Abstract
In all organisms, the protein machinery responsible for the replication of DNA, the replisome, is faced with a directionality problem. The antiparallel nature of duplex DNA permits the leading-strand polymerase to advance in a continuous fashion, but forces the lagging-strand polymerase to synthesize in the opposite direction. By extending RNA primers, the lagging-strand polymerase restarts at short intervals and produces Okazaki fragments1,2. At least in prokaryotic systems, this directionality problem is solved by the formation of a loop in the lagging strand of the replication fork to reorient the lagging-strand DNA polymerase so that it advances in parallel with the leading-strand polymerase. The replication loop grows and shrinks during each cycle of Okazaki-fragment synthesis3. Here, we employ single-molecule techniques to visualize, in real time, the formation and release of replication loops by individual replisomes of bacteriophage T7 supporting coordinated DNA replication. Analysis of the distributions of loop sizes and lag times between loops reveals that initiation of primer synthesis and the completion of an Okazaki fragment each serve as a trigger for loop release. The presence of two triggers may represent a fail-safe mechanism ensuring the timely reset of the replisome after the synthesis of every Okazaki fragment.
The ‘trombone model’ as proposed by Bruce Alberts3 provides an elegant model for coupling many rounds of Okazaki-fragment synthesis on the lagging strand of the replication fork to the continuous production of DNA on the leading strand. The looping back of the lagging strand onto the replisome allows both leading- and lagging-strand DNA polymerases to synthesize in the same direction and facilitates recycling of the lagging-strand DNA polymerase by virtue of its proximity to the RNA primers newly synthesized at the fork. The visualization of replication intermediates of the T4 and T7 bacteriophage replication systems by electron microscopy (EM) demonstrated the existence of replication loops and allowed a characterization of their length distributions4,5. Biochemical studies revealed a number of molecular scenarios that explain how formation and release of replication loops may be regulated6–14. However, no dynamic characterization has been reported and the timeline controlling the various enzymatic activities at the fork is not entirely understood.
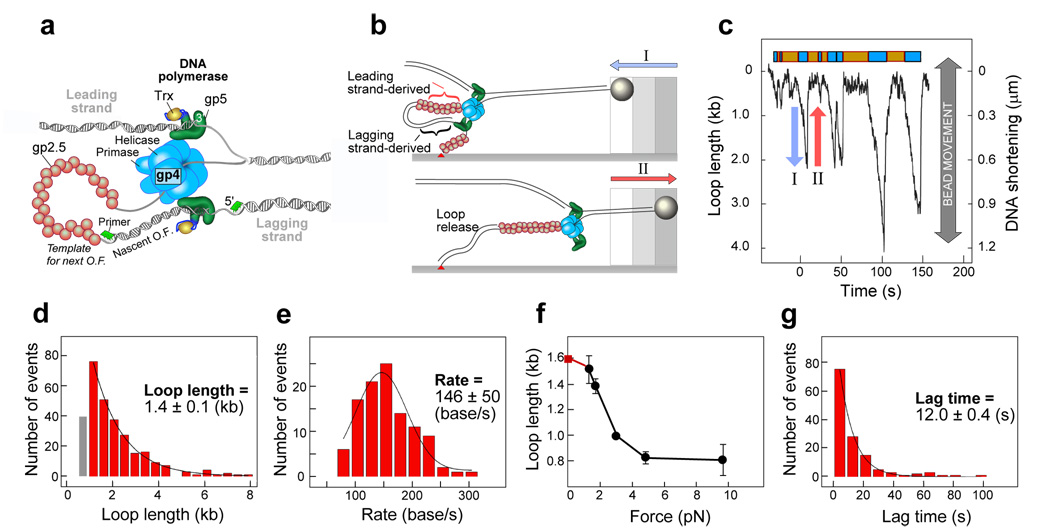
In this study, we report the reconstitution of T7 replisomes and the observation of coordinated leading- and lagging-strand synthesis at the single-molecule level. The T7 replisome consists of only four proteins and it displays all the important features of more complicated replication systems15 (Fig. 1a). These proteins are: the T7 DNA polymerase, a 1:1 complex of T7 gene 5 protein (gp5) and Escherichia coli thioredoxin processivity factor; T7 gene 4 helicase/primase protein (gp4); and T7 gene 2.5 single-stranded DNA binding protein (gp2.5). In previous work, we used single-molecule techniques to study the activity of partially assembled replisome proteins mediating only leading-strand synthesis in both T7 and Escherichia coli16–18. Here, we describe the observation of T7 replication complexes undergoing coordinated leading- and