SUMMARY
After translational termination, mRNA and P site deacylated tRNA remain associated with ribosomes in post-termination complexes (post-TCs), which must therefore be recycled by releasing mRNA and deacylated tRNA and by dissociating ribosomes into subunits. Recycling of bacterial post-TCs requires elongation factor EF-G and a ribosome recycling factor RRF. Eukaryotes do not encode a RRF homologue and their mechanism of ribosomal recycling is unknown. We investigated eukaryotic recycling using post-TCs assembled on a model mRNA encoding a tetrapeptide followed by a UAA stop codon and report that initiation factors eIF3, eIF1, eIF1A and eIF3j, a loosely associated subunit of eIF3, can promote recycling of eukaryotic post-TCs. eIF3 is the principal factor that promotes splitting of post-termination ribosomes into 60S subunits and tRNA- and mRNA-bound 40S subunits. Its activity is enhanced by eIF3j, eIF1 and eIF1A. eIF1 also mediates release of P-site tRNA, whereas eIF3j ensures subsequent dissociation of mRNA.
Keywords: ribosome, translation, recycling, eIF3, eIF1, eIF3j
INTRODUCTION
Protein synthesis occurs in four stages: initiation, elongation, termination and recycling. In eukaryotes, initiation requires at least 11 initiation factors (eIFs) and can be divided into two steps: formation of a 48S initiation complex and its joining with a 60S subunit (Pestova et al., 2007). First, eIFs 3, 1, 1A and eIF2•GTP•Met-tRNAiMet bind to the 40S ribosomal subunit to form a 43S preinitiation complex, which initially attaches to the 5’-proximal region of mRNA after it is unwound by eIFs 4A, 4B and 4F, and then scans to the initiation codon, where it stops and forms a 48S complex with P site codon-anticodon base-pairing. eIFs 5 and 5B mediate subsequent joining of 48S complexes with 60S subunits. During the elongation cycle, elongation factor (eEF) 1A delivers cognate tRNA to the A-site, after which the nascent peptide chain is transferred to the amino acid of the A-site aminoacyl-tRNA. Finally, eEF2 promotes translocation of peptidyl-tRNA from A to P, and of deacylated tRNA from P to E sites. When a stop codon enters the A-site, release factors (eRFs) eRF1 and eRF3 induce hydrolysis of the ester bond of the P-site peptidyl-tRNA (Alkalaeva et al., 2006). The mechanism of the final step, recycling of eukaryotic post-termination complexes (post-TCs), is completely unknown.
During prokaryotic termination, RF1 and RF2 promote peptide release, whereas RF3 mediates release of RF1/RF2 from post-termination ribosomes, and dissociates after hydrolyzing GTP, yielding post-TCs that comprise 70S ribosomes, mRNA and P site deacylated tRNA (Zavialov et al., 2001). Recycling of post-TCs requires EF-G, RRF and initiation factor IF3. EF-G and RRF dissociate post-TCs into free 50S subunits and 30S subunits bound to mRNA and P site deacylated tRNA, and IF3 induces release of tRNA from 30S subunits, after which mRNA dissociates spontaneously (Peske et al., 2005; Zavialov et al., 2005). RRF, formed by two domains, interacts with segments of 23S rRNA that are involved in forming inter-subunit bridges B2a and B3 (Wilson et al., 2005). Ribosome binding sites for RRF and EF-G•GTP overlap, and the simultaneous presence of both factors is allowed only if the head domain of RRF is rotated. It was therefore proposed that EF-G•GDP binds RRF-associated 70S ribosomes and exchanges GDP for GTP, and EF-G•GTP induces a rotational movement of the head domain of RRF, which after hydrolysis of GTP by EF-G promotes subunit separation by disrupting B2a and B3 bridges (Gao et al., 2005).
Eukaryotes do not encode a RRF homologue, and the mechanisms of the preceding termination stage also differ between the two kingdoms. Thus, whereas RF3 increases the rate of RF1/RF2 dissociation from post-TCs, eRF3 ensures rapid and efficient hydrolysis of peptidyl-tRNA by eRF1 (Alkalaeva et al., 2006). Binding of eRF1 and eRF3•GTP to pre-termination complexes (pre-TCs) induces their rearrangement manifested as a 2-nt forward shift of their toe-print. However, such complexes are inactive in peptide release, and further rearrangement, induced by GTP hydrolysis, is required to properly position the GGQ loop of eRF1 in the peptidyl transferase center. eRF1, eRF3 and GTP form a long-lived high affinity complex (Pisareva et al., 2006) suggesting that they likely bind to pre-TCs as an eRF1•eRF3•GTP ternary complex. On the other hand, the mechanism of post-termination dissociation of eRFs is unknown. eRF3•GDP could either potentially dissociate directly after GTP hydrolysis thereby allowing proper positioning of eRF1, or taking into account that eRF1 and eRF3 form a tight complex irrespective of guanine nucleotides, remain bound until the peptide is released and then dissociate with eRF1. The fact that the toe-print shift persists in post-TCs after peptide release suggests that in contrast to prokaryotes, eukaryotic release factors might even remain bound to post-TCs. If this is indeed the case, the mechanism of ribosomal recycling in eukaryotes would likely not be similar to that in prokaryotes, because binding sites for eRF1/eRF3 and prokaryotic EF-G/RRF overlap.
Two observations suggest that eukaryotic ribosomal recycling might not require a special recycling factor, and that initiation factors could mediate this process. First, eIF3, particularly with eIF1, can dissociate 80S ribosomes in the presence of RNAs that can bind directly to the ribosomal mRNA-binding cleft (Kolupaeva et al., 2005), and could therefore play the principal role in splitting mRNA-containing post-termination ribosomes into subunits. Second, eIF1 can dissociate 48S complexes assembled with initiator tRNA containing mutations in the conserved GC pairs in its anticodon stem (Lomakin et al., 2006). This activity of eIF1 could potentially be employed to dissociate deacylated elongator P-site tRNAs, because only initiator tRNA contains such GC pairs in the anticodon stem.
We investigated the mechanism of eukaryotic ribosomal recycling using post-termination complexes assembled in vitro on a model mRNA encoding a tetrapeptide followed by a UAA stop codon and report that together, eIF3, eIF1, eIF1A and eIF3’s loosely associated 3j subunit promote recycling of eukaryotic post-TCs.
RESULTS
Release factors remain associated with post-termination complexes
Release of eRF1/eRF3 from post-TCs has not been studied and the composition of eukaryotic post-TCs is therefore unknown. Binding of eRF1•eRF3•GTP to pre-TCs induces their rearrangement, which is manifested as a 2-nt forward shift of the corresponding toe-print (Alkalaeva et al., 2006). This shift persists in post-TCs after peptide release, which could be due to the continued presence of eRFs on post-termination ribosomes, or to the irreversible/slowly reversible nature of conformational changes that could therefore persist after dissociation of eRFs. If the first scenario is correct, in contrast to prokaryotes, eukaryotic post-TCs should also contain eRFs.
To investigate the mechanism of eukaryotic ribosomal recycling, pre-TCs were assembled from ribosomal subunits, eIFs 2, 3, 1, 1A, 4A, 4B, 4F, 5 and 5B, eEFs 1H and 2, and aminoacylated tRNAs on MVHL-STOP mRNA, which encodes a MVHL tetrapeptide followed by a UAA stop codon (Figure 1A; Alkalaeva et al., 2006), and purified by sucrose gradient centrifugation. The term eIF3 will be used throughout the text to describe eIF3 lacking its 3j subunit. As previously reported (Alkalaeva et al., 2006), incubation of pre-TCs with eRF1/eRF3/GTP shifted their toeprint forward by 2nt from +16nt from the P-site CUG codon to +15nt from the UAA codon (Figure 1B, lanes 1, 2). However, sucrose gradient centrifugation of post-TCs formed by incubating pre-TCs with eRF1/eRF3/GTP eliminated the shift, so post-TCs that had undergone centrifugation and pre-TCs yielded identical toe-prints (Figure 1, lane 4). Centrifugation eliminated the shift irrespective of whether it was originally obtained in conditions when peptide release and GTP hydrolysis were allowed or disallowed by inclusion of GMPPNP or eRF1(AGQ), which is inactive in peptide release (Figure 1C). Addition of eRF1 and eRF3 together, but not separately, to post-TCs that had been subjected to centrifugation resulted in reappearance of the shift (Figure 1D) suggesting that centrifugation dissociates eRFs that normally remain bound to post-TCs. Consistently, eRF1 or eRF3 were not detected by western blotting in the sucrose gradient peak that corresponded to post-TCs (data not shown) or by monitoring [32P]eRF1 (Figures 1E, F). This indicates that the toe-print shift is reversible and requires the physical presence of eRFs on post-termination ribosomes, which in turn means that eukaryotic post-TCs retain eRF1/eRF3 after peptide release. However, we note that it is possible that only eRF1 remains on post-TCs (see Discussion).
Figure 1. Association of release factors with post-TCs.
