAC80858; Rosa rugosa, ‘RrD1’ AB025643, and ‘Rr BAA90745’; Solanum lycopersicum (tomato), ‘AGL1’ AAM33101, ‘TAG1’ AAM33099, ‘Le Rin’ AAM15775, and ‘Le MC’ AF448521; Antirrhinum majus (snapdragon), ‘AmPLE’ AAB25101, ‘AmFAR’ CAB42988, and ‘AmSQUA’ CAA45228; Vitis vinifera, ‘Vitis MADS 1’ AAK58564; Fragaria×ananassa, ‘FaAG’ AAD45814; Arabidopsis, ‘AG’ NP_567569, ‘SHP1/AGL1’ NP_191437, ‘SHP2/AGL5’ NP_850377, ‘STK/AGL11’ NP_001078364, ‘SEP1/AGL2’ AAA32732, ‘SEP2/AGL4’ AAA32734, ‘SEP3/AGL9’ AAB67832, and ‘AP1’ CAA78909.
The genomic clones encoding the cognate genes were also isolated from a genomic library, and their structure was determined and compared with that of other C-type genes (Supplementary Fig. S1 at JXB online). The two Arabidopsis SHATTERPROOF genes have a structure that sets them apart from the other C-type genes considered in this comparison. In particular, they include eight (SHP1) and seven (SHP2) exons, respectively, while all the other C-type genes contain nine exons. The last exon in the two SHP genes is composed of two large regions that consist of coding and 3′-untranslated region (UTR) sequences, respectively, while the last exon of all the other genes is formed by a 3′-UTR sequence, with only a few base pairs of coding DNA. Finally, the second intron is significantly smaller in the two SHP genes compared with the other genes and lacks the conserved regulatory sequences (the aAGAAT and CCAATCA boxes). Therefore, the gene belonging to the AGAMOUS/FARINELLI lineage was named PpFARINELLI (PpFAR), while the gene belonging to the PLENA/SHATTERPROOF lineage was named PpPLENA (PpPLE).
Expression profile of the peach C-type MADS-box genes
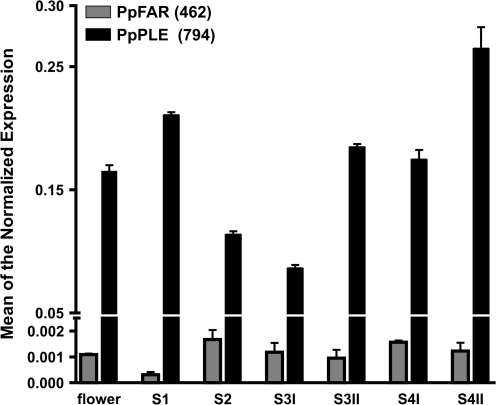
The expression profile of the two genes was determined in peaches at various stages of development by quantitative RT-PCR. As shown in Fig. 2, the expression of PpFAR was generally low compared with that of the PpPLE gene and, except for an increase from stage S1 to stage S2, it did not show relevant variations afterwards.
Fig. 2.
Relative expression profiles of two peach C-type MADS-box genes in flower and fruit. Grey bars represent the values obtained for PpFAR (462), while black bars indicate those of PpPLE (794). Values (means of the normalized expression) have been obtained by real-time qRT-PCR analyses. Stages S1–S4II encompass the development (S1–S3I) and ripening (S3II–S4II) of peach fruits. Bars are the standard deviations from the means.
In contrast, the amount of transcript of PpPLE was high in very young fruits (S1) and decreased afterwards until stage S3I, which corresponds to the beginning of the second stage of fast fruit growth. At the pre-climacteric stage (i.e. S3II), the gene expression showed a relevant increase and remained high throughout the ripening process with a maximum at the stage of full ripening (i.e. S4II).
