Abstract
We present single-molecule studies of the replication machinery of Escherichia coli and describe the visualization of individual E. coli DNA polymerase III (Pol III) holoenzymes engaging in primer extension and leading-strand synthesis. When coupled to the replicative helicase DnaB, Pol III mediates leading-strand synthesis with a processivity of 10.5 kb, 8-fold higher than that of primer extension by Pol III alone. Addition of the primase DnaG to the replisome causes a 3-fold reduction in the processivity of leading-strand synthesis, an effect dependent upon the DnaB-DnaG protein-protein interaction rather than primase activity. A single-molecule analysis of the replication kinetics with varying DnaG concentrations indicates that a cooperative binding of 2–3 DnaG monomers to the propagating DnaB destabilizes the replisome. The modulation of DnaB helicase activity through the interaction with DnaG suggests a mechanism that prevents leading-strand synthesis from outpacing lagging-strand synthesis during slow primer synthesis on the lagging strand.
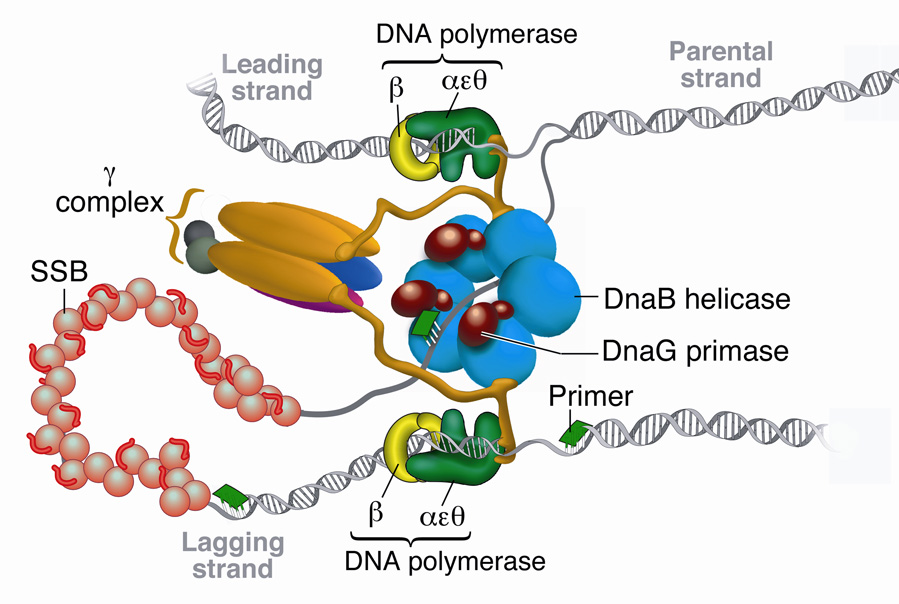
Complete and accurate replication of DNA involves the coordinated activity of a large number of proteins. The replisome, the molecular machinery of DNA replication, unwinds the double-stranded DNA (dsDNA), synthesizes primers to initiate synthesis, and polymerizes nucleotides onto each of the two growing strands1. The replication system of Escherichia coli is ideal for studying the dynamic interplay among the various components at the replication fork. The enzymes of the E. coli replisome duplicate DNA with remarkable efficiency: the replication fork moves at a rate approaching 1000 nucleotides per second while maintaining coordination between continuous synthesis on the leading strand and discontinuous synthesis on the lagging strand1,2. A fully functional replisome that displays all the fundamental enzymatic reactions characterizing DNA replication can be reconstituted in vitro with a limited number of purified key protein components: the DnaB helicase unwinds dsDNA; the DnaG primase synthesizes short oligoribonucleotides for priming of synthesis of the lagging strand; and the DNA polymerase III (Pol III) holoenzyme polymerizes nucleotides onto each nascent strand (Fig. 1)1,3.
Figure 1. E. coli replisome.
Schematic representation of the E. coli replisome depicting coordinated DNA synthesis. Three DnaG primase monomers are shown interacting with the DnaB helicase, adding an RNA primer (green) onto the SSB-coated lagging strand.
The Pol III holoenzyme is composed of three subassemblies: a core polymerase, sliding clamp, and clamp loader complex. The core polymerase is a heterotrimer of three subunits: α, the DNA polymerase; ε, proofreading exonuclease; and θ, which stabilizes ε4. The αεθ core is a poorly processive polymerase that only incorporates <20 nucleotides before dissociating from the primer-template5. However, when tethered to the sliding clamp, a ring-shaped homodimer of β subunits that encircles dsDNA, the processivity of the core increases dramatically to several kilobases (kb) at ~750 bp/s5. The loading of the β2 clamp onto the primer/template strand requires opening of the ring by the γ multiprotein clamp-loading complex6. The γ complex contains a number of different subunits that are required for clamp loading activity and coordination of the different enzymatic activities at the fork. A minimal γ complex that supports clamp loading contains three copies of the γ protein and one copy each of δ and δ’7. To tether the clamp loader to the dual polymerases at the fork, two γ subunits in the clamp loader complex are replaced by τ. γ and τ are products of the same gene, dnaX, with γ smaller by 34 kDa in the C-terminal region due to a programmed translational frameshift8. It is this C-terminal bridge domain of τ that is responsible for core-τ and τ-DnaB interactions9,10 (Fig. 1). Both τ and γ hydrolyze ATP and bind δ and δ’, modulating binding and opening of β27,11. Additionally, χ and ψ proteins associate with the γ complex to allow recognition of the single-stranded DNA (ssDNA)-binding protein SSB to facilitate lagging strand primase-polymerase switching12,13.
The DnaB helicase is a hexameric ATPase and is loaded onto ssDNA by DnaC and the primosomal proteins14. DnaB encircles the lagging strand and translocates towards the fork in a 5’ to 3’ direction, thereby displacing the complementary leading strand. DnaB catalyzes unwinding with poor processivity, but in the context of the replisome a single DnaB hexamer supports extensive synthesis of tens of kb15,16. Serving as the central anchoring component of the replisome, DnaB couples polymerase activity to fork propagation through τ-DnaB interaction and also binds the DnaG primase to regulate the production and deposition of primers on the lagging strand17. DnaG alone catalyzes the synthesis of ‘overlong’ ribonucleotide primers at a rate of ~1 primer/1000s, insufficient for proper replisome function18. Much like the other replisomal components, however, its efficiency increases considerably in the presence of the other replication proteins. When bound to DnaB, DnaG priming efficiency increases 1000 fold, synthesizing a primer every 1–2 seconds, resulting in a deposition of primers onto the lagging strand at ~1–2 kb intervals19–21.