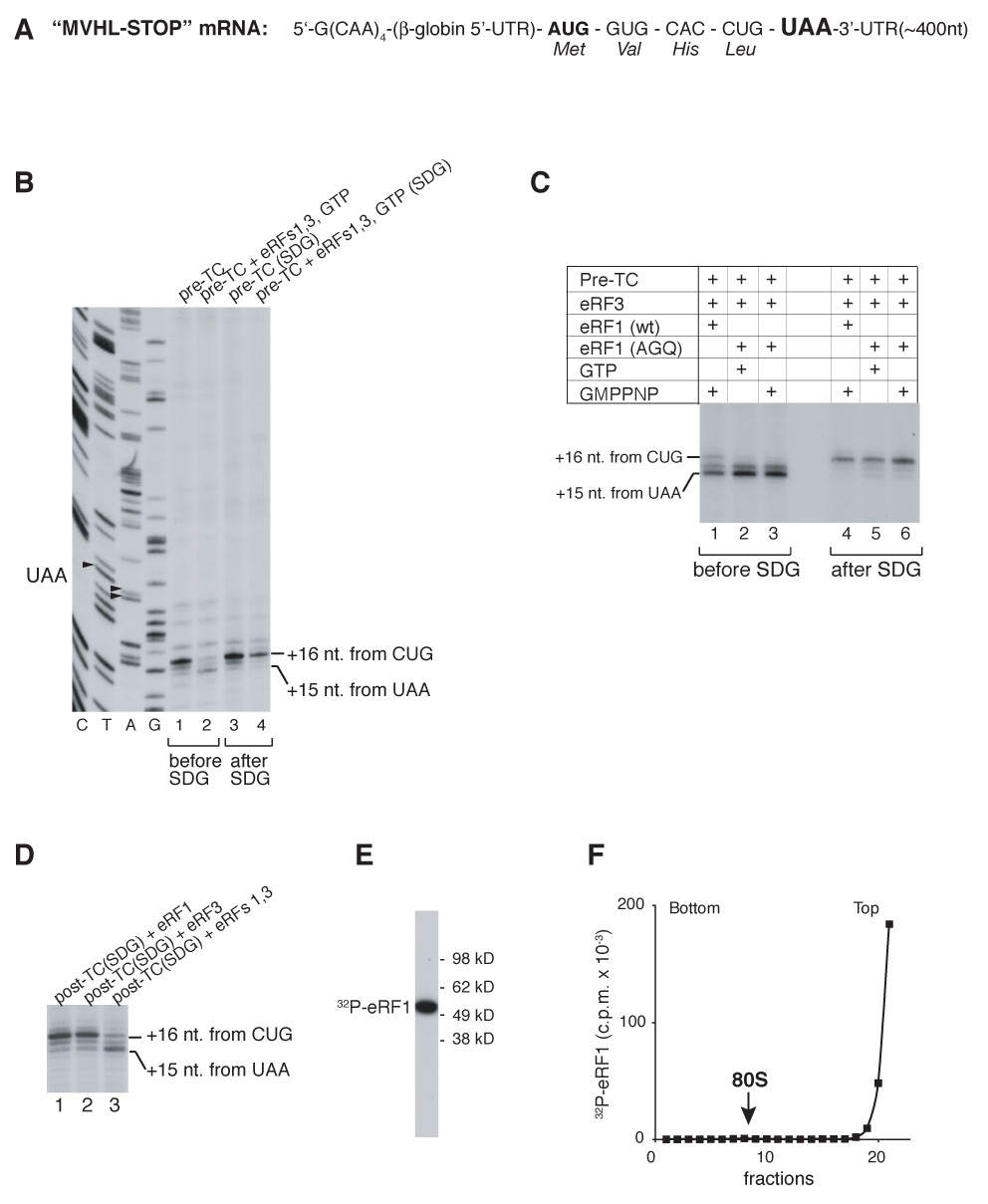
(A) Structure of MVHL-STOP mRNA. (B–D) Toe-printing analysis of ribosomal complexes obtained by incubating pre-TCs with eRFs, GTP and GMPPNP in different combinations, before and after sucrose gradient centrifugation, “SDG” (B, C) and of post-TCs, obtained by incubating pre-TCs with eRF1/eRF3/GTP, subjecting them to sucrose gradient centrifugation, and incubating them again with eRFs, as indicated (D). Lanes C, T, A, G depict cDNA sequences corresponding to MVHL-STOP mRNA. The positions of toe-prints that correspond to ribosomal complexes are indicated. (E) Autoradiograph of [32P]eRF1 phosphorylated by cAMP-dependent kinase. MW markers are indicated. (F) Association of [32P]eRF1 with post-TCs assayed by sucrose gradient centrifugation. The position of 80S ribosomes is indicated.
eIFs 2, 3, 1, 1A, 4A, 4B and 4F promote recycling of post-termination complexes and subsequent formation of 48S initiation complexes on recycled mRNA
The finding that eRF1/eRF3 remain on eukaryotic post-TCs implies that the mechanism of ribosomal recycling in eukaryotes is likely not similar to that in prokaryotes, because binding sites for eRF1/eRF3 and prokaryotic EF-G/RRF overlap. As discussed above, we hypothesized that eukaryotic ribosomal recycling might not need a dedicated factor, and that initiation factors could be sufficient. To test this hypothesis, pre-TCs were assembled on [32P]MVHL-STOP mRNA and purified by sucrose gradient centrifugation. Incubation of pre-TCs with eRF1/eRF3 and eIFs 2, 3, 1, 1A, 4A, 4B and 4F resulted in almost all mRNA associating with 40S subunits (Figure 2A). Toe-printing of the 40S subunit peak fraction yielded stops 15–17nt from the AUG triplet, which is characteristic of 48S initiation complexes (Figure 2B). Formation of 48S complexes required prior termination: no 48S complexes formed in the presence of eIFs without eRF1/eRF3 or if eRF1(AGQ) replaced eRF1(wt) (Figure 2C). Thus, consistent with our hypothesis, eIFs were sufficient to mediate ribosomal recycling and subsequent 48S complex formation on recycled mRNA.
Figure 2. Recycling of post-TCs and assembly of 48S initiation complexes on recycled mRNA promoted by initiation factors.
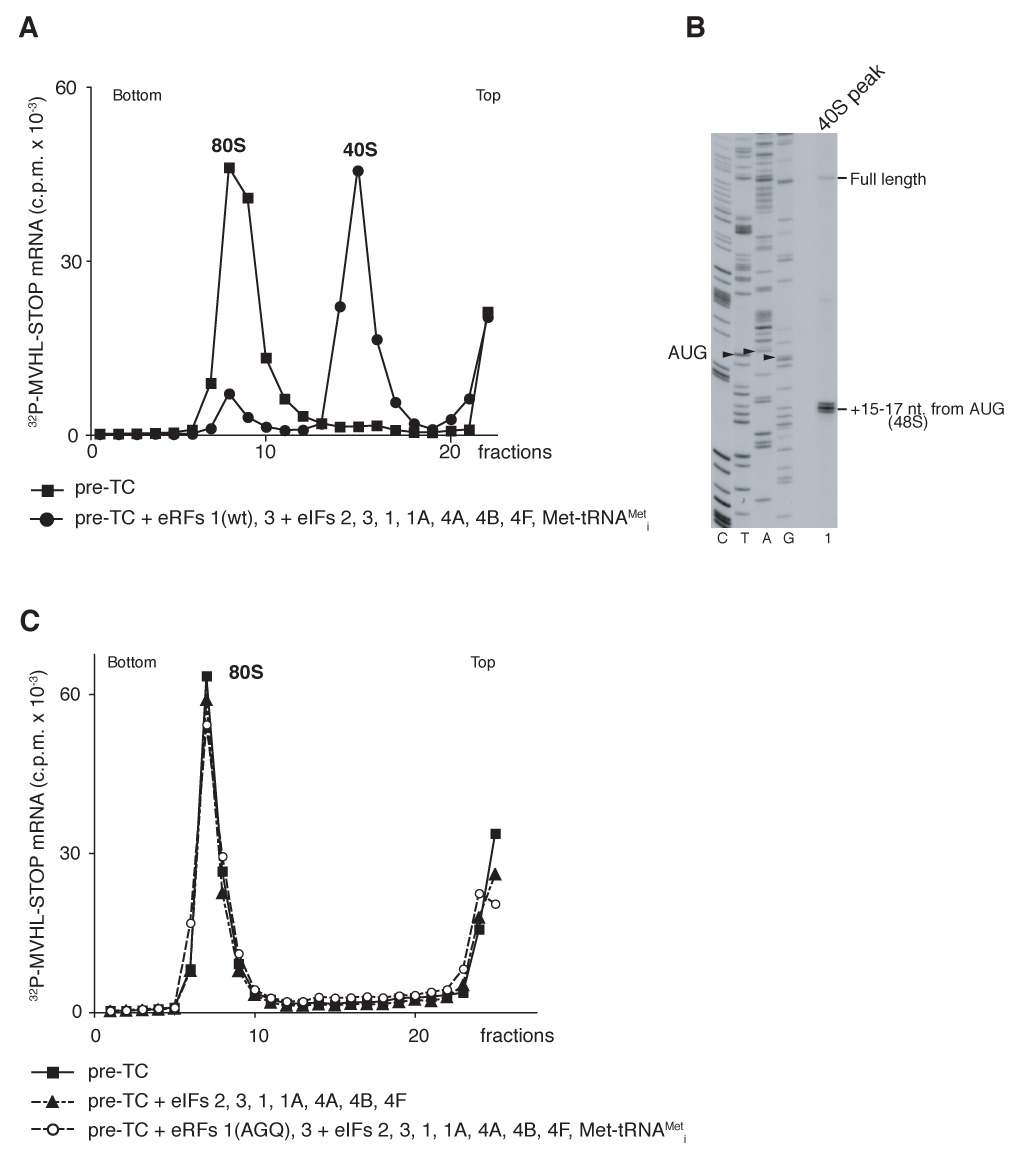
(A, C) Ribosomal association of [32P]MVHL-STOP mRNA after incubation of pre-TCs with eRFs and eIFs, as indicated, assayed by sucrose gradient centrifugation. The positions of 40S subunits and 80S ribosomes are indicated. (B) Toe-printing analysis of the 40S-containing fraction (panel A, circles) obtained after incubating pre-TCs with eRFs and eIFs. The positions of full-length cDNA and of toe-prints that correspond to 48S complexes are indicated. Lanes C, T, A, G depict cDNA sequences corresponding to MVHL-STOP mRNA.