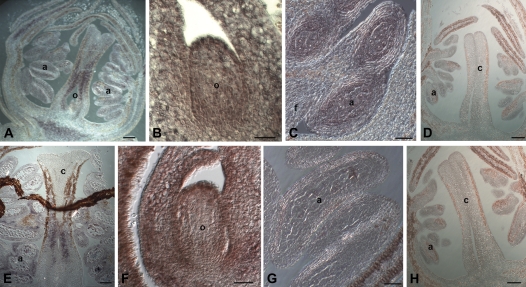
To investigate the expression profile in more detail, in situ hybridization was performed using pre-anthesis flower buds. The analysis has shown that both genes are expressed only in the third and fourth whorls, with overlapping patterns, as shown in Fig. 3A and E. PpPLE and PpFAR are both expressed in the anthers and in the developing pistil. In the stamen in particular, the two genes are expressed at the level of tapetum and sporogenous tissue, whereas they are not expressed in the filament and connecting tissue (Fig. 3C, G).
Fig. 3.
In situ analysis of PpFAR and of PpPLE expression in peach flower buds. Panels in the top half of the figure show hybridization with a PpFAR antisense probe (A–C) and sense probe (D), while those in the bottom half show hybridisation with a PpPLE antisense probe (E–G) and sense probe (H). Scale bars in A, D, F and I represent 100 μm; in B and G, 20 μm; and in C and E, 50 μm. o, ovule; c, carpel; a, anther; f, filament.
In pistil, PpPLE and PpFAR are expressed in the placenta, in the transmitting tract of the style (Fig. 3A, E), and in the ovule (Fig. 3B, F).
Analysis of tomato plants expressing a peach C-type MADS-box gene
In peach fruit, the molecular ripening starts at the S3II pre-climacteric stage (Trainotti et al., 2003), therefore the increasing expression of the PpPLE gene in the fruit mesocarp through stages S3II–S4II suggested that it might play some role in the development of the ripening process. As peach is a woody and recalcitrant species, the fleshy fruit-producing tomato was used to obtain information about the activity of the PpPLE gene.
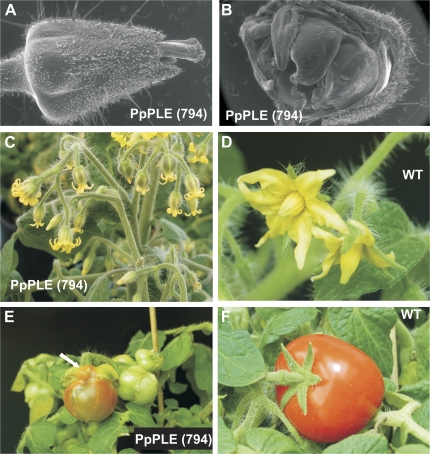
Transformation of tomato yielded 13 different transgenic plants containing the PpPLE coding region under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Interestingly, a mutated phenotype could only be observed at the level of flowers. Three lines produced flowers and fruits that were similar to those of the wild type (Fig. 4D, F). Only one such line produced seeds that yielded kanamycin-resistant plants similar to the wild type. Eight primary transformants produced flowers with a mild mutated phenotype that consisted mostly of a calyx where the various sepals tended to be joined together to various degrees throughout their subsequent development (Fig. 4C). The fused calyx was, however, partly opened at the blossom end, thus allowing a limited anthesis to be visible (Fig. 4C). Four of the above mild lines produced seeds that, in two cases, yielded plants with a stronger phenotype. The remaining transgenic clones showed a particularly severe phenotype, with flower buds that did not open at anthesis because the sepals formed a tube-like structure that allowed only the style/stigma to become visible (Fig. 4A). The reproductive organs were present in all transformants, although in the case of the most severe phenotype the stamens looked partially deformed (Fig. 4B), probably due to mechanical constraints. Later during development, the fused calyx was partially opened by the outgrowth of the ovary-derived fruit which ripened normally (Fig. 4E) like that of the wild-type plants (Fig. 4F), although seeds were never produced and plants had to be propagated vegetatively. Contrary to what occurred in wild-type plants where the sepals maintained a leaf-like structure throughout the entire life of the fruit (Fig. 4F), in transgenic plants with a strong phenotype the sepals developed a fleshy structure and became reddish (Fig. 4E, white arrow), thus behaving like ectopic fruits.
Fig. 4.
Tomato plants constitutively expressing the peach PpPLE (794) cDNA. (A) An ESEM (environmental scanning electron microscopy) picture of a transgenic flower bud with sepals completely fused up to the blossom end of the calyx from which only the style/stigma emerges. (B) Partial removal of the fused calyx permits the partially deformed anthers to be seen. (C) An inflorescence of a transgenic line with a mild phenotype. As in A, the calyx is fused, although not as far as the top, thus allowing the petal tips to become visible at anthesis. For comparison, a wild-type flower is shown in D. Transgenic fruits at various stages of development are shown in E, where a white arrow indicates the fleshy sepals undergoing ripening. A wild-type ripe fruit with leafy sepals still attached is shown in F.