As many as 3 DnaG monomers bind to a DnaB hexamer21. Several models have been posited to explain this stoichiometry and its functional relevance to replisome activity. Many structural and biochemical studies suggest that interactions between adjacent DnaG subunits docked to the DnaB hexamer regulate primase activity, though no definitive picture has yet emerged22,23. To understand how the DnaG-DnaB interaction modulates replisome dynamics requires a kinetic and quantitative characterization of the many transient intermediates involved in replication. With the ensemble averaging inherent in bulk biochemical methods, a population of reactions loses its synchronicity quickly after initiation of the reaction. This dephasing makes observation of any intermediates that occur during replication challenging. Recent advances in imaging and molecular manipulation techniques have made it possible to observe individual proteins and record “molecular movies” that provide new insight into protein dynamics and reaction mechanisms24,25. Here we report the use of single-molecule techniques to observe, in real time, the replication of individual DNA molecules by the E. coli replication machinery, thereby substantially extending the reach of in vitro single-molecule methods to the study of large (>10 proteins, >1 MDa) multiprotein complexes. The results detailed here suggest that the cooperative binding of three DnaG subunits to a DnaB hexamer destabilizes the replication fork. This modulation of leading-strand replication through the interaction of primase with DnaB suggests a mechanism that prevents leading-strand synthesis from outpacing lagging-strand synthesis during slow primer synthesis on the lagging strand.
RESULTS
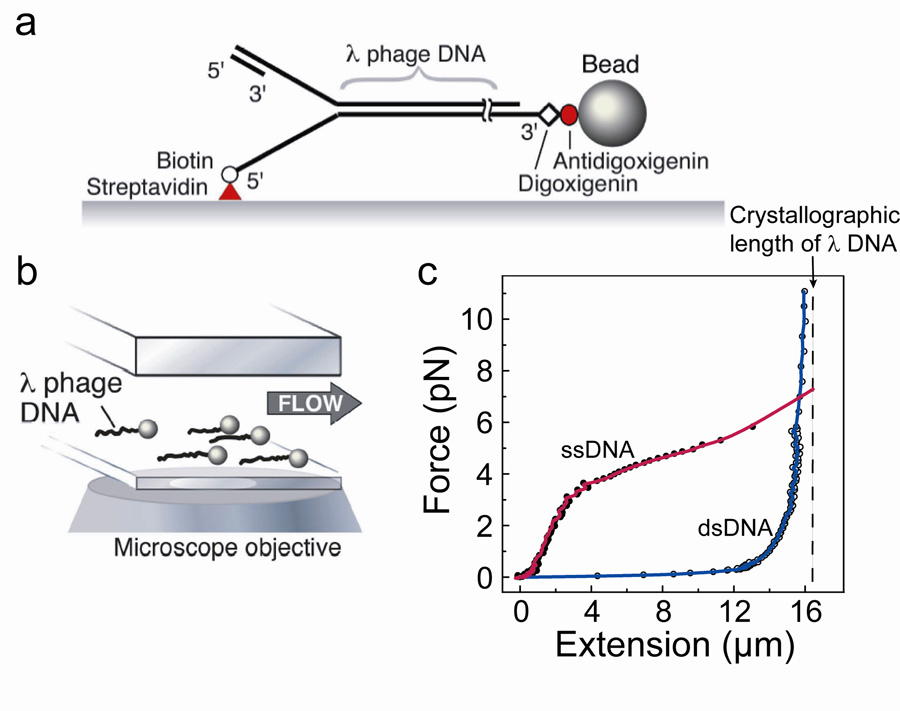
We characterize the kinetics of replication reactions at the single-molecule level by stretching individual DNA molecules and monitoring their lengths in the presence of the various E. coli replication proteins. The 5’ end of one strand of a 48.5 kb-long duplex λ phage DNA molecule is attached to the bottom surface of a glass flow cell via a biotin/streptavidin linker. The opposite 3’ end is linked using a digoxigenin/anti-digoxigenin interaction to a 2.8 µm-diameter bead (Fig. 2a). When a laminar flow is applied above the surface, a force proportional to the flow rate and the diameter of the polymer bead stretches the DNA molecules (Fig. 2b). We measure changes in the lengths of the individual DNA molecules by imaging the beads and tracking their positions. An intrinsic structural property of DNA is that ssDNA is substantially shorter than dsDNA at forces lower than 6 pN (Fig. 2c)26–28. As a consequence, conversion from dsDNA to ssDNA can be monitored through a decrease in total DNA length. Conversely, the production of dsDNA from an ssDNA template will result in a net lengthening of the polymer. We utilize these well-defined differences in length to characterize the activity of enzymes that convert one form of DNA into the other26,27,29–31.
Figure 2. Single-molecule experimental setup.
Single-molecule experimental setup. (a) Duplex λ DNA (48.5 kb) is attached to the surface of the flow cell via the 5′ end of the fork using a biotin-streptavidin interaction, and the 3′ end is attached to a paramagnetic bead using a digoxigenin-anti-digoxigenin interaction. A primed replication fork is formed at the end opposite the bead to allow loading and initiation of the replication proteins. (b) Bead-DNA assemblies are stretched using laminar flow of buffer and imaged using wide-field optical microscopy, permitting simultaneous observation of multiple individual replication reactions. (c) Extension profile of ssDNA (filled circle) and dsDNA (open circle) under low forces. Dashed line shows crystallographic length of fully ds-λ DNA, 16.3 µm29.