lagging-strand synthesis, allowing for kinetic characterization of many rounds of replication loop formation and release. Replication is studied at the single-molecule level by monitoring the length of individual, tethered DNA molecules. Briefly, the lagging strand of a forked-duplex λ phage DNA molecule (48.5 kilo bases (kb)) is attached via one end to the surface of a glass flow cell and the other end to a bead19 (Fig. 1b) (Supplementary Information). A laminar flow exerts a well-controlled drag force on the beads and stretches the DNA molecules. Fig. 1c shows the length of an individual DNA molecule through time while flowing a buffer containing purified gp4, T7 DNA polymerase, gp2.5, Mg2+, four dNTPs, and the ribonucleotides ATP and CTP required for primase activity13. The single-molecule trajectories show repeated cycles of DNA shortening (Fig. 1c, blue arrow) followed by rapid lengthening to the original DNA length (Fig. 1c, red arrow).
Fig. 1.
Observation of replication loops. (a) Organization of the T7 replication fork. Gene 4 protein (gp4) encircles the lagging strand and mediates both the unwinding of double-stranded DNA via its helicase domain and the synthesis of RNA primers via the primase domain. The T7 DNA polymerases are stably bound to gp4 and incorporate nucleotides on the leading and lagging strand. The DNA polymerase is a 1:1 complex of the T7 gene 5 protein (gp5) and E. coli thioredoxin (trx). The ssDNA extruded behind the helicase is coated by the ssDNA-binding protein gp2.5. A replication loop is formed in the lagging strand to allow the polymerase to synthesize in the same direction. The lagging-strand DNA polymerase initiates the replication of Okazaki fragments (O.F.) using RNA primers (green segment) synthesized by the primase domain of gp4. (b) Schematic depiction of the overall length change of tethered DNA as a result of replication loop formation and release. (c) Time course of the length of a single DNA molecule during replication (see Supplementary Information for more examples). The loop growth and lag phases are shown as cyan and orange boxes, respectively. (d) Histogram of the replication-loop length (bars; n=288) fitted with a single-exponential decay (solid line). The gray bar represents loop lengths below 1 kb that are under sampled due to noise in the length measurements. These loops were not included in the fitting of loop-size distributions. (e) Histogram of rates of loop growth (bars; n=107) fitted with a normal distribution (solid line). The rate of loop growth is determined from the slope of the DNA shortening phase. (f) Dependence of replication-loop length on stretching force (n= 108, 288, 158, 64, and 26 at forces of 1.4, 1.7, 3.0, 4.8, and 9.5 pN, respectively). The replication loop length at zero force (red square) is derived from the Okazaki fragment length as measured in bulk-phase experiments (Supplementary Information). (g) Histogram of lag times (bars; n=135) fitted with a single-exponential decay (solid line). All reported mean values are obtained using the maximum likelihood estimation method. Error bars correspond to error in fitting the rate distribution (panel (e)) with a Gaussian function and both loop-length (panels (d) and (f)) and lag-time (panel (g)) distributions with exponential decay functions.
