Abstract
Homozygous ataxia (axJ) mice have reduced expression of ubiquitin-specific protease 14 (Usp14), resulting in severe neuromuscular defects and death by 2 months of age. Transgenic expression of Usp14 exclusively in the nervous system of axJ mice (axJ-Tg) prevents early lethality and restores motor system function to the axJ mice, enabling an analysis of the reproductive capabilities of Usp14-deficient mice. Although female axJ-Tg mice had a 75% reduction of Usp14 in the ovaries, they were able to produce normal litters. Ovary transfer experiments also demonstrated that the ovaries of axJ mice were capable of producing viable pups. In contrast, male axJ and axJ-Tg mice displayed a 50% reduction in testicular Usp14 levels and were infertile, indicating that Usp14 is required for development and function of the male reproductive system. Immunohistochemistry experiments showed that Usp14 is found in the redundant nuclear envelope and cytoplasmic droplet of epididymal spermatozoa. Analysis of axJ testes demonstrated a 50% reduction in testis weight, a 100-fold reduction in sperm number and the presence of abnormal spermatozoa in the epididymis. Histological examination of the Usp14-deficient testes revealed abnormal spermatogenesis and the presence of degenerating germ cells, indicating that Usp14 and the ubiquitin proteasome system are required for spermatid differentiation during spermiogenesis.
Keywords: ubiquitin, proteasome, testis, Usp14, fertility
Introduction
The ubiquitin-proteasome pathway provides the cell with a mechanism to control protein levels by regulated protein degradation (Glickman and Ciechanover, 2002). A poly-ubiquitin chain, which is attached to a substrate protein by a series of enzymatic reactions that involve an E1 activating enzyme, an E2 conjugating enzyme and an E3 ligase, targets a protein for degradation. The regulated degradation of proteins by the ubiquitin proteasome system is essential for many cellular processes including synaptic plasticity, embryonic development and gamete formation. Alterations in the ubiquitin proteasome pathway have been shown to result in neurological disorders, cancer and infertility (An et al., 2006, Reinstein and Ciechanover, 2006).
Deubiquitinating enzymes act antagonistically to protein ubiquitination and are thought to regulate three general processes: the production of monomeric ubiquitin by cleaving ubiquitin from ubiquitin-fusion proteins, recycling ubiquitin from ubiquitin-protein conjugates at the proteasome, and editing ubiquitin chains to regulate ubiquitin chain length. Although more than 90 deubiquitinating enzymes are encoded in the mouse genome, only Poh1, Uch-L5 and Usp14 have been shown to associate with proteasomes (Borodovsky et al., 2001; Lam et al., 1997; Leggett et al., 2002; Papa et al., 1999; Verma et al., 2002; Hu et al., 2005). In addition to regulating the entry of proteins into the proteolytic core of the proteasome, these three deubiquitinating enzymes are thought to help maintain the levels of monomeric ubiquitin by recycling ubiquitin chains from proteins targeted to the proteasome (Guterman and Glickman, 2004).
In axJ mice, loss of the deubiquitinating enzyme Usp14 leads to developmental abnormalities and death by 2 months of age (D’Amato and Hicks, 1965; Wilson et al., 2002). Although Usp14 is ubiquitously expressed (Wilson et al., 2002), only neurological defects have been reported in the axJ mice (D’Amato and Hicks, 1965). Early reports indicated that both axJ male and female mice were sterile (Lyon, 1955). However, it is not clear if the axJ mice suffer from a fertility defect or if they fail to reproduce due to their severe neuromuscular deficits. In our studies of axJ mice, transgenic expression of Usp14 specifically in the nervous system of the axJ mice was able to rescue the neurological defects and enabled the mice to have a normal lifespan (Crimmins et al., 2006). As a result, these transgenic rescue mice enabled us to explore possible non-neuronal functions for Usp14 and to determine if Usp14 was important for fertility. While most non-neuronal organ systems in the axJ-Tg mice did not demonstrate any gross signs of disease, we did observe male-specific fertility defects that included reduced testes size, decreased sperm production, morphologically abnormal spermatozoa and infertility. The results described in this study indicate that Usp14 is required for the development and function of both the nervous system and the male reproductive system.
Materials and Methods
Animals
Wild type C57BL/6J, Usp14axJ (Jackson laboratories, Bar Harbor, ME, USA), Usp14rrk114, and axJ-Tg mice have been maintained in our breeding colony at the University of Alabama at Birmingham, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Homozygous Usp14axJ mice (which we refer to as axJ mice) were generated by intercrossing heterozygous axJ/+ siblings and could be phenotypically identified by 2 weeks of age. Genotypes of each animal were confirmed using MIT markers flanking the axJ mutation. The construction of the axJ-Tg mice, which express Usp14 from the neuronal-specific Thy1.2 promoter, has been described previously (Crimmins et al., 2006). Heterozygous Usp14rrk114/+ mice were generated by injecting C57BL/6J blastocysts with the Bay Genomics RRK114 clone (Mutant Mouse Regional Resource Center, Davis, CA USA) and gene-trap mice were maintained as heterozygotes. All research complied with the United States Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals, and adhered to principles stated in the Guide for the Care and Use of Laboratory Animals, United States National Research Council.
Fertility studies
Eight-week old male and female mice were placed into mating for 4 months. Litter size was determined by counting the number of offspring at birth. A total of 10 matings were analyzed for each genotype examined.