Pol III holoenzyme extends primers in short steps
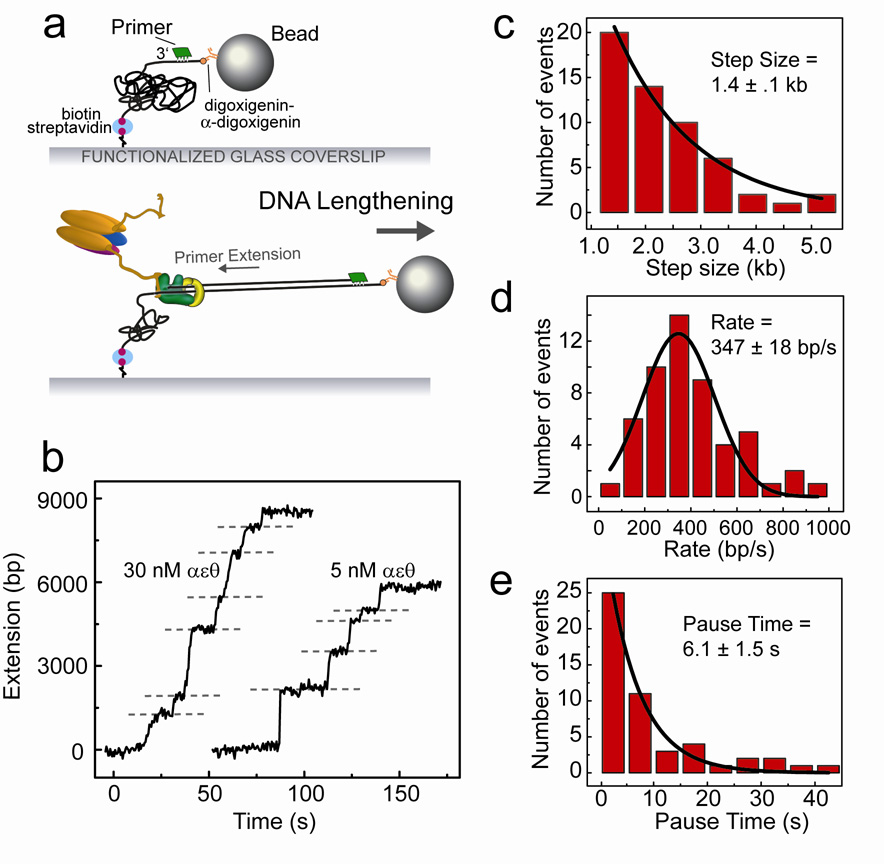
To demonstrate the effectiveness of DNA length measurements as an activity probe for E. coli replication proteins, we study DNA synthesis by individual Pol III holoenzymes. Polymerase activity can be visualized by attaching ssDNA molecules between the glass surface and the bead (Fig. 3a). To prepare ssDNA, surface-attached dsDNA was treated in situ with the replication proteins of bacteriophage T7, which catalyze extensive strand-displacement synthesis on our substrate29. After conversion of long stretches of dsDNA into ssDNA, the flow cell was washed extensively to remove excess proteins, and a 30-base DNA primer was introduced to anneal to the surface-attached ssDNA (Fig. 3a). E. coli Pol III holoenzyme was reconstituted in the flow cell by introduction of τ2γ1δδ’, αεθ, and β2 at equimolar (30 nM) concentration, together with the necessary ATP and dNTPs. (For information on protein purification and reconstitution, see Supplementary Material online.) The positions of DNA-attached beads were recorded on a CCD camera during primer extension by the Pol III holoenzyme and tracked over the course of the experiment. Bead position was then plotted versus time to generate single-molecule trajectories of enzymatic activity (Fig. 3b and Supplementary Fig. 1).
Figure 3. Primer extension by the Pol III holoenzyme.
Primer extension by the DNA polymerase III holoenzyme. (a) ssDNA is generated in the flow cell and an oligonucleotide primer is annealed (see text). Proteins are introduced with dNTPs and ATP, and primer-extension activity is measured as a lengthening of the tethered DNA. (b) Examples of lengthening traces, with extension at 30 nM αεθ on the left and 5 nM αεθ on the right. Dashed lines represent points where pauses in the DNA extension were identified. (c) Distribution of lengthening step sizes, fit with a single-exponential decay. (d) Distribution of rates, fit with a Gaussian distribution. (e) Distribution of lengths of pauses between lengthening steps, fit with a single-exponential decay. Data shown are from experiments performed with 30 nM Pol III holoenzyme.
Template-directed nucleotide incorporation at the 3’ terminus of the primer converts ssDNA to dsDNA and is observed as a lengthening of the DNA. The presence of a low concentration of holoenzyme in solution (30 nM) gives rise to short bursts of enzymatic activity, corresponding to repeated cycles of single extension events: association of the polymerase with the DNA; processive synthesis; and polymerase dissociation from the DNA primer-template (Fig. 3b). No length change was observed in the absence of dNTPs or with the αεθ core polymerase alone. The height of the individual steps is a measure of processivity and obeys a single-exponential distribution with a decay constant of 1.4 ± 0.1 kb (Fig. 3c). The single-exponential nature of the processivity distribution is consistent with a single rate constant determining the kinetics of the enzyme dissociating from the primer-template DNA32. The slopes of the DNA lengthening steps report the instantaneous polymerization rate and show a distribution with a mean of 347 ± 18 bp/s (Fig. 3d). If indeed every burst corresponds to the activity of a new holoenzyme complex associating from solution, the pauses between synthesis events should increase in duration upon decreasing the polymerase concentration. Repeating the experiment with limiting concentration of αεθ (5 nM) results in identical step sizes (1.3 ± 0.3 kb) and rates (505 ± 26 bp/s), but an increased pause time between steps, from 6.1 ± 1.5 s at 30 nM αεθ to 21.1 ± 2.6 s at 5 nM (Fig. 3b, e and Supplementary Figure 2). This increase in pause duration reflects the lower association rate of the Pol III holoenzyme complex with the primer at the lower concentration and confirms that the lengthening steps we observe are indeed true single-enzyme events. We obtain only a 3- to 4-fold increase in pause duration at a 6-fold decrease in concentration. This nonlinearity can be explained by the fact that only the concentration of αεθ is changed while the γ-complex/β2 concentration remains constant.
Early bulk-phase biochemical studies demonstrated a processivity of the Pol III holoenzyme of one hundred to several hundreds of nucleotides in the absence of SSB, though it has been difficult to obtain precise values5. The use of ensemble-averaging techniques to measure processivity values is often complicated by the occurrence of multiple, consecutive events of binding, synthesis, and dissociation on one and the same DNA molecule, a situation that is difficult to distinguish from a single, highly processive event. The real-time observation of primer extension by individual Pol III holoenzymes provides direct information on when a single enzyme starts and finishes synthesis, thus presenting a tool that allows for the analysis of polymerization kinetics with increased quantitative precision.