Dissociation of post-termination ribosomes into subunits
To investigate the activities of individual eIFs in splitting post-termination ribosomes into subunits, pre-TCs were assembled with 60S subunits 32P-phosphorylated by casein kinase II, which strongly phosphorylated two ~14 kD ribosomal proteins (rp) and weakly phosphorylated a ~38 kD rp (Figure 3A, lane 1). Phosphorylation did not affect the activity of 60S subunits in any stage of translation (data not shown). Pre-TCs were purified by sucrose gradient centrifugation, incubated with different combinations of eRFs, eIFs and puromycin and subjected to another round of sucrose gradient centrifugation. Incubation of pre-TCs with eRF1/eRF3 did not cause ribosomal dissociation (Figure 3B, black triangles). However, incubation of pre-TCs with eRF1/eRF3 and eIF3 led to ~45% dissociation of post-termination ribosomes into subunits (Figure 3B, red circles). eIF3 was the only factor that alone had dissociating activity. In its absence, other eIFs (2, 1, 1A, 3j) that interact with 40S subunits and participate in 43S complex formation even together could not promote dissociation (Figure 3B, black circles). eIFs 1 and 1A modestly (~15%) enhanced eIF3’s dissociating activity (Figure 3C, blue squares and green triangles), whereas eIF3j, which stimulates 40S/eIF3 association (Fraser et al., 2004), had a much stronger effect, and 70–75% of pre-TCs were dissociated in the presence of eIF3 and eIF3j (Figure 3C, red circles). Pairwise combinations of eIFs 1, 1A, 3j had a higher stimulatory effect on eIF3’s dissociating activity (Figure 3D, blue squares, magenta diamonds, green triangles), and together, eIFs 3, 1, 1A and 3j promoted nearly complete splitting of post-TCs into subunits (Figure 3D, red circles). eIF2/GTP/Met-tRNAMeti did not influence dissociation by eIF3 alone or in any combination with eIFs 1, 1A, and 3j (Figure 3E; data not shown). Although yeast eEF2 has been reported to promote transient dissociation of empty yeast 80S ribosomes in the presence of ATP (Demeshkina et al., 2007), no influence of eEF2 on dissociation of mammalian post-TCs was observed. Thus, in the absence of initiation factors, eEF2 did not dissociate post-TCs irrespective of the presence of ATP or GTP and did not influence their dissociation by eIF3 alone or with different combinations of eIFs 1, 1A and 3j (Supplemental Figures S1A, S1B). Dissociation of ribosomal complexes by eIFs required termination: only trace amounts of 60S subunits were obtained after incubating pre-TCs with eIFs in the absence of eRF1/eRF3 (Figure 3B, open circles), which could be due to dissociation of 80S complexes, in which peptidyl tRNA had hydrolyzed spontaneously. Dissociation occurred to a similar extent when puromycin replaced eRFs (Figure 3F, red circles and magenta triangles).
Figure 3. Dissociation of post-TCs into subunits.
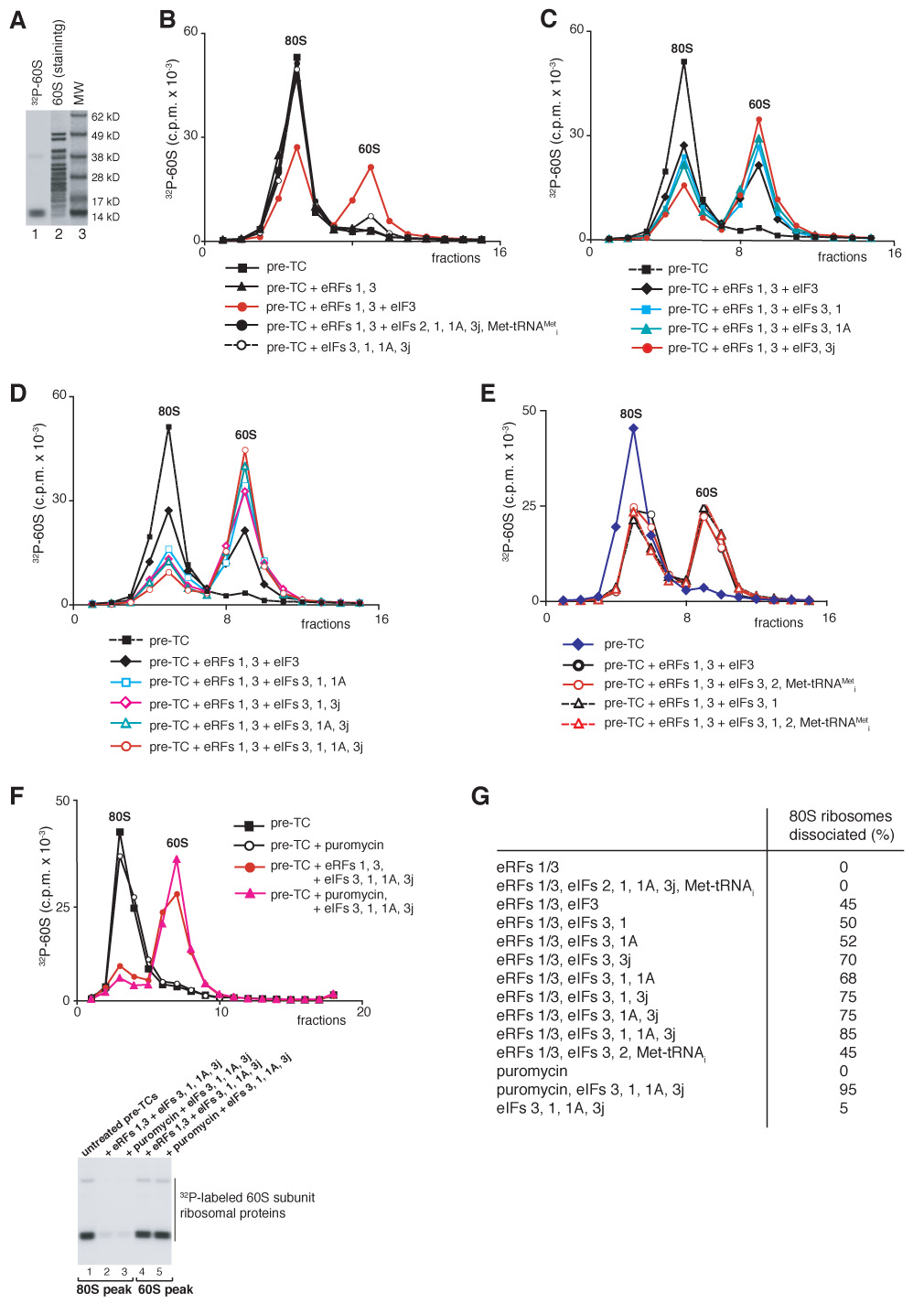
(A) Coomassie staining of 60S subunit proteins (lane 2) and autoradiography of [32P]60S subunits phosphorylated by CKII (lane 1). MW markers are indicated. (B–F) Dissociation of pre-TCs, assembled on MVHL-STOP mRNA with [32P]60S subunits, after incubation with eRFs, eIFs and puromycin, as indicated, assayed after sucrose gradient centrifugation by Cerenkov counting and Pisarev, gel electrophoresis (F, right panel). The positions of 60S subunits and 80S ribosomes are indicated. (G) Summary of dissociation of post-termination ribosomes into subunits by different combination of eIFs (panels B–F).
To verify that recycled 40S subunits remain associated with eIF3 that protects them from reassociation with 60S subunits, pre-TCs were incubated with combinations of factors that included [32P]eIF3 phosphorylated by cAMP-dependent protein kinase, which phosphorylates eIF3a exclusively if eIF3 lacks eIF3j (Unbehaun et al., 2004). As expected, eIF3 was bound to recycled 40S subunits derived by incubating pre-TCs with eRF1/eRF3 and eIFs 3/1/1A/3j (Figure 4A, red triangles). ~40% less eIF3 was associated with recycled 40S subunits obtained in eIF3j’s absence (Figure 4A, blue circles). Because only slightly fewer recycled 40S subunits were obtained in 3j’s absence (Figure 3D, blue squares and red circles), we assume that in the absence of 3j, some eIF3 dissociated from 40S subunits due to the stringency of centrifugation.
Figure 4. Association of eIF3 with recycled 40S subunits and dissociation of P site deacylated tRNA.