Expression profiles of genes related to tomato fruit ripening
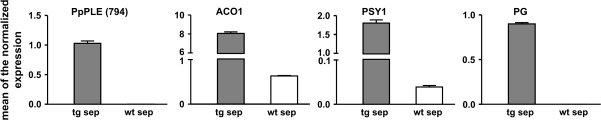
A molecular analysis comparing wild-type and transgenic fleshy sepals confirmed the morphological observations. The selected genes are usually regarded as markers of tomato fruit ripening: ACO1 codes for the ACC oxidase involved in the synthesis of climacteric ethylene (Hamilton et al., 1990; Köck et al., 1991), PSY1 codes for the phytoene synthase exclusively expressed in tomato chromoplasts (Fraser et al., 1994; Bramley 2002), while PG encodes the endopolygalacturonase highly expressed during softening (DellaPenna et al., 1986; Bird et al., 1988). In the red fleshy sepals where the peach PpPLE gene is expressed (Fig. 5), all three marker genes showed a higher transcript amount compared with mature control sepals (Fig. 5).
Fig. 5.
Relative expression profiles of ripening-related genes in ripe fleshy sepals of tomato plants overexpressing the peach PpPLE cDNA (794). Grey bars represent the values obtained for transgenic tomato sepals, while white bars indicate those of the wild type. The analysed genes are: PpPLE (794) (peach PpPLE cDNA), ACO1 (climacteric ACC oxidase), PSY1 (tomato chromoplast phytoene synthase), and PG (tomato ripening endopolygalacturonase). Values (means of the normalized expression) have been obtained by means of real-time qRT-PCR. Bars are the standard deviations from the means.
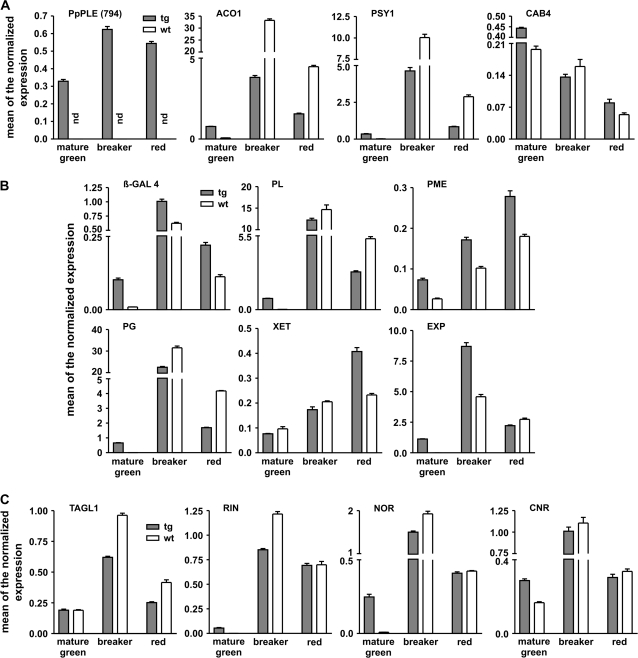
The transgenic fruits expressing the PpPLE gene (Fig. 6A) ripened normally, and this was also confirmed by the expression of the ACO1 and PSY1 genes previously used to analyse the fleshy sepals. However, very low amounts of both ACO1 and PSY1 transcripts could already be detected in mature green transgenic fruits, in contrast to controls (Fig. 6A). This finding was a surprise since the mature green tomatoes were sampled by visually checking their colour and by considering the days from anthesis. Therefore, the expression of a CAB gene encoding a chlorophyll a/b-binding protein (Pichersky et al., 1987) was also measured. This protein is an important component of photosystem II, and can therefore be regarded as a good indicator of the ‘green’ state of the fruit. Interestingly, the amount of the CAB transcripts was higher in the mature green transgenic fruits than in those of the mature green wild type (Fig. 6A). However, while the expression of the CAB gene decreased slowly during the passage to the breaker stage in wild-type fruits, in transgenic fruits the gene expression showed a sharp drop and was more than halved at the breaker stage.