In the presence of a saturating concentration of gp2.5 that is required to coordinate leading- and lagging-strand synthesis13,20, ssDNA is equal in length to dsDNA (Supplementary Information) excluding a scenario in which conversion between the two forms of DNA is responsible for the observed length changes. Since the bead is attached to the surface through the lagging strand, the formation of a replication loop in this strand will decrease the apparent DNA length by an amount equal to the DNA length stored in the loop. Several lines of evidence support the notion that the observed DNA shortening is a result of coupled DNA replication and loop formation: first, the loop length is comparable to that observed in EM studies. Figure 1d shows the wide distribution of loop lengths observed for 115 individual replisomes. This distribution can be described with a single-exponential distribution with a decay constant of 1.4 ± 0.1 kb, close to the average length of 2 kb as obtained in previous EM studies5. Second, inhibition of either the primase or DNA polymerase activity abolishes all DNA shortening events (Supplementary Information). Third, the average rate of DNA shortening observed during loop growth (146 ± 50 b/s; Fig. 1e) is nearly twice the rate observed for leading-strand polymerization alone under similar experimental conditions (80 bp/s) (Supplementary Information)16, consistent with the notion that loop growth is supported by both leading-and lagging-strand synthesis with a net rate that contains the contributions of the two polymerases. Fourth, we observe multiple DNA shortening events (an average of 3 ± 2 loops per replication event with 25% of replisomes displaying more than 4 loops; Supplementary Information) that occur in rapid succession on a small fraction of surfacete-thered DNA molecules (5%). This pattern indicates the presence of stably assembled and processive replisomes, as opposed to the distributive activity of replication proteins leading to isolated looping events. Fifth, loop length and the number of successive loops formed per replisome are reduced by increasing force (Fig. 1f). This observation is consistent with the prediction that the applied force will be exerted directly on the protein interactions that hold the loop together.
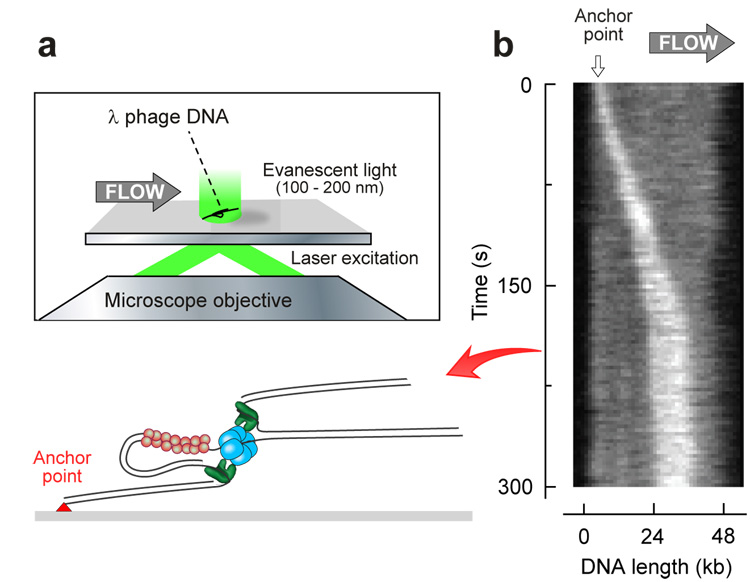
Finally, fluorescence time-lapse microscopy on individual, replicating DNA molecules demonstrates that DNA is synthesized on both the leading and the lagging strand (Fig. 2). In these experiments λ DNA templates were stained with a fluorescent, dsDNA-specific dye and the fluorescent DNA was imaged while flow-stretched and replicated by the T7 replisome. The growth of a leading-strand product, its continuous movement along DNA, and the double-stranded nature of the lagging-strand product demonstrate that both leading and lagging strands are being replicated. Further confirmation of the presence of both leading- and lagging-strand synthesis is obtained by a bulk-phase analysis of replication products obtained under identical conditions as used in the single-molecule experiments (Supplementary Information). The length of Okazaki fragments (0.8 kb) corresponds well with half of the mean loop length as measured in the single-molecule experiments (1.4 kb/2 = 0.7 kb), in agreement with the fact that half of the replication loop consists of the nascent Okazaki fragment produced by the lagging-strand DNA polymerase and the other half of ssDNA extruded by the helicase5.
Fig. 2.
Fluorescence imaging of coordinated DNA replication. (a) Schematic depiction of experimental design. Individual DNA molecules are fluorescently stained by means of intercalating dye, stretched by flow and imaged through Total Internal Reflection Fluorescence (TIRF) microscopy. (b) Kymograph of an individual DNA molecule undergoing coordinated replication (see Supplementary Movie).