Leading-strand synthesis by the E. coli replisome is highly processive
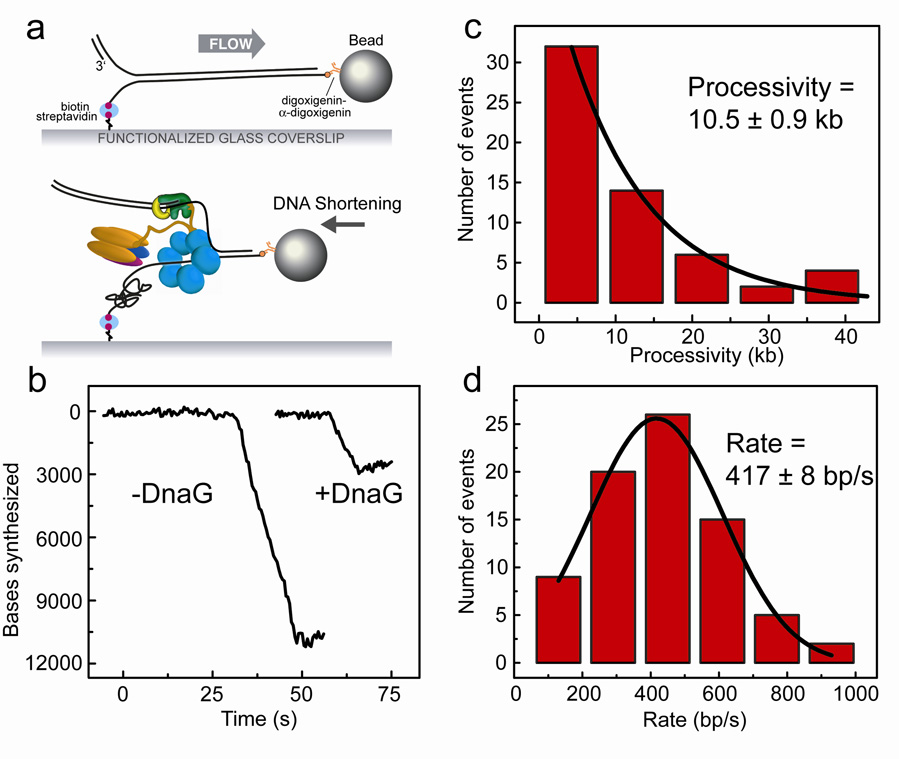
After establishing primer extension by the Pol III holoenzyme alone, we next examined DNA replication by Pol III together with DnaB. By flowing the Pol III holoenzyme and DnaB into the flow cell, we were able to initiate leading-strand synthesis on tethered λ dsDNA containing a pre-made replication fork (Fig. 4a). To facilitate loading of the hexameric DnaB onto the pre-made fork, we included the DnaB loader protein DnaC33. DNA synthesis catalyzed by the Pol III holoenzyme converts the ssDNA arising from DnaB helicase activity into dsDNA. To restrict replicative activity to the leading strand, we used a clamp loading complex of stoichiometry τ1γ2δδ’, allowing the assembly of only one Pol III core34. In addition, the absence of χ and ψ from the clamp-loading complex, and the omission of DnaG and SSB assure the preclusion of lagging-strand synthesis. Proteins were introduced at equimolar concentrations into a 37°C-heated flow chamber in the presence of ATP and dNTPs. In the presence of leading-strand synthesis and absence of synthesis on the lagging strand, the leading strand will be converted into dsDNA while the lagging strand remains in the single-stranded form. By attaching the DNA to the surface of the flow cell by the 5’ biotin-labeled lagging strand, leading-strand synthesis can be detected by an effective shortening of the DNA (Fig. 4a).
Figure 4. Leading-strand synthesis by Pol III holoenzyme with DnaB.
Leading-strand synthesis by Pol III holoenzyme coupled with DnaB helicase. (a) Duplex λ DNA with a premade replication fork is stretched under laminar flow, and Pol III holoenzyme, DnaB and DnaC are introduced into the chamber. Synthesis is observed as a shortening of the tethered DNA. (b) Examples of leading-strand synthesis traces, in the absence (left) and presence (right) of DnaG. (c) Distribution of leading-strand synthesis processivities, fit with a single-exponential decay. Leading-strand synthesis was observed as single shortening events with an average processivity of 10.5 kb. (d) Distribution of synthesis rates, fit with a Gaussian distribution.
Leading-strand synthesis was observed as highly processive DNA shortening events (Fig. 4b and Supplementary Fig. 4). No change in DNA length was seen when the experiment was performed without dNTPs or with the Pol III holoenzyme alone (Supplementary Fig. 4), which is not capable of strand-invasion synthesis9. In addition, no DNA length change was observed in the presence of only DnaB and its loader DnaC, which is consistent with the inability of the DnaB helicase to unwind stretches of dsDNA longer than 30 bp in bulk phase experiments15. The interaction between DnaB and τ is thought to increase helicase processivity9. However, when DnaBC and either the τ1γ2δδ’ or τ2γ1δδ’χψ complex was added to the flow cell, and the αεθ core and β2 were omitted from the reaction, no DNA length change could be seen. We therefore conclude that our observation of DNA shortening is due to DNA synthesis activity of a single E. coli leading-strand synthesis complex.
Analyzing 65 individual leading-strand synthesis reactions resulted in a measured processivity of 10.5 ± 0.9 kb and a mean synthesis rate of 417 ± 8 bp/s (Fig. 4c, d). This processivity value was lower than expected based on previously reported bulk-phase biochemical studies, which determined the processivity of the Pol III holoenzyme complexed to DnaB to be >50 kb16. A possible explanation is that the single-molecule experiments allow for an unequivocal discrimination between one processive event and multiple successive ones. The length of final DNA products as observed in bulk-phase experiments could contain the contributions of multiple synthesis events on an individual DNA molecule. Also, the topological challenge faced by DnaB to assemble on the surface-immobilized 5’ terminus of the pre-made fork in our experiments could result in a lower probability of reinitiation of leading-strand synthesis after dissociation of a complex from the primer template. Consistent with previous observations that χ and ψ function outside of leading-strand synthesis13, we found that the processivity and rate values are similar for reactions performed with τ2γ1δδ’ (8.5 ± 2.4 kb at 380 ± 51 bp/s) or τ2γ1δδ’χψ clamp loader complexes (7.5 ± 2.8 kb at 300 ± 73 bp/s). The force used in this experiment acts to stretch the DNA but is sufficiently low as to not affect local DNA-protein or basepair interactions35. As a control, reducing the DNA stretching force from 3 pN to 1 pN resulted in similar processivities and rates (7.4 ± 0.3 kb at 528 ± 133 bp/s). We therefore conclude that our experimentally observed values are accurate measures of single Pol III-DnaB leading-strand synthesis events.
DnaG binding to DnaB destabilizes the replisome
We next aimed to examine the effects of DnaG primase activity on leading-strand synthesis. Earlier single-molecule experiments on the bacteriophage T7 replication system revealed that primase activity resulted in a pausing of leading-strand synthesis29. We proposed that this tight coupling between primase activity and fork progression serves as a molecular brake that prevents leading-strand synthesis from surpassing lagging-strand synthesis while a primer is being synthesized. However, with the E. coli system, the addition of DnaG (300 nM) and rNTPs (200 µM each) to the leading-strand synthesis reactions resulted in a drastic reduction of the processivity, and no discrete pausing events were observed (Fig. 4b and Supplementary Figure 4). On average, the replication reaction stopped after synthesis of 2.9 ± 0.5 kb. The rate of leading-strand synthesis (340 ± 28 bp/s) was similar to that obtained in the absence of primase activity. Adding only rNTPs without DnaG did not result in a reduction of the processivity (10.8 ± 1.4 kb), indicating that the lower processivity of leading-strand synthesis observed with DnaG was not due to rNTP affecting polymerase activity (Fig. 5b).
Figure 5. Replicative abortion is independent of DnaG primase activity.