Fig. 6.
Relative expression profiles of ripening-related genes in fruit of tomato plants either wild type or overexpressing the peach PpPLE cDNA (794). Grey bars represent values obtained for the transgenic fruit, white bars indicate those of the wild type. (A) pPLE (794) (peach PpPLE cDNA), ACO1 (climacteric ACC oxidase), PSY1 (tomato chromoplasts phytoene synthase), CAB (a chlorophyll a/b-binding protein-encoding gene used as a marker of the fruit ‘green’ state). (B) Softening-related genes: β-GAL (β-galactosidase), PL (pectate lyase), PME (pectin methyl esterase), PG (endopolygalacturonase), XET (xyloglucan endotransglycosylase), EXP (expansin). (C) Tomato transcription factor-encoding genes: TAGL1 (a C-type MADS-box gene), RIN (a SEPALLATA-type MADS-box gene), NOR (a NAM-like gene), and CNR (an SBP-box gene). Values (means of the normalized expression) have been obtained by means of real-time qRT-PCR. Bars are the standard deviations from the means of three independent replicates.
Transgenic ripe fruits were softer than those of the wild type (data not shown), so the expression was analysed for a number of tomato genes involved in the softening process (reviewed in Brummell and Harpster, 2001, and references therein), and whose orthologues are also active in the softening of peaches (Trainotti et al., 2003). In particular, the selected genes encode proteins specifically involved (i) in the degradation of the lateral branches of parietal polysaccharides [i.e. β-galactosidase (β-GAL)]; (ii) in the degradation of pectins [i.e. pectate lyase (PL), pectin methyl esterase (PME), and PG]; and (iii) in the destabilization of the cellulose–hemicellulose network [i.e. expansin (EXP) and xyloglucan endotransglycosylase (XET)].
The pattern of β-GAL gene expression was the same in both control and transgenic fruits, with a maximum at the breaker stage, although the amount of transcript was generally higher in the transgenic fruits (Fig. 6B). However, at the mature green stage, transcripts were barely detectable in control fruits and were much higher in transgenic fruits.
In control fruits, PL transcripts were not detected at the mature green stage, reached a maximum at the breaker stage, and decreased to a much lower amount in red fruits (Fig. 6B). In transgenic plants, the PL transcripts were significantly lower in red fruits compared with controls, but transcription of the PL gene started earlier in transgenics, and transcripts were already present in the mature green fruit (Fig. 6B).
The demethylating activity of PME is preliminary to the pectin degradation carried out by PG. In control fruits, a very low amount of PME transcript was present at the mature green stage, and a continuous increase occurred afterwards up to a maximum in red fruits (Fig. 6B). A similar pattern of expression was observed in transgenic fruits, although in this case the amount of transcript was always much higher compared with control fruits.
The pattern of expression of PG was similar in control and transgenic fruits, although the latter showed a low amount of transcripts in mature green fruits not observed in the corresponding control fruits (Fig. 6B).
XET activity is involved in the rearrangement of xyloglucans, therefore it can cause a destabilization of the cell walls. In control fruits, XET transcripts were already high in mature green fruits and increased further during ripening, up to a maximum in red fruits (Fig. 6B). In transgenic fruits, XET expression was comparable with that of the wild type until the breaker stage, but reached much higher values at the red stage.
EXP mRNA was already present in mature green transgenic fruits; subsequently its level became extremely high in breaker fruits, and dropped to lower values in red fruits (Fig. 6B). In control plants, no EXP transcripts were detected in mature green fruits. Afterwards, the EXP gene expression showed a maximum at the breaker stage and decreased in red fruits.
The expression profile was also determined for a number of transcription factor-encoding genes, among them TAGL1 (Busi et al., 2003), which is the tomato orthologue of the PpPLE gene. Overall, the pattern of expression was the same, with a maximum in breaker fruit, although a smaller amount of transcript was present in both breaker and red transgenic fruits compared with control fruits (Fig. 6C).