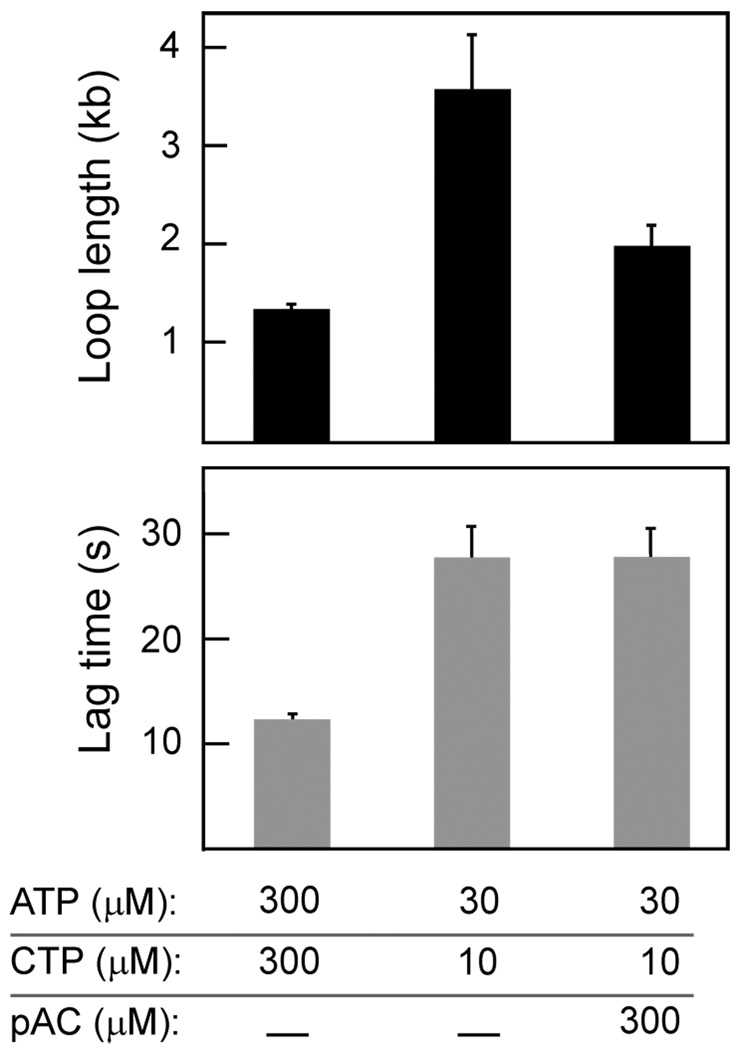
Observation of replication-loop formation and release provides us with kinetic information that is inaccessible using bulk-phase assays. The stochastic and sequential nature of the many enzymatic processes involved causes the synchronicity of a population of reactions to be lost quickly and will obscure kinetic information on the short-lived, intermediate steps. Our single-molecule experiments reveal the presence of lag times between the release of one replication loop and formation of the next, a step unobservable in bulk-phase assays. The distribution of the lag times follows a single-exponential dependence with a decay constant of 12.0 ± 0.4 seconds (Fig. 1g). The appearance of a lag time after replication-loop release suggests the requirement for intermediate steps prior to the formation of a new replication loop. At least three steps are necessary for a loop to be formed: the recognition by the primase of a priming site in the lagging strand, the synthesis of a primer, and its hand-off to the lagging-strand DNA polymerase. The importance of primer synthesis in regulating the timing of the events at the replication fork is clear from several studies that found an effect of primase activity on the size of Okazaki fragments in T7, T4, and E. coli replisomes10,12,13. However, the timing of primer synthesis and its causal relation with loop release is not established. By changing the concentration and nature of the ribonucleotides required by the primase, we can modulate the observed loop dynamics and extract a timeline of enzymatic events during the replication cycle. T7 DNA primase recognizes four sequences 5’-GGGTC, 5’-TGGTC, 5’-GTGTC, and 5’-TTGTC and requires only ATP and CTP to synthesize primers (5’-ACCC, 5’-ACCA, 5’-ACAC, and 5’-ACAA) complementary to the four nucleotides at the 5’ end of the recognition sequence21. Reducing the ATP and CTP concentrations from the optimal 300 µM to 30 and 10 µM, respectively, shows an increase in loop length from 1.4 ± 0.1 kb to 3.6 ± 0.5 kb (Fig. 3). The lag time between loops also increased significantly, from 12.0 ± 0.4 sec to 28.6 ± 2.8 sec (Fig. 3). The observation that changing the kinetics of primer synthesis alters the loop lengths as well as the length of the lag times led us to consider how the different steps in primer synthesis play a role in the timing of the events that lead to both loop release and loop formation. Primer synthesis takes place in two distinct steps22–24. First, the primase condenses ATP and CTP to form pppAC, the diribonucleotide that is present as the starting nucleotide pair in all four possible primers. Subsequently, pppAC is extended in a much slower step to a full-length, tetra-ribonucleotide primer in a sequence-dependent manner22–24. T7 DNA primase can utilize pAC and extend it efficiently using only ATP and CTP23. Therefore, by providing the coupled replication reaction with a pre-made pAC, we can bypass the requirement of the condensation step. At low ATP (30 µM) and CTP (10 µM) concentrations, the presence of 300 µM pAC nearly restored the loop length to that observed at optimal ATP and CTP concentrations (Fig. 3). The lag time, however, remains unaffected by the presence of pAC. These results demonstrate that the first step in primer synthesis, condensation of ATP and CTP to form pppAC, triggers loop release. As a consequence, the lag time has to include the slow extension step of pppAC to a full tetra-ribonucleotide.
Fig. 3.