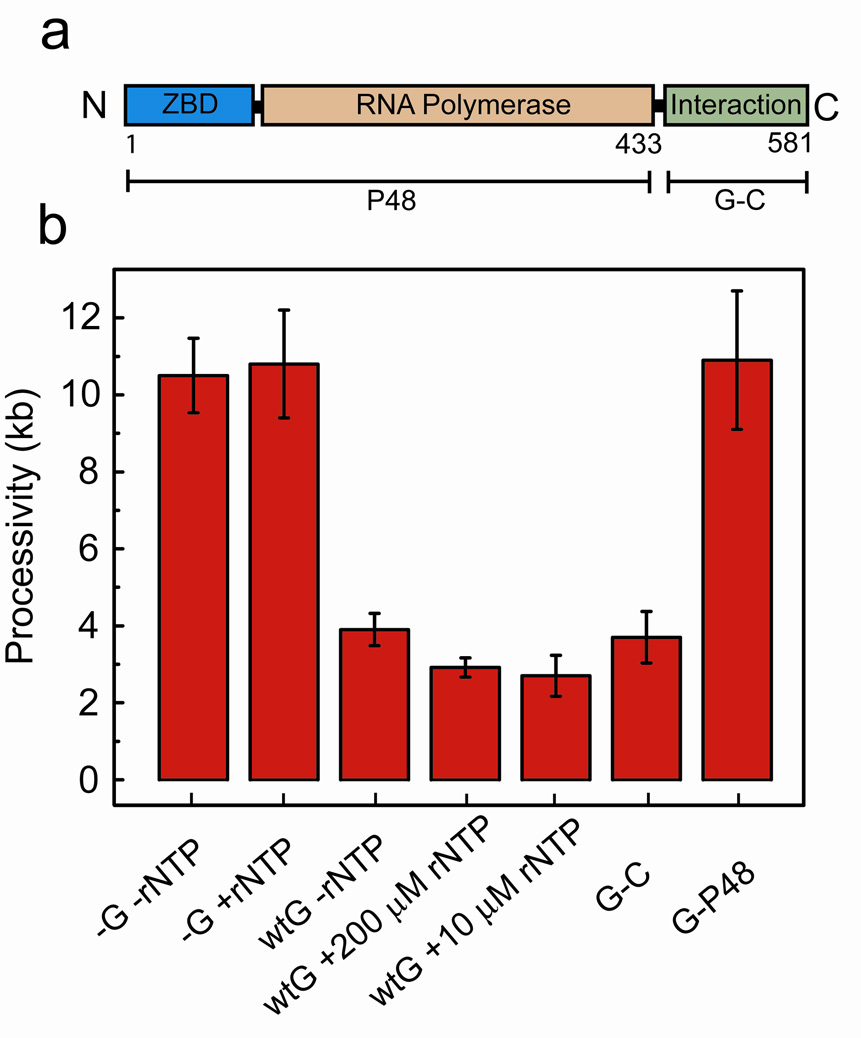
Replicative abortion is independent of DnaG primase activity. (a) DnaG is composed of three major domains: Zn-binding (ZBD), RNA polymerase, and DnaB-interaction domains. Mutant DnaG-P48 is a deletion of the C-terminal 148-residue interaction domain, and DnaG-C is a deletion of the N-terminal 433-residue primase domains as indicated by the brackets. (b) Processivities of leading-strand synthesis as obtained from single-molecule experiments under indicated conditions.
Based on these results, we sought to examine more fully the two aspects of DnaG function, primer synthesis and DnaG-DnaB interaction, in causing shortening of leading-strand synthesis. To reduce or inhibit primase activity while allowing DnaG to interact with DnaB, we carried out the experiment both with a decreased rNTP concentration and without rNTPs entirely. We observed consistently decreased processivities of 2.7 ± 0.5 kb and 3.9 ± 1 kb respectively, indicating that more frequent fork stalling is independent of rNTP concentration (Fig. 5b). Additionally, we repeated the experiments using a truncated DnaG that lacks the C-terminal 16 kDa domain responsible for the interaction with DnaB, but retains full primase activity (DnaG-P48, Fig. 5a)36,37. We observed a high processivity value (10.9 ± 1.8 kb), indicating that DnaG-DnaB protein-protein interactions are necessary to cause replicative stall (Fig. 5b).
To determine whether DnaG-DnaB protein-protein interactions are indeed sufficient to modulate leading-strand synthesis, we introduced a truncated DnaG comprised of only the 148-residue domain responsible for interaction with DnaB (DnaG-C, Fig. 5a)37. In the presence of 300 nM DnaG-C the processivity of leading-strand synthesis was measured to be 3.6 ± 0.5 kb, consistent with the value measured with the full-length primase (Fig. 5b). In summary, these results indicate that the DnaG-DnaB protein-protein interaction is the cause of premature abortion of leading-strand synthesis in the presence of DnaG.
Replisome disassembly is caused by the cooperative binding of multiple DnaG monomers to DnaB
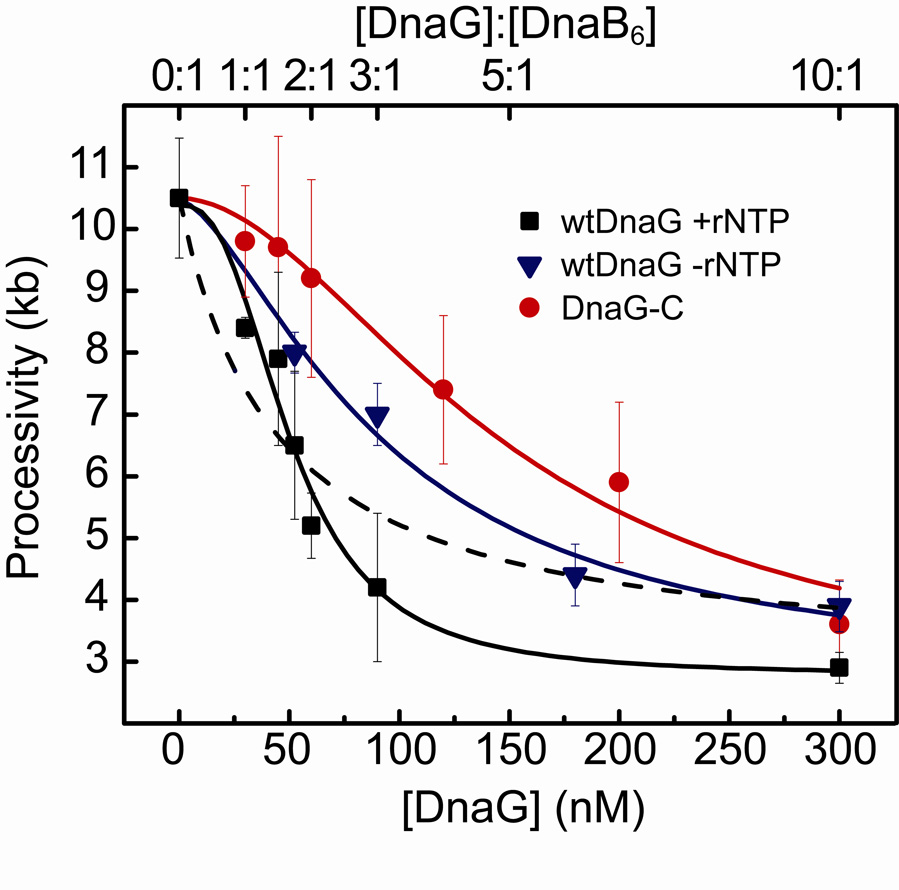
Previous studies have shown that up to three DnaG monomers can bind to a single DnaB hexamer21,37. Models of interactions between adjacent DnaB-bound DnaG monomers have been proposed as a mechanism of regulating primase activity22,23, and a recent crystal structure of DnaB with the interaction domain of DnaG from Bacillus stearothermophilus shows three DnaG domains in complex with a single DnaB hexamer38. To investigate the effect of oligomerization of DnaG on DnaB on replication fork progression, we performed the single-molecule leading-strand synthesis experiment with various concentrations of DnaG in the presence of rNTPs (200 µM each) and measured the processivity of leading-strand synthesis (Fig. 6, squares). We analyzed the data by fitting the processivities with a cooperative binding equation (Fig. 6) to examine the effect of DnaG stoichiometries on replication stability. If a simple hyperbolic dependence on the DnaG concentration were seen, we could conclude that the DnaG monomers bind to DnaB stochastically and independently, with a Hill coefficient of 1. By constraining the fit equation to a Hill coefficient of 1, a poor fit is obtained (Fig. 6, dashed line), with a χ2 value of 0.96. Any cooperativity would be evident as a sigmoidal curve with Hill coefficient >1. Indeed, a much better fit to the data is obtained (χ2=0.32) by using the Hill coefficient as a free fitting parameter, resulting in a Hill coefficient of 2.6. These data therefore suggest that cooperative binding of as many as 2 to 3 DnaG molecules to the DnaB hexamer during active DNA replication leads to a reduced processivity of the replisome. This cooperativity is different from earlier equilibrium binding studies of the interaction between DnaG and DnaB, which observed a non-cooperative interaction37. Additionally, we determined an effective KD of 50 ± 6 nM, significantly lower than the ~2 µM obtained from previously published binding studies37.