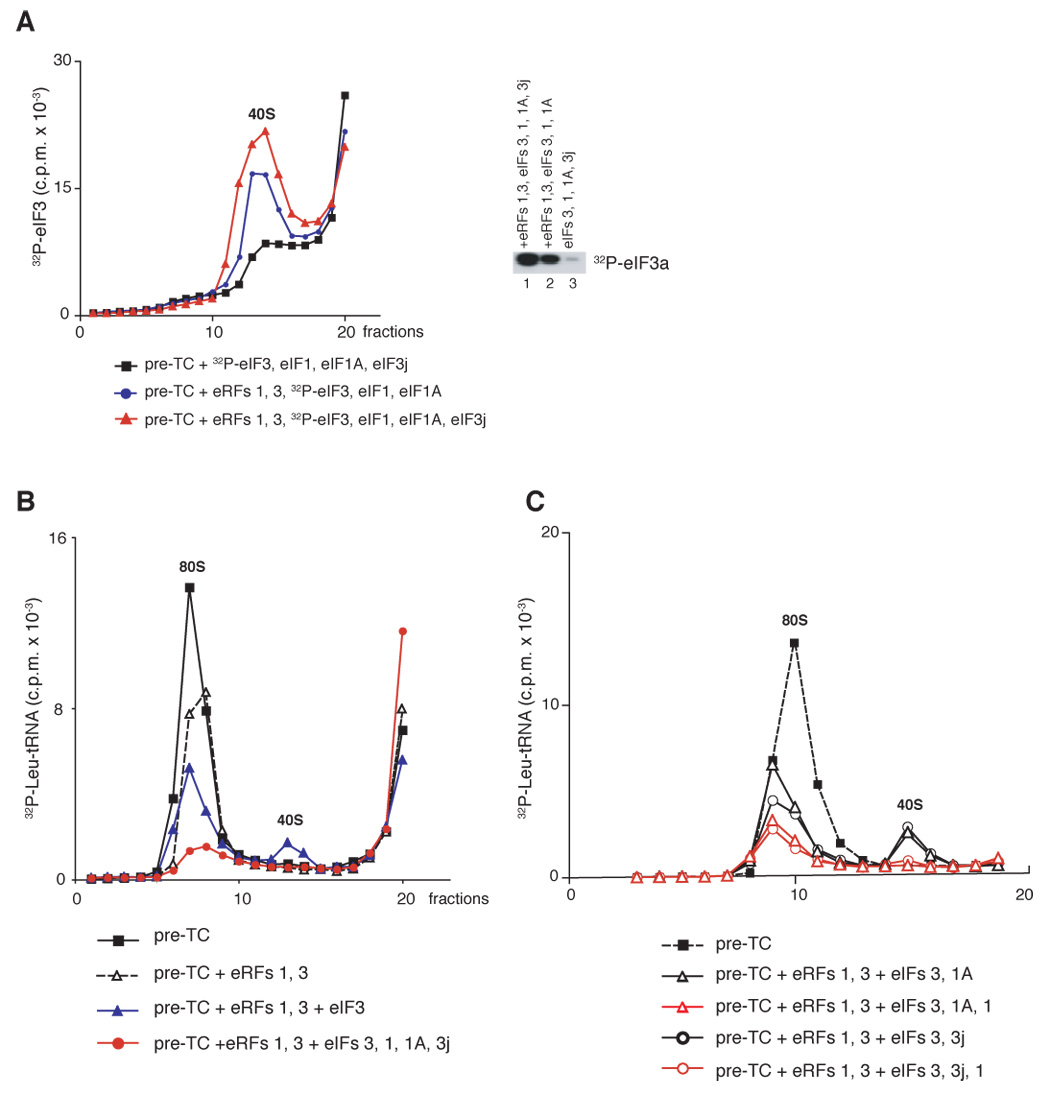
(A) Ribosomal association of [32P]eIF3 after incubation of pre-TCs assembled on MVHL-STOP mRNA with different combination of eRFs and eIFs, as indicated, assayed after sucrose gradient centrifugation by Cerenkov counting and gel electrophoresis (right panel). (B, C) Ribosomal association of [32P]tRNALeu after incubation of pre-TCs assembled in the presence of Leu-[32P]tRNALeu with eRFs and eIFs, as indicated, assayed after sucrose gradient centrifugation by Cerenkov counting. The positions of 40S subunits and 80S ribosomes are indicated.
These data show that eIF3 is the principal factor that promotes splitting of post-TCs into subunits and remains bound to recycled 40S subunits preventing their re-association with 60S subunits; its activity is strongly enhanced by eIFs 3j, 1 and 1A. The fact that equally efficient dissociation of post-TCs occurred in the presence of eRF1/eRF3 or puromycin indicates that eRFs are not essential for recycling.
Dissociation of P site deacylated tRNA from post-termination complexes
To investigate the dissociation of P site deacylated tRNA, pre-TCs were assembled with Leu-[32P]tRNALeu, purified by sucrose gradient centrifugation, incubated with different combinations of eRFs, eIFs and puromycin, and subjected again to sucrose gradient centrifugation. Incubation of pre-TCs with either eRFs or puromycin led to release of ~40% of Leu-tRNALeu, which migrated at the top of the gradient, indicating that peptide release destabilizes binding of P site tRNA (Figure 4B, open triangles; data not shown). After incubating pre-TCs with eRF1/eRF3 and eIF3, ~30% of tRNA remained bound to post-termination ribosomes, ~10% was associated with 40S subunits, and the remainder migrated at the top of the gradient (Figure 4B, blue triangles). After incubation of pre-TCs with eRF1/eRF3 and eIFs 3/1/1A/3j, very little tRNA remained bound to 80S ribosomes and none was associated with recycled 40S subunits (Figure 4B, red circles). To determine which factor was responsible for dissociating tRNA from 40S subunits, recycling was studied in the presence of different combinations of eIFs 1, 1A and 3j (Figure 4C). Whereas in the presence of eIFs 1A/3j ~15% tRNA was still bound to 40S subunits, no tRNA was associated with 40S subunits in the presence of eIF1.
These data indicate that in addition to enhancing eIF3’s dissociating activity, eIF1 also promotes release of P site deacylated tRNA from recycled 40S subunits. We note, that because P site initiator tRNA binds 40S subunits unstably in the absence of eIF2 (Unbehaun et al., 2004), it is possible that some tRNA dissociated from recycled 40S subunits due to the stringency of centrifugation, so the true proportion of tRNA-bound recycled 40S subunits might be higher.
Dissociation of mRNA from post-termination complexes
To investigate mRNA release, pre-TCs were assembled on [32P]MVHL-STOP mRNA, purified by sucrose gradient centrifugation, incubated with different combinations of eRFs, eIFs and puromycin, and subjected again to sucrose gradient centrifugation. Incubation of pre-TCs with eRF1/eRF3 led to release of 35–40% of mRNA from 80S ribosomes, which migrated at the top of the gradient (Figure 5A, black diamonds). Dissociation of mRNA was most likely the result of destabilization of its ribosomal binding after dissociation of P site tRNA. eIFs 1, 1A, 3j and eIF2/Met-tRNAi/GTP did not exacerbate the destabilizing effect of eRF1/eRF3 (Figure 5A, black circles), which was consistent with their inability to influence the integrity of post-TCs in the absence of eIF3. However, after incubating pre-TCs with eRF1/eRF3 and eIF3, only 25–30% of mRNA was associated with 80S ribosomes, whereas ~20% became bound to 40S subunits and the remainder formed mRNPs that migrated near the top of the gradient (Figure 5A, magenta triangles). Consistent with the ability of eIF1 and eIF1A to enhance eIF3’s dissociating activity, their inclusion in reaction mixtures separately or together reduced the amount of mRNA associated with 80S ribosomes to 10–15%, but ~25% was still bound to recycled 40S subunits (Figure 5B). Consistently, toe-printing of 80S ribosome-containing fractions showed a proportional specific decrease in the intensity of the toe-print 16nt from the P-site CUG codon corresponding to post-TCs (Figure 5D). Neither eIFs 4A/4B/4F, nor eIF2/Met-tRNAMeti influenced the ribosomal distribution of mRNA caused by eIFs 3/1/1A (data not shown).
Figure 5. Dissociation of mRNA from post-TCs.
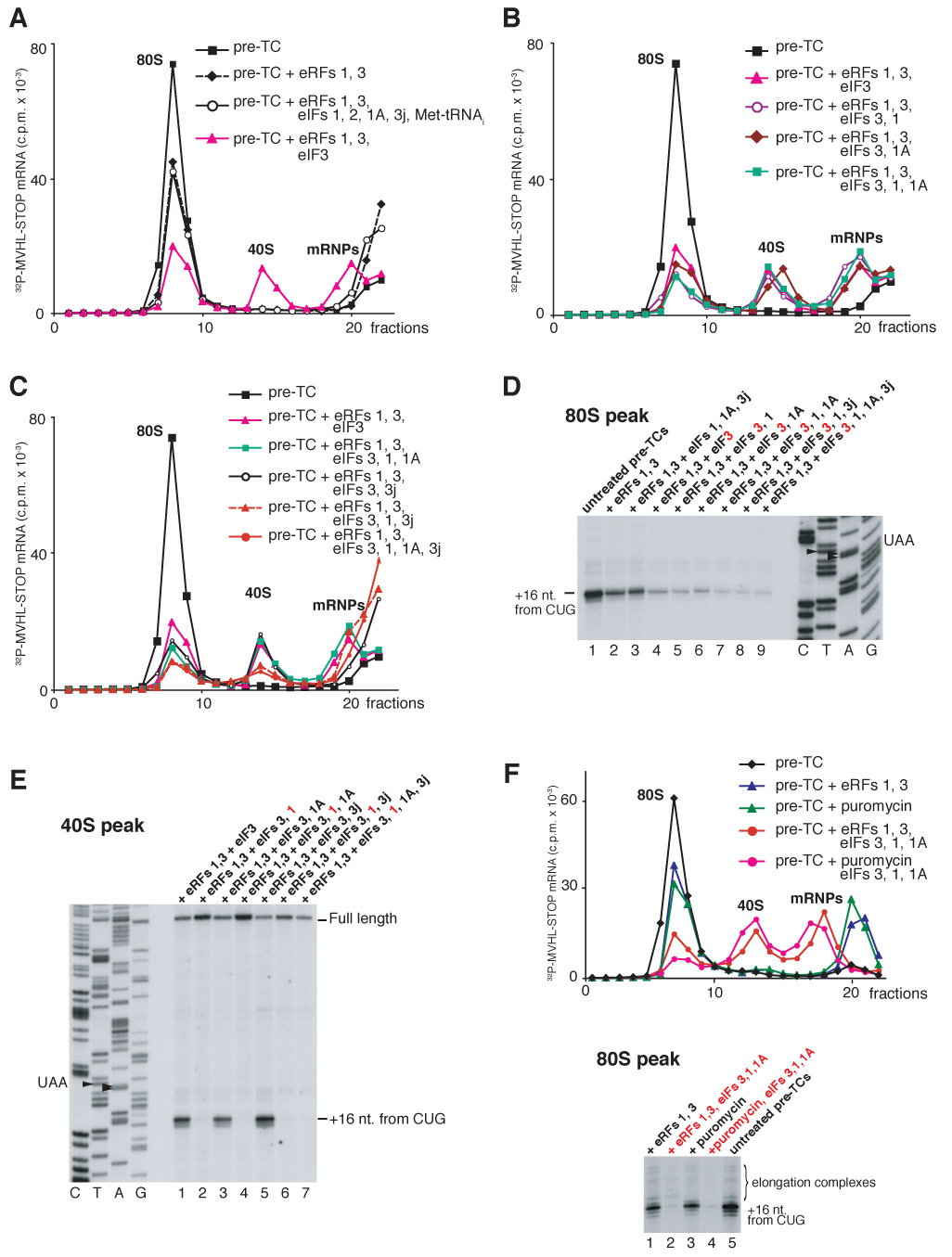
(A–C, F) Ribosomal association of [32P]MVHL-STOP mRNA after incubation of pre-TCs with eRFs, eIFs and puromycin, as indicated, assayed after sucrose gradient centrifugation by Cerenkov counting. The positions of 40S subunits, 80S ribosomes and mRNPs are indicated. (D,E, lower panel of F) Toe-printing analysis of 40S-containing fractions shown on panels A–C (E), and of 80S-containing fractions shown on panels A–C and F (D and F, respectively). The positions of full-length cDNA and of toe-prints that correspond to ribosomal complexes are indicated. Lanes C, T, A, G depict cDNA sequences corresponding to MVHL-STOP mRNA.