Replication-loop dynamics depend on primase activity. Loop growth and lag times are compared for different concentrations of ribonucleotides. Replication-loop lengths and lag times were determined as described in Figure 1. For loop lengths, sample size n=288 at 300 µM ATP, 300 µM CTP; n=61 at 30 µM ATP, 10 µM CTP; and n=72 at 30 µM ATP, 10 µM CTP, 300 µM pAC. For lag times, n=135 at 300 µM ATP, 300 µM CTP; n=25 at 30 µM ATP, 10 µM CTP; and n=30 at 30 µM ATP, 10 µM CTP, 300 µM pAC.
Our observation that replication-loop length is determined by primer synthesis and previously reported dependencies of Okazaki fragment length on primase activity10,12,13 suggests a signaling mechanism by which the primase controls the timing of the cycle of enzymatic events at the fork. The observation that the average Okazaki-fragment length as observed in the bulk phase does not decrease upon introduction of pAC (Supplementary Information) demonstrates that pAC triggers loop release before the nascent Okazaki fragment is completed, explaining previous observations of ssDNA gaps in EM and bulk-phase assays10,11. Primer synthesis and loop release before completion of the Okazaki fragment will result in a length decrease of the ssDNA template available for the next Okazaki fragment. Consequently, a gradual decrease in replication loop length is predicted as the replisome progresses. However, a length comparison between subsequent replication loops formed by individual replisomes suggests no apparent trend in loop size (mean length difference is 0.17 ± 0.3 kb).
In an alternative model, the encounter of the lagging-strand DNA polymerase with the 5’ terminus of the previously synthesized Okazaki fragment triggers dissociation of the polymerase and subsequent loop release6–9. In this collision model, leading-strand synthesis needs to continue after loop release to allow the primase to search for its recognition sequence. In this case, the size of the subsequent Okazaki fragment is expected to increase by the extra amount of ssDNA generated during the primase search. Therefore, the collision model predicts a gradual increase in Okazaki fragment size as the replisome progresses. To address the possibility that both signaling and collision models are operative, preventing a net change in loop length, we test a number of predictions for the behavior of the lag times between loops in the two different mechanisms. In both mechanisms, the lag time will contain the primer extension step and handoff of the primer to the lagging-strand DNA polymerase. In the collision mechanism, however, the lag time will also include additional leading-strand synthesis mediating the search of the primase for a priming sequence. A lack of length contrast between ssDNA and dsDNA prevents us from directly observing leading-strand synthesis during the lag phase. Since further leading-strand synthesis will give rise to a lengthening of the Okazaki fragment produced during the next cycle and collision-mediated loop release requires completion of the fragment, we predict a positive correlation between the lag time and the increase in the length of a loop. Similarly, we predict no correlation between lag time and loop length change in the case of a signaling mechanism. Inefficient primer utilization and a subsequent increase in single-stranded DNA template size may result in an increase in Okazaki fragment size, but loop release will still be triggered by the stochastic condensation of ATP and CTP and will remain uncorrelated with lag time.