Figure 6. Cooperative DnaG-DnaB interaction is dependent upon RNA pol domain.
Cooperative DnaG-DnaB interaction is dependent upon the RNA polymerase domain of primase. Processivity values for increasing concentrations of wild-type DnaG with rNTPs (squares), of wild-type DnaG without rNTPs (triangles), and of DnaG-C without rNTPs (circles). DnaG concentration and DnaG:DnaB6 concentration ratio in solution is indicated at the bottom and top axis, respectively. Data are fit with the binding equation: , where h is the Hill coefficient, and A and B are scaling parameters. Fit lines are shown for each condition: DnaG+rNTPs (black; KD=50.7 ± 5.8 nM, h= 2.6 ± 0.8), DnaG–rNTPs (blue; KD= 93 ± 40 nM, h= 1.5 ± 0.8), and DnaG-C (red; KD= 150 ± 9 nM, h= 1.8 ± 0.2). The dashed line (blue) represents a fit to the data obtained with DnaG and rNTPs with the Hill coefficient h fixed at 1. Its poor fitting emphasizes the presence of cooperativity in DnaG-DnaB binding in the presence of ribonucleotides and RNA polymerase domains.
N-Terminal domain of DnaG is required for cooperative DnaG-DnaB interaction
Even though high concentrations of the DnaG mutant containing only the C-terminal DnaB interaction domain (DnaG-C, Fig. 5a) result in a reduced processivity, it is not clear whether DnaB-DnaG interactions are sufficient to mediate the cooperativity that we observed with the wild-type DnaG. By measuring the processivity of leading-strand synthesis with various concentrations of the DnaG-C mutant, we were able to examine the effect of the absence of Zn-binding (ZBD) and RNA polymerase (RNAP) domains on cooperativity between DnaG monomers in replication fork destabilization. Previous structural and biochemical work suggested the existence of interactions between the ZBD of one DnaG subunit and the RNAP domain of another. This cross talk between neighboring DnaG monomers while bound to a DnaB hexamer is thought to introduce cooperativity and play a role in primer synthesis22,23. Figure 6 shows a significant decrease in cooperativity over increasing amounts of DnaG-C (circles), with a lower Hill coefficient of 1.8. The curve is also noticeably shifted to the right, corresponding to a 3-fold increase in our apparent KD (150 ± 9 nM for DnaG-C versus 50 ± 6 nM for the full-length DnaG). This is consistent with a difference in KD of a similar magnitude observed in surface plasmon resonance studies of binding of DnaG and DnaG-C to ssDNA-bound DnaB37. The reduced cooperativity of the interaction between DnaG-C and DnaB emphasizes the importance of the ZBD and RNAP domains for the cooperative DnaG-DnaB interaction. To examine whether this effect is dependent upon primase activity, we performed the experiments with full-length DnaG protein in the absence of rNTPs. The resulting curve is similar to that with DnaG-C, with a Hill coefficient of 1.5 and a KD of 93 ± 40 nM (Fig. 6, triangles), indicating a role for rNTP binding in primer synthesis in the cooperative DnaG-DnaB interaction that mediates stalling of the replication fork. Thus the N-terminal ZBD and RNAP domains as well as primer synthesis all appear to be important to achieve cooperativity in the interaction between DnaG and DnaB. These results are consistent with models of inter-monomer interactions22,23 between the ZBD and RNAP domains of adjacent DnaG molecules bound to DnaB to facilitate cooperative interactions in this regulation of primase activity.
DISCUSSION
The E. coli replisome has long served as a paradigm for studying the various activities of DNA replication. Though classic studies have elucidated most of the numerous protein functions and interactions, several aspects of the replisome can be addressed only through direct examination of the components and activities of individual complexes. By using single-molecule DNA replication techniques, we report here the first single-molecule studies of functioning E. coli replication proteins. The direct observation of the different enzymatic activities during replication allows us to individually examine these protein complexes engaged in DNA synthesis, and by calculating effective stoichiometries and parameters of DnaG-DnaB interaction responsible for destabilization of the replisome, we demonstrate our ability to quantitatively describe protein-protein interactions in an active replisome.
The activities of the various proteins within the replisome are known to be coupled via several critical protein-protein interactions: α binds β2 for processive DNA synthesis; α binds τ, which binds DnaB, thereby coupling nascent strand synthesis to the propagating helicase at the fork; DnaG monomers bind to DnaB for efficient primer synthesis. Here, we have demonstrated the importance of two of these interactions for effective replication. The Pol III holoenzyme alone synthesizes approximately 1.4 kb on a ssDNA template and is incapable of strand-invasion synthesis, but when tethered to DnaB a single complex can synthesize stretches of DNA greater than 10 kb by coupling nucleotide polymerization to strand separation and fork propagation. We argue that the processivity of a single DnaB-Pol III holoenzyme leading-strand complex is only 10.5 kb, as varying the clamp loader complex stoichiometry or force of the experiment had no effect, and predominantly single shortening-event traces were observed (i.e., reinitiation on a synthesized DNA was readily observable but rare).