Dissociation of post-TCs by eIFs 3/1/1A led to a high proportion of mRNA (20–25%) being bound to 40S subunits. Post-TCs could either be dissociated into 60S subunits and mRNA-bound 40S subunits or mRNA could first have been released and then rebound to recycled 40S subunits. In the first scenario, mRNA would occupy the same position on 40S subunits as it did in pre-TCs, whereas in the second, it would rebind randomly. Toe-printing of 40S subunit-containing fractions, obtained by incubating pre-TCs with eRFs and eIF3 or eIFs 3/1A, yielded a prominent stop +16 nt. from the P-site CUG codon (Figure 5E, lanes 1, 3). The toe-print’s position and the fact that reverse transcriptase displaces 40S subunits from mRNA in the absence of P site codon-anticodon base-pairing suggest that incubation of post-TCs with eIF3 or eIFs 3/1A yields a high proportion of 40S/mRNA/tRNA complexes, consistent with the observation that some tRNA remains bound to 40S subunits if recycling occurs in the absence of eIF1 (Figures 4B, C). Consistently, toe-printing of 40S-containing fractions obtained in the presence of eIF1 yielded only full-length cDNA (Figure 5E, lanes 2, 4). mRNA’s position in such complexes was determined by site-specific UV cross-linking using [32P]MQQ-STOP mRNA containing 4-thiouridine (4SU) at only two positions, in the initiation and stop codons, which were separated by two CAA triplets and flanked by CAA repeats (Figure 6A). In pre-TCs assembled on this mRNA (Figure 6B, lane 3), the U of the AUG codon is at position −5 and the U in the UAA stop codon is at +4. Nucleotides at different positions relative to the initiation codon in initiation complexes cross-link to characteristic ribosomal proteins (Pisarev et al., 2006; Pisarev et al., in preparation). Thus in 48S and 80S initiation complexes, 4SU at −5 and +4 positions specifically cross-linked to rpS5 and rpS28 (Figure 6C, lane 2; Pisarev et al., in preparation) and to rp15, respectively (Figure 6C, lane 3; Pisarev et al., 2006). In pre-TCs, MQQ-STOP mRNA would be expected to cross-link to rpS5/rpS28 (via U of the AUG codon at position −5) and to rpS15 (via U of the UAA codon at position +4), and if post-TCs are dissociated into 60S subunits and mRNA-bound 40S subunits, the same cross-linking pattern should be observed for 40S/mRNA complexes. Exactly this pattern was observed for 40S subunit complexes obtained by incubating pre-TCs with eRF1/eRF3 and eIFs 3/1/1A (Figure 6C, lane 1), which indicates that on 40S subunits recycled in the presence of eIF1, at least some mRNA continues to occupy the same position as it did in pre-TCs.
Figure 6. The position of mRNA on recycled 40S subunits and the influence of initiation factors on peptide release.
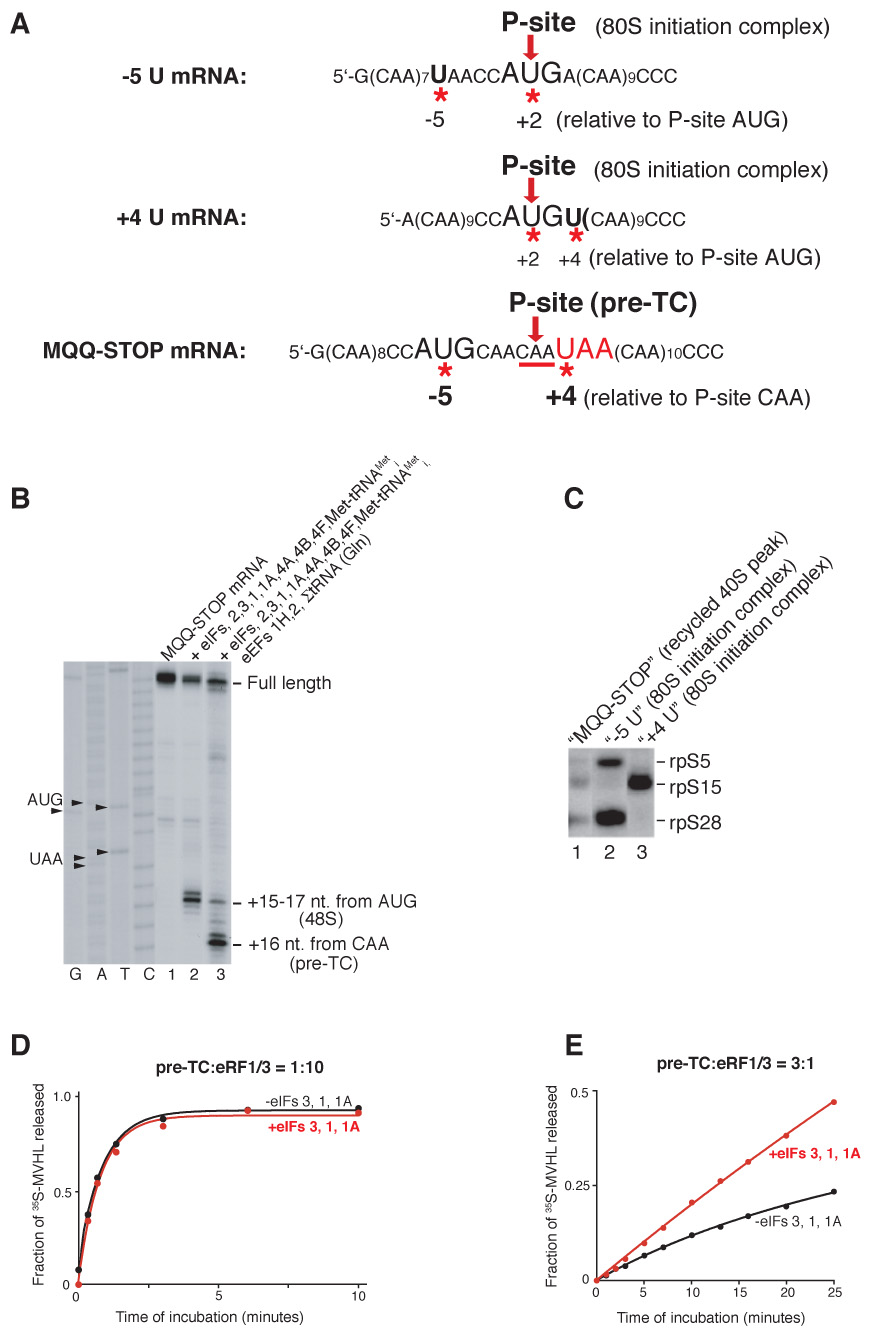
(A) Sequences of −5U, +4U and MQQ-STOP mRNAs. The positions of Us (indicated by red asterisks) relative to the P-site codons (red arrows) in 80S initiation complexes or in pre-TCs are indicated. (B) Toe-printing analysis of 48S complexes and pre-TCs assembled on MQQ-STOP mRNA. The components of reaction mixtures are indicated. The position of ribosomal complexes are shown relative to the mRNA codon in the P site. Lanes C, T, A, G depict cDNA sequences corresponding to MQQ-STOP mRNA. (C) UV cross-linking of 32P-labeled −5U, +4U and MQQ-STOP mRNAs containing 4-ThioU with ribosomal proteins in recycled 40S subunit-containing complexes or in 80S initiation complexes, as indicated, assayed by SDS-PAGE and autoradiography. (D, E) Kinetics of [35S]MVHL tetrapeptide release in the presence (red circles) and in the absence (black circles) of eIFs at different ratios of eRFs and pre-TCs.