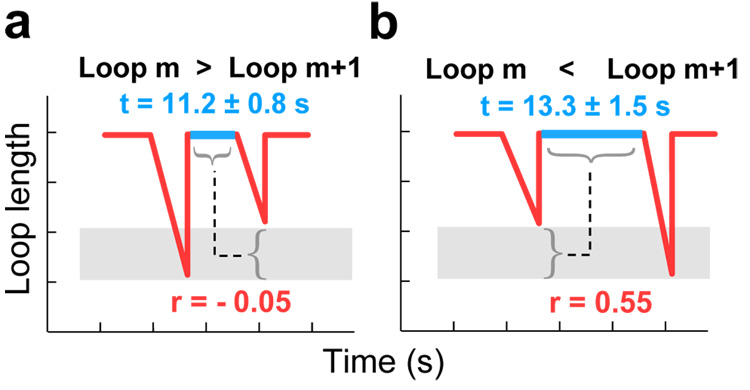
To test the presence of both signaling and collision mechanisms, we divide all observed loop pairs into two categories, solely based on whether the second loop in a pair was longer or shorter than the first loop of the pair, and determine the correlation for each of the two groups between lag time and loop growth. Indeed, we see a correlation between lag time and loop length change in the data set that showed loop growth (r=0.55) (n=50; the probability that a similarly sized data set of two uncorrelated variables would produce this correlation coefficient is less than 0.05%), but not in the group of loop pairs that showed a decrease in length (r=−0.05 with n=49) (Figure 4a and 4b). A slightly longer lag time between loop pairs that showed an increase in length (collision-controlled; 13.3 ± 1.5 seconds) versus pairs that showed a length reduction (signaling controlled; 11.2 ± 0.8 seconds) lends further support to the notion that after collision-mediated release, extra leading-strand synthesis is required to facilitate the search for a primer site. Additionally, experiments done under conditions that disfavor the collision mechanism by increasing the Okazaki fragment size while maintaining the pAC synthesis kinetics as a loop release signal result in the absence of any correlation between lag time and loop length (Supplementary Information). Taken together, these results suggest that both the signaling and collision mechanism are operative during coupled replication.
Fig. 4.
Signaling and collision mechanisms both serve to trigger loop release. (a) Schematic depiction of the correlation (r=−0.05; sample size n=49) between the lag time and the size change of the following loop for the sets of loops where loop m+1 is smaller than loop m. (b) Schematic depiction of the correlation (r=0.55; n=50) between the lag time and the size change of the following loop for the sets of loops where loop m+1 is larger than loop m.
In previous single-molecule studies, we have demonstrated that primase search is a stochastic process, with a limited probability of the primase recognizing and utilizing a priming sequence during its scanning of the lagging strand16. The random nature of primase activity poses a fundamental challenge to coordinating the timing of primer synthesis with the release of the replication loop. The utilization of both the signaling and collision mechanisms to release a replication loop and initiate formation of a new one provides an elegant mechanism to allow the replisome to deal with the stochastic nature of the primase activity. The signaling mechanism will release the replication loop if the primase locates one of its sequences before the nascent Okazaki fragment is finished. On the other hand, if the nascent Okazaki fragment is finished and the primase did not engage a recognition sequence, then the collision mechanism acts as a fail-safe mechanism to trigger loop release and ensure a proper reset of the cycle of enzymatic events at the replication fork.