The RIN, NOR, and CNR tomato genes have been shown to control fruit ripening in such a strict manner that, when mutated, each of them causes a block of the ripening process. Under the present experimental conditions, all three genes showed a similar expression pattern with a maximum at the breaker stage followed by a decrease at the red stage (Fig. 6C). However, a very important difference was found at the mature green stage where, in the case of both RIN and NOR, some expression was present in transgenic fruits in contrast to an undetectable expression in control fruits, while in the case of the CNR gene the expression was higher in transgenic compared with control fruits.
Discussion
Recently, the sequences of two peach cDNAs that have been regarded as the orthologues of AGAMOUS (PpAG1, Martin et al., 2006) and SHATTERPROOF (PPERSHP, Tani et al., 2007) have been published. The same cDNAs had also been independently obtained in the authors' laboratory from an EST cDNA library (LT and GC, unpublished data) and used as a starting point for the present study. In particular, the cognate genes have been isolated from a peach genomic library, and the resulting structure (Supplementary Fig. S1 at JXB online), together with protein alignments (Fig. 1), indicate that the PPERSHP gene (Tani et al., 2007) might actually be an orthologue of PLENA, while PpAG1 (Martin et al., 2006) appears to be orthologous to FARINELLI, reproducing to some extent the situation of Anthirrinum majus (snapdragon). Such a conclusion also appears to be supported by a comparison of the percent identity values of the C-type MADS-box proteins of Arabidopsis, snapdragon, and peach, respectively (Supplementary Table S2 at JXB online). Expression analysis and in situ hybridization demonstrated that both genes are expressed during carpel and stamen development, as expected for C-class genes, thus confirming the RT-PCR data of Tani et al (2007). In contrast, real-time experiments carried out with whole flowers (Fig. 2) seem to indicate that the PpPLENA (PpPLE) is more abundant in the carpel, even though its expression domains are overlapping with those of MADS 462 (Fig. 3, and Martin et al., 2006). Furthermore PpPLE expression increases during fruit ripening, while that of PpFAR does not. Because of this, PpPLE has been studied in more detail. Interestingly, even though it is known that peach belongs to the rosid subclass of eudicotyledons, as does Arabidopsis, while snapdragon (A. majus) falls in the asterids subclass, as does tomato, the peach genes are more similar to those of snapdragon and tomato than to those of Arabidopsis (see Supplementary data at JXB online for gene structures and proteins comparisons). The two peach C-type MADS box genes described here have a gene structure that is conserved in the single Arabidopsis AG gene and in the two snapdragon FAR and PLE genes, but differs substantially from that of the two SHP Arabidopsis genes. Recently, SHP1/SHP2 and PLE as well as AG and FAR were shown to be orthologues, respectively (Causier et al., 2005). Based on gene structure and protein similarity, peach MADS 462 can reasonably be considered to be the peach gene orthologous to both AG and FAR and thus it might also be named PpAG, as Martin et al. (2006) proposed. In contrast, MADS 794 could be orthologous to either SHP or AG. Should the first hypothesis be true, SHP should have undergone a deep structural rearrangement while MADS 794 would have retained its ancient AG-like gene structure. In contrast, should the second hypothesis be correct, MADS 794 would correspond to the second AG gene that Causier et al. (2005) have hypothesized as lost in Arabidopsis, therefore the peach gene should be named PpAG2. In this case, peach orthologues to SHP would be either not discovered yet or deleted from its genome. A similar situation (two AG genes and the apparent lack of SHP) has been described for poplar (Leseberg et al., 2006), a tree of the rosid subclass. The correct orthology assignment will be clarified when the peach genome sequence becomes available, thus allowing the analysis of syntenic loci conservation, beside the comparison of gene structure and protein similarities. Meantime, the name PpPLE is proposed for the peach MADS 794 gene.
Because of its high level of expression during the ripening of peach fruit, a construct for constitutive expression of PpPLE has been introduced in tomato. Tomato is an optimal model system for this study since it can be transformed and, like peach, it produces fleshy fruits, although peaches are drupes while tomatoes are berries. Nevertheless, the general ripening process is very similar, both of them being climacteric fruits.
The constitutive expression of the PpPLE in tomato caused a homeotic conversion of the flower first whorl organs into carpels, and the molecular data indicate that the resulting ectopic fruits also underwent normal ripening. The fact that constitutive expression of the PpPLE gene caused a homeotic conversion of the first whorl but not of the second whorl seems in agreement with the results of Causier et al. (2005) who found that ectopic expression of PLENA in Antirrhinum resulted in a conversion of sepals into carpels, while transformation of petals into male organs was less apparent.