We observe a reduction in processivity of leading-strand synthesis upon addition of DnaG and ribonucleotides. In previous single-molecule studies on bacteriophage T7 leading-strand synthesis, we reported that during primer synthesis on the lagging strand, leading-strand synthesis pauses29. We proposed that this tight coupling serves as a molecular brake that prevents leading-strand synthesis from outpacing lagging-strand synthesis while a primer is being synthesized. However, the experiments described here demonstrate that the addition of the DnaG C-terminal domain that is responsible for interaction with DnaB but does not contain the RNA polymerase domain necessary for primase activity to the E. coli leading-strand synthesis reaction is sufficient to cause abortion of replication. This observation demonstrates that the termination of leading-strand synthesis is solely dependent on the interaction between DnaG and DnaB, and not on the synthesis of a primer. In the presence of primase activity, the interaction between DnaG and DnaB as observed in our single-molecule experiments is highly cooperative, whereas little cooperativity is observed when using the DnaG mutant unable to synthesize primers. This observation supports previously proposed interactions between the zinc-binding and RNA polymerase domains of adjacent DnaB-bound DnaG monomers22,23. Crystal structures of Bacillus stearothermophilus DnaB and helicase-binding domain of DnaG show three DnaG monomers interacting with a DnaB hexamer in conformations favoring close proximity, supporting the hypothesis that the primase monomers interact in trans38. These and similar interactions between primase monomers in the T7 replication system have been proposed to serve as a mechanism of regulating primase activity22,23,39. Our experiments demonstrate that even though primase activity is not essential for the premature abortion of leading-strand synthesis, it does modulate the kinetics of the fork halting.
If cessation of leading-strand synthesis is not caused by primase activity but solely by the interaction between DnaG and DnaB, then what is the underlying molecular mechanism? The fact that the processivity remains nonzero at high DnaG concentrations (Fig. 6) suggests that after association of a trimer of DnaG to the DnaB, an additional rate-limiting step needs to take place before replication is stopped. The observation that DnaG-DnaB interactions alone, as opposed to primase activity, are sufficient for this behavior suggests a model where DnaG destabilizes or stalls the replisome. A possibility is that cooperative binding of DnaG to DnaB during active DNA replication weakens the interaction between τ and DnaB, leading to more rapid disassembly of the replisome and a reduced processivity. From the single-molecule experiments, we obtained an effective KD of the interaction between DnaG and DnaB that is significantly lower than observed in bulk-phase binding studies (50 nM compared to 2 µM)37. This reduction most likely reflects the true strength of the DnaB-DnaG interaction in the context of the replisome, where association of DnaB with the τ subunit of Pol III strengthens its interaction with DnaG, whereas simultaneous interaction of 2 or 3 DnaG molecules with DnaB weakens its interaction with τ. The primase-mediated reduction in leading-strand processivity in the absence of SSB may indicate a regulatory mechanism to insure coupling of leading-strand synthesis to the normal sequence of events during primer synthesis on the lagging strand; primase is believed normally to dissociate promptly from DnaB following primer synthesis by association with SSB flanking the primer terminus13. In the persistent absence of SSB and lagging-strand synthesis, leading-strand replication could stall due to easier dissociation of the helicase from the polymerase.
These experiments nicely emphasize the strengths of real-time single-molecule observations of functional interactions at the replication fork. Subsequent studies will address dynamic interactions in coordinated DNA replication by complete E. coli replisomes. By introducing lagging-strand synthesis in the replication reactions29 we will be able to characterize the mechanisms that coordinate leading- and lagging-strand synthesis. Measuring timing and sizes of formed replication loops will allow us to examine primer handoff to the lagging-strand core polymerase, DnaG-DnaB interaction after primer synthesis, the role of χ, ψ, and SSB in coordination, and the dynamics of Pol III holoenzyme on the lagging strand. Examination of these and other questions will move us further towards the goal of direct observation of complete replisomes and full elucidation of the myriad aspects of DNA replication.
MATERIALS AND METHODS
Replication proteins
Methods for preparation of the following proteins from overproducing strains were essentially as described previously: DnaB helicase and DnaC helicase loader40, DnaG primase41, DnaG-P48 and DnaG-C37, and the Pol III holoenzyme subunits, α, δ and δ'42, ε and θ43, β244, γ and χ45. Subunit ψ was produced using strain BL21(λDE3)/pLysS/pET-ψ and refolded in the presence of χ to form the ψχ complex, essentially as described46,47. Plasmid pJC491, which directs overproduction of the τ, but not the γ subunit10 was used to produce τ in the ompT strain BL21(λDE3)recA48; purification of τ based on a published method49 is described in Supplementary Fig. 5. The αεθ core subassembly was reconstituted by mixing of purified α with excess ε and θ, then purified by chromatography on a column of DEAE-Sephacel (GE Healthcare), as described in Supplementary Fig. 6. The clamp loader assemblies τ1γ2δδ', τ2γ1δδ', and τ2γ1δδ'χψ were reconstituted by sequential mixing of purified subunits and separated by chromatography on a MonoS column (GE Healthcare), using modifications of the methods of Pritchard et al.50, as described in Supplementary Fig. 7.
Single-molecule DNA replication assay
Phage λ DNA molecules were annealed and ligated to modified oligonucleotides to introduce a biotinylated fork on one end of the DNA and a digoxigenin moiety on the other end as described previously29. The resulting DNA molecules were attached with the 5’ terminus of the bifurcated end to the streptavidin-coated glass surface of a flow cell and with the 3’ end of the same strand to a 2.8 µm diameter anti-digoxigenin-coated paramagnetic bead (Dynal). To prevent nonspecific interactions between the beads and the surface, a 1.7 pN magnetic force on the bead was applied upward by positioning a permanent magnet above the flow cell. Experiments were performed at 37 °C. Beads were imaged with a CCD camera (Q-Imaging Rolexa Fast) at a time resolution of 500 ms and their positions were determined by particle-tracking software (Semasopht). Traces were corrected for residual instabilities in the flow by subtracting traces corresponding to tethers that were not enzymatically altered, as described previously29 (Supplementary Figure 3). Pauses of primer extension were selected as a minimum of 5 data points (images taken at 2 Hz) with amplitude fluctuations less than three times the standard deviation of the noise (σ=30 nm). All error bars represent the error in fitting an exponential (processivity and pause time) or Gaussian distribution (rate).
Unless specified otherwise, E. coli proteins were introduced as: τ1γ2δδ’ (15 nM), αεθ (30 nM), β2 (30 nM), DnaB (30 nM, as hexamer), DnaC (180 nM, as monomer) and DnaG (various) in E. coli replication buffer (50 mM HEPES-KOH pH 7.9, 12 mM Mg(OAc)2, 80 mM KCl, 0.1 mg/ml bovine serum albumin with 5 mM dithiothreitol, 760 µM dNTPs and 1 mM ATP, added immediately prior to introduction to the flow cell). CTP, GTP and UTP (rNTPs) were added at 200 µM if desired. All proteins were present continuously during observation of single-molecule replication events.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Joseph Loparo, Harvard Medical School, for construction of a flow cell heating apparatus and critical reading of the manuscript and Drs. Ah Young Park and Mark Mulcair, Australian National University, for preparation of several proteins. We thank Steven Moskowitz, Advanced Medical Graphics for illustrations. This work was supported in part by funding from the National Institution of Health and National Science Foundation (A.M.v.O.) and by a grant from the Australian Research Council (N.E.D.)