METHODS SUMMARY
Single-molecule DNA stretching
A biotinylated fork was introduced on one end of phage λ DNA to attach it to the streptavidin-coated glass surface of a flow cell. The other DNA end was funtionilized with a digoxigenin moiety to couple it to a 2.8-µm diameter anti-digoxigenin-coated bead16. A laminar flow was introduced to exert a well-defined drag force on the bead, resulting in a stretching of the DNA molecules. Coordinated DNA synthesis was carried out by flowing purified gp4 helicase/primase25, T7 DNA polymerase26, and gp2.527 with nucleotides. Beads were imaged onto a CCD using dark-field microscopy and their positions tracked using particle-tracking software. For a detailed description, see Methods and Supplementary Information.
Fluorescence imaging
DNA was tethered at the forked end to functionalized cover slips as described above, but beads were omitted. In the presence of dsDNA-specific stain, the hydrodynamically stretched DNA was imaged using through-objective TIRF microscopy (Methods and Supplementary Information).
METHODS
Single-molecule stretching and length measurements
Phage λ DNA molecules were annealed and ligated to modified oligonucleotiodes to introduce a biotinylated fork on one end of the DNA and a digoxigenin moiety on the other end. The resulting DNA molecules were attached with the 5’terminus of the bifurcated end to the streptavidin-coated glass surface of a flow cell and with the 3’ end of the same strand to a 2.8 µm diameter anti-digoxigenin-coated paramagnetic bead (Dynal) (Supplementary Information). To prevent nonspecific interactions between the beads and the surface, a 1-pN magnetic force on the bead was applied upward by positioning a permanent magnet above the flow cell. Beads were imaged with a CCD camera at a time resolution of 500 ms and their positions were determined by particle-tracking software (Semasopht). Coordinated DNA synthesis in the flow cell was carried out by flowing gp4 helicase/primase (10 nM hexamers), T7 DNA polymerase (a purified 1:1 complex of gp5 and thioredoxin) (80 nM), and gp2.5 (750 nM), in buffer A (40 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, 50 mM potassium glutamate (pH 7.5), 0.1 mg/ml BSA), 600 µM each of dATP, dCTP, dGTP, dTTP) and 300 µM of ATP and CTP. Traces were corrected for instabilities in the flow by subtracting traces corresponding to tethers that were not enzymatically altered. Brownian motion and residual fluctuations resulted in a 300 bp error in DNA length measurement.
Fluorescence imaging
DNA was tethered at the forked end to functionalized cover slips as described above, but beads were omitted. Instead, the hydrodynamic drag on the DNA itself was used to extend the molecule. In the presence of 100 nM Sytox Orange dsDNA-specific stain (Invitrogen), the stretched DNA was imaged using through-objective TIRF microscopy (Olympus IX-71; 60× 1.45 N.A.). A CW 532 nm diode laser (Crystalaser) was used to excite stain at power densities sufficiently low to minimize photo-induced cleavage of stained DNA over the time scale of an experiment. Protein and nucleotide concentrations were identical to those used to replicate the bead-tethered DNA molecules (see above). Single-molecule bead-tethering assays demonstrated that the kinetics of leading-strand synthesis and coordinated replication were not influenced by the presence of the stain (Supplementary Information).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jong-Bong Lee for technical advice and Steven Moskowitz (Advanced Medical Graphics) for illustrations. This work was supported by the National Institutes of Health (GM-077248 to A.M.v.O. and GM-54397 to C.C.R.) and the National Science Foundation (CAREER grant 0543784 to A.M.v.O.). J.J.L. acknowledges the Jane Coffin Childs Memorial Fund for a postdoctoral fellowship.