As well as in flowers, the PpPLE gene is normally expressed in increasing amounts during ripening (i.e. stages S3II–S4II) of the peach fruit mesocarp, thus suggesting that it might also play a role in the ripening process. In the transgenic tomatoes harbouring the peach PpPLE cDNA, the expression of genes related to softening appears particularly indicative of a faster and enhanced softening process, since all the genes showed either an anticipated or an increased expression in the mature green transgenic tomatoes compared with the controls. The precocious expression of the β-GAL and EXP genes is worth mentioning. β-GAL activity renders cell walls more porous to other cell wall-degrading enzymes due to the degradation of the pectin lateral branches. EXP has the ability to destabilize the cellulose–hemicellulose network (Cosgrove, 2000), and it has been shown that tomatoes expressing an antisense transgene for EXP had reduced softening, while other clones overexpressing the same EXP cDNA yielded fruits with enhanced softening (Brummell et al., 1999).
Particularly interesting are the data relating to genes encoding transcription factors known to be involved in tomato fruit ripening. The LeMADS RIN gene has expression that is barely detectable in mature green fruits but shows a relevant increase during the ripening phase (Vrebalov et al., 2002; Bartley and Ishida, 2007). In the present conditions, the expression of RIN followed the same pattern in both cases, except for a difference in the mature green fruit where its expression was already measurable in transgenic, in contrast to wild-type, fruit.
The NOR gene has been cloned and patented (Giovannoni et al., 2004) but, to our knowledge, its pattern of expression has only been published for tomato sepals grown in vitro and induced to become fleshy and red like the real berries. In that work it was shown that NOR expression was undetectable at the mature green stage, peaked at the orange stage, and had a slight decrease at the red stage (Bartley and Ishida, 2007). In the present conditions NOR expression followed the same pattern except for an anticipated expression in the mature green transgenic berries.
The CNR gene is expressed throughout development and ripening of the tomato fruit, although the amount of transcript shows a sharp increase at the breaker stage and a subsequent decrease at lower levels in red fruits (Manning et al., 2006). In agreement with such a pattern, in the present experimental conditions CNR transcripts were present in both transgenic and control green fruits, but the amount of transcript measured in transgenic berries was ∼2-fold that in the controls. Also in the transgenic tomatoes the maximum transcript amount was observed at the breaker stage.
The general anticipated expression of all the analysed genes in the mature green transgenic berries might suggest that those fruit were actually less green, hence at a more advanced stage of maturation compared with wild-type fruit. Yet, the use of a CAB gene as a marker of the greenness state of the fruit demonstrated that the green transgenic berries had a higher amount of CAB transcripts, and could therefore be regarded as greener than those of the wild type. Interestingly, the amount of transcript of the CAB gene also showed a marked drop already in fruits at the breaker stage, thus confirming the faster ripening of the transgenic fruits.
The fact that so many different genes related to ripening showed an altered expression in transgenic fruits suggests that the genes responsible for such a change should be those for one (35S-PpPLE) or more (direct and indirect targets of 35S-PpPLE) transcription factors. However, the fact that the ripening process was not particularly changed from a qualitative point of view suggests that PpPLE might actually have modified the activity of other endogenous ‘factors’. A likely candidate might be TAGL1, the tomato orthologue of PpPLE, whose expression was first described by Busi et al. (2003). They observed TAGL1 expression in both flowers and fruits, although they only analysed fruits at early stages of development. In a more recent study, Hileman et al. (2006) showed that both TAG1 and TAGL1 transcripts could be recovered from tomato flowers and fruits, although an RT-PCR analysis demonstrated that TAGL1 has a much higher level of expression than TAG1 in both reproductive organs (i.e. stamens and carpels) and fruits, thus mimicking the situation observed in peach for the PpPLE and PpFAR genes, respectively. In the present experiments, TAGL1 showed a comparable expression pattern in both transgenic and control fruits. The proteins encoded by TAGL1 and PpPLE share a significant similarity (72%), therefore the peach protein might have the same activity as the endogenous TAGL1 protein, and this might cause a misregulation of the regulatory activity normally performed by TAGL1. Thus, the PpPLE and TAGL1 proteins might participate in the formation of the same complexes and/or compete for the same targets.