Thus, dissociation of post-TCs by eIFs 3/1A and eIFs 3/1A/1 yields relatively high amounts of 40S/mRNA/tRNA and 40S/mRNA complexes, respectively. Taking the stringency of sucrose gradient centrifugation into account, the true proportion of these complexes could be even higher. This raises the question of how mRNA is released from recycled 40S subunits. eIF3j and mRNA bind to eIF3-associated 40S subunits with negative cooperativity (Unbehaun et al., 2004; Fraser et al., 2007). Consistently, inclusion of eIF3j in reaction mixtures that contained eIFs 3/1 or eIFs 3/1/1A led to ~75% dissociation of mRNA from recycled 40S subunits (Figure 5C, red triangles and red circles). However, eIF3j did not reduce association of mRNA with recycled 40S subunits obtained with eIF3 or with eIFs 3/1A (Figure 5C, open circles; Figure 5E, lane 5; data not shown), which indicates that the P-site codon-anticodon interaction, which is retained on recycled 40S subunits in the absence of eIF1, protects mRNA from dissociation by eIF3j.
Similar levels of mRNA dissociation from 80S ribosomes were observed after incubating pre-TCs with eRF1/eRF3 or puromycin, and even less mRNA was bound to 80S ribosomes after incubation of pre-TCs with eIFs 3/1/1A and puromycin than with eIFs and eRF1/eRF3 (Figure 4F) due to puromycin-mediated dissociation of the trace amounts of elongation complexes present in pre-TC preparations (Figure 4F, lower panel). This again confirms that eRFs are not essential for recycling.
In conclusion, dissociation of post-TCs by eIFs 3/1A or by eIFs 3/1A/1 yields relatively high proportions of 40S/mRNA/tRNA and 40S/mRNA complexes, respectively. Dissociation of mRNA from recycled 40S subunits can be promoted by eIF3j after eIF1-induced release of tRNA. We found that eIFs 3, 1, 1A and 3j acted identically on post-TCs assembled on mRNA with a 12 amino acid-long open reading frame (Supplemental Figure S2).
Initiation factors stimulate peptide release when pre-termination complexes are in excess over release factors
The findings that eRF1/eRF3 remain bound to post-TCs and that post-TCs can be dissociated by eIFs 3/1/1A/3j imply that when pre-TCs are in excess over eRFs, these eIFs should enhance peptide release by recycling eRFs. To test this, pre-TCs assembled with [35S]Met-tRNAMeti were incubated with either excess or substoichiometric amounts of eRF1/eRF3 and with or without eIFs. When the concentration of eRF1/eRF3 exceeded that of pre-TCs, eIFs did not influence release of 35S-MVHL tetrapeptide (Figure 6D). However, when pre-TCs were in excess over eRF1/eRF3, eIFs strongly stimulated peptide release (Figure 6E). These results confirm that eRFs remain bound to post-TCs and can be recycled by eIFs as a part of post-TCs.
DISCUSSION
The mechanism of post-termination recycling is a long-standing unresolved question in eukaryotic protein synthesis. The ability to reconstitute all prior stages in vitro (Alkalaeva et al., 2006) has now enabled us to investigate this final stage in translation. We report here that eukaryotic recycling can be promoted by the combination of eIF3, eIF1, eIF1A and eIF3j.
Composition of post-termination complexes
Binding of eRF1•eRF3•GTP to pre-TCs induces their rearrangement, which is manifested as a 2-nt forward shift of the corresponding toe-print and persists in post-TCs after peptide release (Alkalaeva et al., 2006). Sucrose gradient centrifugation of post-TCs eliminated the shift, but it reappeared on addition of eRF1/eRF3 suggesting that centrifugation dissociates eRFs that normally remain bound to post-TCs. Theoretically, the rearrangement of ribosomal complexes induced by eRF1(wt)•eRF3•GTP that results in this shift could be maintained by eRF1 alone after eRF3•GDP dissociation and peptide release, because eRF1(AGQ) induces the shift without eRF3 (Alkalaeva et al., 2006). eRF1 could therefore remain bound to post-TCs alone, whereas eRF3 could dissociate upon GTP hydrolysis. However, because eRF1 and eRF3 form a tight complex with GDP and without guanine nucleotides (Mitkevich et al., 2006; Pisareva et al., 2006), eRF3 could also remain on post-TCs due to its tight binding to eRF1. The toe-print shift obtained with eRF1(wt)•eRF3•GTP was slightly less complete than when peptide release was not allowed (in the presence of eRF1(AGQ) or GMPPNP (Alkalaeva et al., 2006) suggesting that eRFs may dissociate slowly from post-TCs.
eIF3 dissociates post-termination ribosomes into subunits in a manner that is enhanced by eIF3j, eIF1 and eIF1A
eIF3 has the principal role in recycling. It was the only factor that could split post-termination ribosomes on its own. eIF3’s dissociating activity was strongly enhanced by eIF3j and less so by eIF1 and eIF1A. Together, eIFs 3/3j/1/1A mediated near-complete dissociation of post-termination ribosomes. After dissociation, eIF3 (and likely other eIFs) remained bound to recycled 40S subunits, protecting them from reassociation. The primary role of eIF3 in dissociating mRNA-containing post-TCs is consistent with its reported activity in dissociating 80S ribosomes in the presence of RNAs that can bind directly to the mRNA-binding cleft (Kolupaeva et al., 2005). Interestingly, eIF1 and eIF1A stimulated dissociation of post-TCs less than dissociation of empty 80S ribosomes in the presence of RNA (Kolupaeva et al., 2005), whereas conversely, eIF3j’s stimulatory effect was higher. Post-TCs formed by incubating pre-TCs with eRFs or puromycin were recycled equally efficiently, which indicates that eRFs are not essential for recycling. However, kinetic analysis was outside the scope of this study, so it cannot be excluded that eRFs bound to post-TCs and/or eRF-induced conformational changes in post-TCs might influence the dissociation rate.
How do eIFs 3, 3j, 1 and 1A split post-termination ribosomes into subunits? The ~800 kDa eIF3 is a five-lobed particle that binds to the solvent side of the 40S subunit (Siridechadilok et al., 2005). Its left leg binds below the platform near the 60S subunit interface and covers rpS13, which interacts with helix 34 of the 60S subunit and thus contributes to intersubunit bridge B4 (Spahn et al., 2001). It was suggested that eIF3’s dissociating activity could at least in part be due to disruption of this bridge (Siridechadilok et al., 2005). Binding of eIF3 likely also induces conformational changes in 40S subunits that might contribute to dissociation. Both eIF1 and eIF1A bind to the intersubunit surface of 40S subunits. eIF1 was modeled onto the 40S subunit platform near the P site (Lomakin et al., 2003), where it would block access of 60S subunits to elements of 18S rRNA involved in forming intersubunit bridges B2b and B2d (Spahn et al., 2001). By analogy with its prokaryotic homologue IF1 (Carter et al., 2001), eIF1A likely binds to the 40S subunit A site. The mechanism of stimulation by eIF1 and eIF1A of eIF3’s dissociating activity would therefore be consistent with a model in which eIF3 binds to the 40S subunit’s solvent side and causes changes in the subunit interaction that allow eIF1 and eIF1A to access their binding sites on the 40S subunit interface. eIF1 and eIF1A would then occlude elements of 40S subunits that are involved in interaction with 60S subunits and possibly also induce conformational changes in 40S subunits that impair their binding to 60S subunits. The fact that eIF3’s moderate dissociating activity in the absence of eIF3j could be enhanced by increasing eIF3’s concentration three-fold from 250 nM (Figure 3) to 750 nm (Supplemental Figure S1C) suggests that stimulation by eIF3j of eIF3’s dissociating activity could at least in part be due to its stimulation of 40S/eIF3 association (Fraser et al., 2004). However, the recent report that the C-terminal portion of eIF3j binds directly to the mRNA-binding channel and A site at the intersubunit side of 40S subunits (Fraser et al., 2007) suggests that the mechanism by which eIF3j stimulates eIF3’s dissociating activity could be more complicated.
eIF1 mediates dissociation of P site deacylated tRNA
Incubation of pre-TCs with eRF1/eRF3 or puromycin resulted in similar levels (30–40%) of tRNA dissociation, as assayed by sucrose gradient centrifugation, suggesting that as in prokaryotes, peptide release destabilizes binding of P site tRNA. After recycling of post-TCs by either eIF3 alone or in combination with eIF1A and eIF3j, ~15% of P site deacylated tRNA remained bound to recycled 40S subunits in 40S/tRNA/mRNA complexes. Taking the stringency of sucrose gradient centrifugation into account, the true proportion of tRNA-bound recycled 40S subunits might be even higher. However, no tRNA was associated with recycled 40S subunits in the presence of eIF1, which therefore in addition to enhancing eIF3’s dissociating activity, also promotes tRNA release from recycled 40S subunits. Dissociation of P site elongator tRNA from 40S/mRNA/tRNA complexes by eIF1 is consistent with eIF1’s ability to dissociate 48S complexes assembled with initiator tRNA containing mutations in the three conserved GC pairs in its anticodon stem (Lomakin et al., 2006). In prokaryotes, dissociation of P site deacylated elongator tRNA from recycled 30S subunits is mediated by IF3 (Karimi et al., 1999), which discriminates against tRNAs that do not have three consecutive GC pairs in the anticodon stem, a unique feature of initiator tRNA (Laursen et al., 2005). Binding of IF3 induces conformational changes in the 30S subunit that enable GA1338-9 of 16S rRNA to monitor tRNA identity through minor groove interaction with these GC pairs (Dallas and Noller, 2001; Lancaster and Noller, 2005). eIF1 and the functional C-terminal domain of IF3 occupy the same ribosomal regions (Dallas and Noller, 2001; Lomakin et al., 2003), play similar roles in initiation codon and initiator tRNA selection and even perform some functions in heterologous systems in a reciprocal manner (Pestova and Kolupaeva, 2002; Lomakin et al., 2006). It is thus likely that the same mechanism underlies eIF1’s tRNA-dissociating activity.