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
REFERENCES
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Yuzhakov A, Turner J, O'Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 3.Wu CA, et al. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. IV. Reconstitution of an asymmetric, dimeric DNA polymerase III holoenzyme. J Biol Chem. 1992;267:4064–4073. [PubMed] [Google Scholar]
- 4.Kelman Z, O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 5.Fay PJ, Johanson KO, McHenry CS, Bambara RA. Size classes of products synthesized processively by DNA polymerase III and DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1981;256:976–983. [PubMed] [Google Scholar]
- 6.Bloom LB. Dynamics of loading the Escherichia coli DNA polymerase processivity clamp. Crit Rev Biochem Mol Biol. 2006;41:179–208. doi: 10.1080/10409230600648751. [DOI] [PubMed] [Google Scholar]
- 7.Jeruzalmi D, et al. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 8.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 10.Jergic S, et al. The unstructured C-terminus of the τ subunit of Escherichia coli DNA polymerase III holoenzyme is the site of interaction with the α subunit. Nucleic Acids Res. 2007;35:2813–2824. doi: 10.1093/nar/gkm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 12.Glover BP, McHenry CS. The χψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 13.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA--a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 14.Konieczny I. Strategies for helicase recruitment and loading in bacteria. EMBO Rep. 2003;4:37–41. doi: 10.1038/sj.embor.embor703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the Unzipping mechanism of the of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 16.Mok M, Marians KJ. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J Biol Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- 17.Schaeffer PM, Headlam MJ, Dixon NE. Protein--protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- 18.Swart JR, Griep MA. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 1995;34:16097–16106. doi: 10.1021/bi00049a025. [DOI] [PubMed] [Google Scholar]
- 19.Lu YB, Ratnakar PV, Mohanty BK, Bastia D. Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc Natl Acad Sci U S A. 1996;93:12902–12907. doi: 10.1073/pnas.93.23.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SK, Bhattacharyya S, Griep MA. DnaB helicase stimulates primer synthesis activity on short oligonucleotide templates. Biochemistry. 2000;39:736–744. doi: 10.1021/bi991554l. [DOI] [PubMed] [Google Scholar]
- 21.Mitkova AV, Khopde SM, Biswas SB. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J Biol Chem. 2003;278:52253–52261. doi: 10.1074/jbc.M308956200. [DOI] [PubMed] [Google Scholar]
- 22.Corn JE, Pease PJ, Hura GL, Berger JM. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol Cell. 2005;20:391–401. doi: 10.1016/j.molcel.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Corn JE, Berger JM. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 2006;34:4082–4088. doi: 10.1093/nar/gkl363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toprak E, Selvin PR. New fluorescent tools for watching nanometer-scale conformational changes of single molecules. Annu Rev Biophys Biomol Struct. 2007;36:349–369. doi: 10.1146/annurev.biophys.36.040306.132700. [DOI] [PubMed] [Google Scholar]
- 25.Greenleaf WJ, Woodside MT, Block SM. High-resolution, single-molecule measurements of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci U S A. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 28.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 29.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 30.van Oijen AM. Honey, I shrunk the DNA: DNA length as a probe for nucleic-acid enzyme activity. Biopolymers. 2007;85:144–153. doi: 10.1002/bip.20624. [DOI] [PubMed] [Google Scholar]
- 31.van Oijen AM, et al. Single-molecule kinetics of λ exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 32.Schnitzer MJ, Block SM. Statistical kinetics of processive enzymes. Cold Spring Harb Symp Quant Biol. 1995;60:793–802. doi: 10.1101/sqb.1995.060.01.085. [DOI] [PubMed] [Google Scholar]
- 33.Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J Biol Chem. 1989;264:2469–2475. [PubMed] [Google Scholar]
- 34.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Bustamante C, Smith SB, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Curr Opin Struct Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 36.Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 37.Oakley AJ, et al. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J Biol Chem. 2005;280:11495–11504. doi: 10.1074/jbc.M412645200. [DOI] [PubMed] [Google Scholar]
- 38.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 39.Lee SJ, Richardson CC. Interaction of adjacent primase domains within the hexameric gene 4 helicase-primase of bacteriophage T7. Proc Natl Acad Sci U S A. 2002;99:12703–12708. doi: 10.1073/pnas.202471499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.San Martin MC, Stamford NPJ, Dammerova N, Dixon NE, Carazo JM. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995;114:167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 41.Stamford NPJ, Lilley PE, Dixon NE. Enriched sources of Escherichia coli replication proteins. The dnaG primase is a zinc metalloprotein. Biochim Biophys Acta. 1992;1132:17–25. doi: 10.1016/0167-4781(92)90047-4. [DOI] [PubMed] [Google Scholar]
- 42.Wijffels G, et al. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides from β2-binding proteins. Biochemistry. 2004;43:5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- 43.Hamdan S, et al. Hydrolysis of the 5'-p-nitrophenyl ester of TMP by the proofreading exonuclease (ε) subunit of Escherichia coli DNA polymerase III. Biochemistry. 2002;41:5266–5275. doi: 10.1021/bi0159480. [DOI] [PubMed] [Google Scholar]
- 44.Oakley AJ, et al. Flexibility revealed by the 1.85 Å crystal structure of the β sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr D Biol Crystallogr. 2003;59:1192–1199. doi: 10.1107/s0907444903009958. [DOI] [PubMed] [Google Scholar]
- 45.Ozawa K, et al. Cell-free protein synthesis in an autoinduction system for NMR studies of protein-protein interactions. J Biomol NMR. 2005;32:235–241. doi: 10.1007/s10858-005-7946-4. [DOI] [PubMed] [Google Scholar]
- 46.Xiao H, Crombie R, Dong Z, Onrust R, O'Donnell M. DNA polymerase III accessory proteins. III. holC and holD encoding χ and ψ. J Biol Chem. 1993;268:11773–11778. [PubMed] [Google Scholar]
- 47.Gulbis JM, et al. Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur J Biochem. 2004;271:439–449. doi: 10.1046/j.1432-1033.2003.03944.x. [DOI] [PubMed] [Google Scholar]
- 48.Williams NK, et al. In vivo protein cyclization promoted by a circularly permuted Synechocystis sp. PCC6803 DnaB mini-intein. J Biol Chem. 2002;277:7790–7798. doi: 10.1074/jbc.M110303200. [DOI] [PubMed] [Google Scholar]
- 49.Maki S, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. I. Purification and distinctive functions of subunits τ and γ, the dnaZX gene products. J Biol Chem. 1988;263:6547–6554. [PubMed] [Google Scholar]
- 50.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ' with DnaX4 forms DnaX3δδ'. Embo J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.