The investigation of the involvement of MADS-box genes in the development of reproductive structures has mostly dealt with their role in the formation of flowers and, to a lesser extent, with the patterning of dry fruits. In this work, it has been shown that a C-type MADS-box gene, besides its expected role in the formation of carpels, as evidenced by the homeotic conversion of sepals into carpels following its constitutive expression in tomato, is also recruited afterwards during the transformation of the carpel into a ripe fleshy fruit. This is not surprising if one considers that the overall development of a carpel represents a continuum that starts with its determination in the forming flower bud and ends with its transformation into a fruit. Therefore, a parsimonious use of the same regulatory genes, as exemplified here by the peach PLENA-like MADS-box gene, seems to be exploited by higher plants for the development of their reproductive structures.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of the oligonucleotides used in the qRT-PCR experiments.
Table S2. The amino acid sequence identities among various C-type MADS-box transcription factors (Arabidopsis, snapdragon, tomato, and peach).
Fig. S1. Comparison of the structural features of a number of C-type MADS-box genes (Arabidopsis, snapdragon, and peach).
Supplementary Material
Acknowledgments
GC dedicates this paper to the memory of A Messeri, the university professor who introduced him to the pleasure of plant science studies. The authors acknowledge financial support by the Italian Ministry of University and Research (MIUR, grants FIRB and PRIN). The authors would also like to thank Nicola Sassi for help in the isolation of the two peach genes from a genomic library and their partial characterization, and Simona Masiero and Alessia Losa for assisting with in situ hybridization experiments.
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanskley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrándiz C, Martínez-Laborda A. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development. 2007;134:2663–2671. doi: 10.1242/dev.02864. [DOI] [PubMed] [Google Scholar]
- Balanzá V, Navarrete M, Trigueros M, Ferrándiz C. Patterning the female side of Arabidopsis: the importance of hormones. Journal of Experimental Botany. 2006;57:3457–3469. doi: 10.1093/jxb/erl188. [DOI] [PubMed] [Google Scholar]
- Bartley GE, Ishida BK. Ethylene-sensitive and insensitive regulation of transcription factor expression during in vitro tomato sepal ripening. Journal of Experimental Botany. 2007;58:2043–2051. doi: 10.1093/jxb/erm075. [DOI] [PubMed] [Google Scholar]
- Bird CR, Smith CJS, Ray JA, Moureau P, Bevan MW, Bird AS, Hughes S, Morris PC, Grierson D, Schuch W. The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Molecular Biology. 1988;11:651–662. doi: 10.1007/BF00017465. [DOI] [PubMed] [Google Scholar]
- Boss PK, Vivier M, Matsumoto S, Dry IB, Thomas MR. A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Molecular Biology. 2001;45:541–553. doi: 10.1023/a:1010634132156. [DOI] [PubMed] [Google Scholar]
- Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, Kater M, Colombo L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. The Plant Cell. 2007;19:2544–2556. doi: 10.1105/tpc.107.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley PM. Regulation of carotenoid formation during tomato fruit ripening and development. Journal of Experimental Botany. 2002;53:2107–2113. doi: 10.1093/jxb/erf059. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Molecular Biology. 2001;47:311–340. [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. The Plant Cell. 1999;11:2203–2216. doi: 10.1105/tpc.11.11.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi MV, Bustamante C, D'Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E. MADS-box genes expressed during tomato seed and fruit development. Plant Molecular Biology. 2003;52:801–815. doi: 10.1023/a:1025001402838. [DOI] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, Ingram R, Xue Y, Schwarz-Sommer Z, Davies B. Evolution in action: following function in duplicated floral homeotic genes. Current Biology. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. PLENA and FARINELLI: redundancy and regulatory interactions between two Anthirrinum MADS-box factors controlling flower development. EMBO Journal. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Alexander DC, Bennett AB. Molecular cloning of tomato fruit polygalacturonase: analysis of polygalacturonase mRNA levels during ripening. Proceedings of the National Academy of Sciences, USA. 1986;83:6420–6424. doi: 10.1073/pnas.83.17.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. A genetic framework for fruit patterning in Arabidopsis thaliana. Development. 2005;132:4687–4696. doi: 10.1242/dev.02062. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Yanofsky MF. Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. BioEssays. 