eIF3j mediates dissociation of mRNA
Incubation of pre-TCs with eRFs or puromycin led to ~40% mRNA release from post-TCs, most likely as a direct result of destabilization of its ribosomal binding after spontaneous release of tRNA. After dissociation of post-TCs by eIF3 alone or in any combination with eIF1A and eIF1, 20–25% of mRNA remained bound to recycled 40S subunits, indicating stable association of mRNA with eIF3-bound 40S subunits even in the absence of P site tRNA, consistent with the mutual stabilization of mRNA and eIF3 on 40S subunits (Unbehaun et al., 2004; Kolupaeva et al., 2005). Again, regarding the stringency of centrifugation, the true proportion of mRNA-bound recycled 40S subunits might be higher. mRNA dissociation from recycled 40S subunits was mediated by eIF3j but only if eIF1 was present, which suggests that eIF3j can dissociate mRNA only if it is not stabilized by P-site tRNA. This activity of eIF3j was consistent with the negative cooperativity between eIF3j and mRNA in binding 40S subunits (Unbehaun et al., 2004; Fraser et al., 2007) and the fact that a high affinity for mRNA binding to the 40S subunit in the presence of eIF3j can be restored upon recruitment of P-site initiator tRNA (Fraser et al., 2007). Interestingly, toe-print analysis of 40S ribosomal complexes obtained in the presence of eIFs 2, 3, 1, 1A, 4A, 4B and 4F, but in the absence of eIF3j, predominantly yielded stops +15−17nt from the AUG codon that corresponded to 48S complexes, and only a very weak full-length cDNA that could be attributed to 40S/mRNA intermediates (Figure 2B). The lack of 40S/mRNA intermediates in such circumstances might be caused by displacement from mRNA of recycled 40S subunits by scanning 43S complexes. Similar displacement could be mediated by the even more processive elongating ribosomes. It is also worth noting that the yeast eIF3j ortholog HCR1 is not essential in S. cerevisiae (Valasek et al., 1999), which suggests that the mechanisms of recycling of post-TCs in budding yeast and in higher eukaryotes might not completely identical.
A model for ribosomal recycling by initiation factors and its implications
We propose the following model for dissociation of post-TCs (Figure 7). After peptide release, one or both eRFs remain bound to post-TCs. eIF3, eIF1, eIF1A and eIF3j cooperatively dissociate such post-TCs into free 60S subunits and mRNA- and tRNA-bound 40S subunits. eIF1 then promotes dissociation of P-site deacylated tRNA, after which eIF3j mediates release of mRNA. eIF3 clearly initiates recycling, but the order in which other factors join the process is unknown. Although eIF1 and eIF3j enhance dissociation of post-TCs into subunits, to emphasize their specific roles in tRNA and mRNA release, they are shown in Figure 7 to enter the recycling pathway at these stages. Although a high proportion of post-TCs underwent complete recycling even in the presence of eIF3 alone, this is not reflected in Figure 7 because the extent of dissociation of the relatively less stable 40S/mRNA and 40S/mRNA/tRNA intermediates may have been exaggerated by the stringency of centrifugation.
Figure 7. A model for eukaryotic ribosomal recycling.
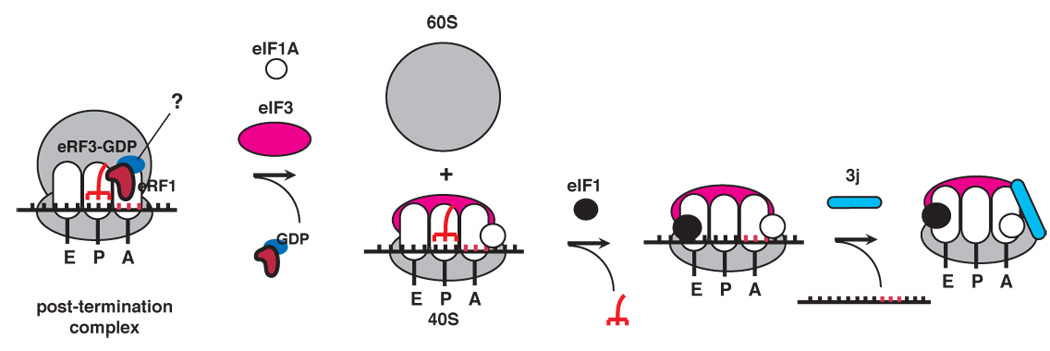
Despite obvious differences, this model has some close parallels with recycling in prokaryotes (Peske et al., 2005; Zavialov et al., 2005). Thus, eukaryotic post-TCs are also initially dissociated into large ribosomal subunits and tRNA-/mRNA-bound small subunits. Although the bulk of eIF3 binds to the 40S subunit solvent side (Siridechadilok et al., 2005) and it therefore seems as if in contrast to prokaryotes, dissociation of eukaryotic post-TCs involves eIF3 acting from this side of 40S subunits rather than from the intersubunit space, binding of part of eIF3j to the intersubunit surface of the 40S subunit (Fraser et al., 2007) suggests that dissociation might also involve eIF3 acting from within the intersubunit space. Moreover, subsequent ejection of deacylated P site tRNA is promoted by eIF1 in the mammalian system and by IF3 in prokaryotes (Karimi et al., 1999), factors that bind to identical regions on small ribosomal subunits and play equivalent roles during initiation.
What are the implications of the proposed mechanism? To explain the stimulatory effect on translation of the poly(A) binding protein (PABP), which binds the eIF4G subunit of eIF4F, eRF3 and the poly(A) tail of mRNA and thus brings the eIF4F-bound 5’-end of mRNA close to the termination site, it was proposed that PABP promotes shunting of terminating ribosomes to the 5’-end of the same mRNA (Uchida et al., 2002). Our finding that eIF3, which also interacts with eIF4G (Pestova et al., 2007), plays the principal role in recycling and remains bound to 40S subunits after this process, provides a missing link for the proposed mechanism of preferential participation of recycled 40S subunits in new rounds of initiation on the same mRNA.
Ribosomal recycling must influence post-termination events such as reinitiation and possibly nonsense mediated decay (NMD). Except in a few special cases, eukaryotic reinitiation occurs efficiently only after translation of short ORFs, following eIF4G-dependent initiation (Pöyry et al., 2004). Reinitiation likely requires prolonged retention of recycled 40S subunits on mRNA, which raises the question of how this is achieved after translation of short but not long ORFs if eIF3 mediates recycling in both cases. It has been suggested that eIF3/eIF4G remain bound to 80S ribosomes during the first few elongation cycles (Pöyry et al., 2004). Thus, after translation of short ORFs, eIF4G could stabilize binding of mRNA to recycled 40S subunits. Alternatively, whereas eIF3 that promotes recycling after translation of long ORFs joins post-TCs de novo and contains eIF3j, recycling after translation of short ORFs may be promoted by bound eIF3 from which eIF3j was displaced during initiation. The latter possibility is supported by a report that if 43S complexes were assembled with eIF3 containing eIF3j, this subunit was nevertheless released from assembled 48S complexes (Unbehaun et al., 2004). Any eIF3 that remains associated with 80S ribosomes derived from such 48S complexes likely still lacks eIF3j. However, although these explanations for the possibility of reinitiation after translation of short but not long ORFs are plausible, and our model is consistent will all current data, we cannot strictly rule out that the proposed recycling mechanism may be preferentially used after translation of short ORFs when eIF3 still remains associated with elongating ribosomes, and recycling after translation of long ORFs occurs by another mechanism. For instance, ATP-dependent ribosomal dissociation by yeast eEF2 has recently been reported (Demeshkina et al., 2007).
An early step in NMD, in which aberrant mRNAs that contain premature stop codons are degraded, involves binding of NMD factors Upf1 and SMG-1 to eRF1/eRF3 on prematurely terminating ribosomes to form a “SURF” complex (Kashima et al., 2006). eIF3 has been implicated in NMD (Morris et al., 2007) and the observations reported here, which implicate eIF3 in recycling, suggest that this process may be linked to NMD, for example in resolution of the SURF/post-TC complex.
EXPERIMENTAL PROCEDURES
Plasmids
Expression vectors for His6-tagged eIFs 1, 1A, 4A, 4B, 5, eRF1, 3j, eRF1(AGQ) mutant and eRF3aC lacking the N-terminal 138 a.a., which is referred to as eRF3 throughout the text, as well as transcription vectors for MVHL-STOP mRNA, +4U mRNA and Met-tRNAi have been described (Alkalaeva et al., 2006 and references therein; Pisarev et al., 2006; Unbehaun et al., 2006 and references therein). Transcription vectors for tRNAVal (GUG) and tRNAHis (CAC) were made by inserting DNA sequences flanked by a T7 promoter and a BstN1 restriction site into pUC57 (GenScript Corp.), and for −5U mRNA was made by inserting DNA flanked by a T7 promoter between PstI and SmaI restriction sites of pUC18 (Picoscript). To construct a vector for MQQ-STOP mRNA, DNA with an upstream T7 promoter was inserted between PstI and SmaI restriction sites in pUC18 (Picoscript). mRNAs and tRNAs were transcribed using T7 polymerase. For UV-cross-linking experiments, 32P-labeled −5U, +4U and MQQ-STOP mRNAs containing 4-thioU (8×106 cpm/mg) were transcribed from SmaI-digested plasmids in the presence of 4-thioUTP and [α32P]CTP (222 Tbq/mmol). For mRNA dissociation experiments, 32P-labeled MVHL-STOP and M(VF)5L-STOP mRNAs (1.5×106 cpm/mg) were transcribed in the presence of [α32P]ATP (222 Tbq/mmol). For toe-printing experiments, MVHL-STOP and MQQ-STOP mRNAs were transcribed from MscI- and SapI-digested plasmids.