REFERENCES
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annual Review of Biochemistry. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annual Review of Biochemistry. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Alberts BM, et al. Studies on DNA-replication in the bacteriophage-T4 invitro system. Cold Spring Harbor Symposia on Quantitative Biology. 1982;47:655–668. doi: 10.1101/sqb.1983.047.01.077. [DOI] [PubMed] [Google Scholar]
- 4.Chastain PD, Makhov AM, Nossal NG, Griffith JD. Analysis of the Okazaki fragment distributions along single long DNAs replicated by the bacteriophage T4 proteins. Molecular Cell. 2000;6:803–814. doi: 10.1016/s1097-2765(05)00093-6. [DOI] [PubMed] [Google Scholar]
- 5.Park K, Debyser Z, Tabor S, Richardson CC, Griffith JD. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. Journal of Biological Chemistry. 1998;273:5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 6.Carver TE, Sexton DJ, Benkovic SJ. Dissociation of bacteriophage T4 DNA polymerase and its processivity clamp after completin of Okazaki fragment synthesis. Biochemistry. 1997;36:14409–14417. doi: 10.1021/bi971423p. [DOI] [PubMed] [Google Scholar]
- 7.De Saro FJL, Georgescu RE, O'Donnell M. A peptide switch regulates DNA polymerase processivity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14689–14694. doi: 10.1073/pnas.2435454100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacker KJ, Alberts BM. The rapid dissociation of the T4 DNA-polymerase holoenzyme when stopped by a DNA hairpin helix: a model for polymerase release following the termination of each Okazaki fragment. Journal of Biological Chemistry. 1994;269:24221–24228. [PubMed] [Google Scholar]
- 9.Li XJ, Marians KJ. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. Journal of Biological Chemistry. 2000;275:34757–34765. doi: 10.1074/jbc.M006556200. [DOI] [PubMed] [Google Scholar]
- 10.Yang JS, Nelson SW, Benkovic SJ. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Molecular Cell. 2006;21:153–164. doi: 10.1016/j.molcel.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 11.Nossal NG, Makhov AM, Chastain PD, Jones CE, Griffith JD. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. Journal of Biological Chemistry. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- 12.Wu CA, Zechner EL, Reems JA, McHenry CS, Marians KJ. Coordinated leading-strand and lagging-strand synthesis at the Escherichia Coli DNA-replication fork: primase action regulates the cycle of okazaki fragment synthesis. Journal of Biological Chemistry. 1992;267:4074–4083. [PubMed] [Google Scholar]
- 13.Lee J, Chastain PD, Griffith JD, Richardson CC. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage T7 replication proteins. Journal of Molecular Biology. 2002;316:19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 14.Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. Journal of Biological Chemistry. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 15.Richardson C, Bacteriophage C. T7: minimal requirements for the replication of a duplex DNA molecule. Cell. 1983;33:315–317. doi: 10.1016/0092-8674(83)90411-7. [DOI] [PubMed] [Google Scholar]
- 16.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 17.Tanner NA, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nature Structural & Molecular Biology. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Molecular Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 19.van Oijen AM, et al. Probing enzymatic activity of a single molecular motor on DNA with a novel flow-stretching technique. Biophysical Journal. 2003;84:141A–142A. [Google Scholar]
- 20.Lee J, Chastain PD, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Molecular Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 21.Frick DN, Richardson CC. DNA primases. Annual Review of Biochemistry. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 22.Qimron U, Lee SJ, Hamdan SM, Richardson CC. Primer initiation and extension by T7 DNA primase. Embo Journal. 2006;25:2199–2208. doi: 10.1038/sj.emboj.7601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusakabe T, Richardson CC. Gene 4 DNA primase of bacteriophage T7 mediates the annealing and extension of ribo-oligonucleotides at primase recognition sites. Journal of Biological Chemistry. 1997;272:12446–12453. doi: 10.1074/jbc.272.19.12446. [DOI] [PubMed] [Google Scholar]
- 24.Frick DN, Kumar S, Richardson CC. Interaction of ribonucleoside triphosphates with the gene 4 primase of bacteriophage T7. Journal of Biological Chemistry. 1999;274:35899–35907. doi: 10.1074/jbc.274.50.35899. [DOI] [PubMed] [Google Scholar]
- 25.Notarnicola SM, Mulcahy HL, Lee J, Richardson CC. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. Journal of Biological Chemistry. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 26.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. Journal of Biological Chemistry. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 27.Hyland EM, Rezende LF, Richardson CC. The DNA binding domain of the gene 2.5 single-stranded DNA-binding protein of bacteriophage T7. Journal of Biological Chemistry. 2003;278:7247–7256. doi: 10.1074/jbc.M210605200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.