2004;27:42–49. doi: 10.1002/bies.20165. [DOI] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky Kater MM, Colombo L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. The Plant Cell. 2003;15:2603–2611. doi: 10.1105/tpc.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Romieu C, Moing A, Bouquet A, Maucourt M, Thomas MR, Torregrosa L. The grapevine fleshless berry mutation. A unique genotype to investigate differences between fleshy and nonfleshy fruit. Plant Physiology. 2006;140:537–547. doi: 10.1104/pp.105.067488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillati JJ, Kiser J, Rose R, Comai L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology. 1987;5:726–730. [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development. Plant Physiology. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. Genetic regulation of fruit development and ripening. The Plant Cell. 2004;16:S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ, Tanksley SD, Vrebalov J, Noensie E. NOR gene for use in manipulation of fruit quality and ethylene response. 2004 US Patent No. 5,234,834 issued 13 July 2004. [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature. 1990;346:284–287. [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Molecular Biology and Evolution. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- Hoefgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Research. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck M, Hamilton A, Grierson D. Eth1, a gene involved in ethylene synthesis in tomato. Plant Molecular Biology. 1991;17:141–142. doi: 10.1007/BF00036816. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg CH, Li A, Kang H, Duvall M, Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378:84–94. doi: 10.1016/j.gene.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Fischer RL. Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiology. 1988;88:370–374. doi: 10.1104/pp.88.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Manning K, Tor M, Poole M, Hong Y, Aj Thompson, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- Martin T, Hu M, Labbé H, McHugh S, Svircev A, Miki B. PpAG1, a homolog of AGAMOUS, expressed in developing peach flowers and fruit. Canadian Journal of Botany. 2006;84:767–776. [Google Scholar]
- Muller PY, JanovjakH Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative realtime RT-PCR. Biotechniques. 2002;32:1372–1379. [PubMed] [Google Scholar]
- Østergaard L, Kempin SA, Bies D, Klee HJ, Yanofsky MF. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnology Journal. 2006;4:45–51. doi: 10.1111/j.1467-7652.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Hoffman NE, Malik VS, Bernatzky R, Tanksley SD, Szabo L, Cashmore AR. The tomato Cab-4 and Cab-5 genes encode a second type of CAB polypeptides localized in photosystem II. Plant Molecular Biology. 1987;9:109–120. doi: 10.1007/BF00015643. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. The Plant Cell. 1994;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Yanofsky MF. Fruit development in Arabidopsis. The Arabidopsis Book. 2005;52:1–50. doi: 10.1199/tab.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin FM, Aharoni A, Salentijn EMJ, Schaart JG, Boone MJ, Hannapel DJ. Expression patterns of a putative homolog of AGAMOUS, STAG1, from strawberry. Plant Science. 2003;165:959–968. [Google Scholar]
- Seymour G, Poole M, Manning K, King GJ. Genetics and epigenetics of fruit development and ripening. Current Opinion in Plant Biology. 2008;11:58–63. doi: 10.1016/j.pbi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Tani E, Polidoros AN, Tsaftaris AS. Characterization and expression analysis of FRUITFULL- and SHATTERPROOF-like genes from peach (Prunus persica) and their role in split-pit formation. Tree Physiology. 2007;27:649–659. doi: 10.1093/treephys/27.5.649. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Tor M, Barry CS, Vrebalov J, Orfila C, Jarvis MC, Giovannoni JJ, Grierson D, Seymour GB. Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiology. 1999;120:383–389. doi: 10.1104/pp.120.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigchelaar EC, McGlasson WB, Buescher RW. Genetic regulation of tomato fruit ripening. HortScience. 1978;13:508–513. [Google Scholar]
- Trainotti L, Zanin D, Casadoro G. A cell wall-oriented genomic approach reveals a new and unexpected complexity of the softening in peaches. Journal of Experimental Botany. 2003;54:1821–1832. doi: 10.1093/jxb/erg198. [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni JJ. A MADS-box gene necessary for fruit ripening at the tomato Ripening-Inhibitor (Rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- Zanchin A, Bonghi C, Casadoro G, Ramina A, Rascio N. Cell enlargement and cell separation during peach fruit development. International Journal of Plant Sciences. 1994;155:49–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.