Purification of factors and ribosomal subunits
Rabbit 40S and 60S subunits, eIFs 2, 3, 4F and 5B, eEF1H and eEF2, and recombinant His6-tagged eIFs 1, 1A, 4A, 4B, 5, 3j, eRF1, eRF1(AGQ) and eRF3 were purified as described (Alkalaeva et al., 2006 and references therein; Unbehaun et al., 2004).
eIF3 and eRF1 were 32P-phosphorylated (3×105 and 2×105 cpm/pmol) using the catalytic subunit of cAMP-dependent protein kinase, followed by purification on mono Q. 60S subunits were 32P-phosphorylated with casein kinase II (9×105 cpm/pmol) and purified by centrifugation through 10–30% sucrose density gradients.
Aminoacylation of tRNA
Rabbit aminoacyl-tRNA synthetases were purified, and native total rabbit tRNA (Novagen) and in vitro transcribed tRNAMeti, tRNAVal and tRNAHis were aminoacylated with Met, Val, His, Leu, Phe, as described (Pestova and Hellen, 2003). For peptide release experiments, transcribed tRNAMeti was aminoacylated using [35S]Met (6×105 cpm/pmol). Native tRNALeu was purified from total tRNA by gel-filtration on Superdex 75 and reverse phase chromatography on a Waters 3.9×300 mm Delta Pak C4 column (Unbehaun et al., 2004). [5’-32P]tRNALeu was obtained by dephosphorylating native tRNALeu with alkaline phosphatase followed by phosphorylation with [γ-32P]ATP by T4 polynucleotide kinase.
Assembly and purification of ribosomal complexes
48S/80S initiation complexes and pre-TCs were assembled on MVHL-STOP, MQQ-STOP, −5U and +4U mRNAs and purified by sucrose density gradient centrifugation essentially as described (Alkalaeva et al., 2006). For mRNA release, UV cross-linking, subunit dissociation and peptide release experiments, pre-TCs were assembled with [32P]MVHL-STOP, 4-thioU-containing [32P]MQQ-STOP, [32P]−5U and [32P]+4U mRNAs, [32P]60S subunits and [35S]Met-tRNAMeti, respectively. For tRNA release experiments, pre-TCs were assembled with in vitro transcribed Met-tRNAMeti, Val-tRNAVal and His-tRNAHis, and native Leu-[32P]tRNALeu.
Association of release factors with post-TCs
Pre-TCs (0.3 pmol) assembled on MVHL-STOP mRNA were incubated in 150 µl buffer A (20mM Tris, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM DTT) with 250 nM eRF1, [32P]eRF1, eRF1(AGQ) and eRF3, as indicated, in the presence of 1 mM GTP or GMPPNP for 10 min at 37°C and subjected to centrifugation through 10–30% sucrose density gradients prepared in buffer A in a Beckman SW55 rotor at 53,000 rpm for 75 minutes. Fractions that corresponded to ribosomal complexes were analyzed by toe-printing (Alkalaeva et al., 2006) using [32P]primer complementary to nt 197–214 of β-globin mRNA.
Post-termination assembly of 48S complexes
Pre-TCs (0.3 pmol) assembled on [32P]MVHL-STOP mRNA were incubated in 150 µl buffer A + 1 mM GTP with 250 nM eRF1, 250 nM eRF1(AGQ), 250 nM eRF3, 250 nM eIF3, 250 nM eIF2, 250 nM eIF1, 250 nM eIF1A, 250 nM eIF4A, 250 nM eIF4B, 125 nM eIF4F and 125 nM Met-tRNAMeti, as indicated, for 10 minutes at 37°C and subjected to sucrose gradient centrifugation as described above. Ribosomal association of mRNA was measured by Cerenkov counting of an aliquot of each fraction. Fractions that corresponded to ribosomal complexes were analyzed by toe-printing.
Dissociation of post-termination 80S ribosomes into subunits
Pre-TCs (0.3 pmol) assembled on MVHL-STOP mRNA with [32P]60S subunits were incubated in 150 µl buffer A + 1 mM GTP with 250 nM eRF1, 250 nM eRF3, 1 mM puromycin, 250 nM eIF3, 250 nM eIF1, 250 nM eIF1A, and 250 nM eIF3j, as indicated, for 10 minutes at 37°C and subjected to sucrose gradient centrifugation for 105 minutes. Association of ribosomal subunits was measured by Cerenkov counting of an aliquot of each fraction.
Association of eIF3 with recycled 40S subunits
Pre-TCs (0.3 pmol) assembled on MVHL-STOP mRNAs were incubated in 150 µl buffer A + 1 mM GTP with 250 nM eRF1, 250 nM eRF3, 250 nM [32P]eIF3, 250 nM eIF1, 250 nM eIF1A, 250 nM eIF3j, as indicated, for 10 minutes at 37°C and subjected to sucrose gradient centrifugation at 53,000 rpm for 75 minutes. Ribosomal association of [32P]eIF3 was measured by Cerenkov counting of an aliquot of each fraction.
mRNA release
Pre-TCs (0.3 pmol) assembled on [32P]MVHL-STOP mRNA were incubated in 150 µl buffer A + 1 mM GTP with 250 nM eRF1, 250 nM eRF3, 1 mM puromycin, 250 nM eIF3, 250 nM eIF1, 250 nM eIF1A, 250 nM eIF3j, 250 nM eIF2, and 125 nM Met-tRNAMeti, as indicated, for 10 minutes at 37°C and subjected to sucrose gradient centrifugation at 53,000 rpm for 75 minutes. Ribosomal association of mRNA was measured by Cerenkov counting of an aliquot of each fraction. Fractions that corresponded to ribosomal complexes were analyzed by toe-printing.
tRNA release
Pre-TCs (0.3 pmol) assembled on MVHL-STOP mRNA with Leu-[32P]tRNALeu were incubated in 150 µl buffer A + 1 mM GTP with 250 nM eRF1, 250 nM eRF3, 250 nM eIF3, and 250 nM eIF1, as indicated, for 10 minutes at 37°C and subjected to sucrose gradient centrifugation at 53,000 rpm for 75 minutes. Ribosomal association of [32P]tRNALeu was measured by Cerenkov counting of an aliquot of each fraction.
UV cross-linking assay
40S subunit-containing ribosomal complexes obtained by incubating pre-TCs assembled on 4-thioU-containing [32P]MQQ-STOP mRNA with eRF1, eRF3, eIF3 and eIF1, and purified by sucrose gradient centrifugation, 80S initiation complexes assembled on 4-thioU-containing 32P-labeled −5U and +4U mRNAs and also purified by sucrose gradient centrifugation were UV-irradiated at 360 nm (Pisarev et al., 2006). Ribosomal fractions were treated with RNase A and subjected to electrophoresis in NuPAGE 4%–12% Bis-Tris gel (Invitrogen) followed by autoradiography.
Peptide release assay
For experiments shown in Figure 6D, 2.5 nM pre-TCs assembled on MVHL-STOP mRNA with [35S]Met-tRNAMeti were incubated with 25 nM eIF1 and 25 nM eRF3, and for experiments shown in Figure 6E, 5 nM pre-TCs were incubated with 1.6 nM eRF1 and 1.6 nM eRF3 in 500 µl buffer A + 1 mM GTP at 37°C with or without 250 nM eIF3, 250 nM eIF1 and 250 nM eIF1A. Peptide release from 40 µl aliquots taken at different times was assayed by TCA precipitation (Zavialov et al., 2001).
Supplementary Material
ACKNOWLEDGEMENTS
We thank L.L. Kisselev for the gift of eRF1 and eRF3 expression vectors. This work was supported by NIH grant GM80623 to TVP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Hirokawa G, Kaji A, Kaji H. Novel activity of eukaryotic translocase, eEF2: dissociation of the 80S ribosome into subunits with ATP but not with GTP. Nucleic Acids Res. 2007;35:4597–4607. doi: 10.1093/nar/gkm468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JWB, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J. Biol. Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfusss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CUT, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 2006;34:3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C, Wittmann J, Jack HM, Jalinot P. Human INT6/eIF3e is required for nonsense-mediated mRNA decay. EMBO Rep. 2007;8:596–602. doi: 10.1038/sj.embor.7400955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T, Lorsch JR, Hellen CUT. Translational control in biology and medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. The mechanism of translation initiation in eukaryotes; pp. 87–128. [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CUT, Pestova TV. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valásek L, Hasek J, Trachsel H, Imre EM, Ruis H. The Saccharomyces cerevisiae HCR1 gene encoding as homologue of the p35 subunit of human translation initiation factor 3 (eIF3) is a high copy suppressor of a temperature-sensitive mutation in the Rpg1p subunit of yeast eIF3. J. Biol. Chem. 1999;274:27567–27572. doi: 10.1074/jbc.274.39.27567. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P. X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