Antibodies
Rabbit anti-USP15 antiserum (PW9795; Biomol, Plymouth Meeting, PA, USA) was raised against a peptide derived from human USP15. Mouse IgG against UCHL1 (Abcam ab20559, Cambridge, MA, USA) was raised against native UCHL1 from brain. Rabbit anti-UCHL3 antiserum (LS-A8724; MBL International, Woburne, MA, USA) was raised against a synthetic peptide from the C-terminus of UCHL3. Rabbit polyclonal antibody against the proteasomal 19S regulatory ATPase subunit PSMC1 (PW 8160; Biomol) was raised by immunization with full length recombinant S. pombe mts2 protein. Rabbit polyclonal antibody against the 20S proteasomal core subunits (PW 8155; Biomol) was raised by immunization of rabbits with proteasomal preparations purified from human red blood cells. Mouse IgG against the 20S proteasomal core subunits α1, 2, 3, 5, 6 and 7 (PW 8195; Biomol) was raised against a peptide corresponding to the prosbox I motif shared by these alpha-type subunits. Mouse IgG anti-ubiquitin FK1 (PW 8805; Biomol) was raised against polyubiquitinated lysozyme, specifically recognizes K29, K48 and K63 linked multi-ubiquitin chains, and does not react with non-conjugated monoubiquitin or with monoubiquitnated proteins. Mouse IgM anti-ubiquitin KM 691 (MC-033; Kamiya, Seattle, WA, USA) was raised against recombinant human ubiquitin, and recognizes unconjugated monoubiquitin as well as a variety of ubiquitinated proteins and multi-ubiquitin chains. All antibodies were used at a 1:100 dilution for overnight incubation. Rabbit anti-Usp14 antibodies were described previously (Anderson et al., 2005).
Isolation of mouse proteins
Mice 4 to 8 weeks of age and of appropriate genotype were sacrificed by CO2 asphyxiation. Tissues were homogenized in 1 to 3 mL of homogenization buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5% SDS, 2 mM N-ethylmalemide, and Complete Protease Inhibitors (Roche, Indianapolis, IN, USA). Following homogenization, tissues were sonicated for 10 s and then centrifuged at 17,000 g for 10 min at 4°C. Supernatants were removed and immediately frozen at −80°C. Protein concentrations were determined by using the Bicinchoninic Acid (BCA) protein assay kit from Pierce (Rockford, IL, USA).
Immunoblotting
Proteins were resolved on either 4-12% Bis-Tris gels or 4-20% Tris-Glycine NUPAGE gels (Invitrogen, Carlsbad, CA, USA) and transferred onto either nitrocellulose or PVDF membranes. Membranes were blocked in PBS containing 2% BSA. Primary antibodies were diluted in PBS containing 2% BSA and incubated at room temperature for 1 h. Primary antibodies were detected using a 1:5,000 dilution of anti-mouse or anti-rabbit HRP-conjugated secondary antibody (Southern Biotechnology Associates, Birmingham, AL, USA) and Luminol reagents (Pierce).
Quantitation of immunoblots
Blots were scanned using a Hewlett Packard Scanjet 3970 and quantitated using UNSCAN-IT software (Orem, UT, USA). Each value represents the average and standard error from six blots using three different wild type and mutant animals.
Histology
Whole testis and epididymis were dissected from the mice and the testis was nicked with a scalpel to facilitate fixation. The whole tissue was then submerged overnight in 4% paraformaldehyde. After fixation and thorough washing in PBS, tissues were embedded in paraffin by conventional procedures and positioned within a paraffin block so that the long axis of the testis and all three major epididymal compartments could be sectioned at the same time. Sections were cut on a microtome, mounted on microscopy slides, deparaffinized by xylene and stained by hematoxylin-eosin, antibody or incubated with X-gal to determine gene expression patterns.
Sperm counts
Epididymides of 6-week old mice were dissected and minced in PBS to liberate sperm. Sperm numbers from six animals were determined using a haemocytometer.
Immunohistochemistry
Mouse testicular and epididymal tissues were excised, washed in PBS, fixed in 4% paraformaldehyde and embedded in paraffin by using conventional methodology. After dewaxing and rehydration, the tissues were blocked with 5% normal goat serum (Sigma, St. Louis, MO, USA) for 25 min, incubated with the primary antibody overnight at 4°C, washed in buffer and incubated with appropriate secondary antibody conjugated to red fluorescent TRITC or green fluorescent FITC fluorochrome. To counterstain cell nuclei, DAPI (Molecular Probes, Carlsbad, CA, USA) was added to the secondary antibody solution at a final concentration of 5 μg/mL. Negative controls were generated by replacing the specific primary antibody with non-immune rabbit and/or mouse sera as needed, followed by the appropriate, species-specific secondary antibody conjugates. All dilutions of antibodies and sera and all washes were performed in PBS containing 0.1 % Triton-X-100 (Sigma), 1% normal goat serum and sodium azide. All secondary antibodies were purchased from Zymed (San Francisco, CA, USA).
Immunocytochemistry
Testicular cells or epididymal spermatozoa were released in TL-Hepes medium by mincing the appropriate tissues and were collected by a 5 min. centrifugation at 350 × g. Cells were attached to poly-L-lysine coated coverslips and fixed in 2% electron microscopy grade formaldehyde (PolyScience, Niles, IL, USA) as described (Sutovsky, 2004). Samples were blocked, and incubated with primary and secondary antibodies as described for immunohistochemistry. Coverslips with spermatozoa or testicular cells were mounted on conventional microscopy slides in VECTASHIELD® mounting medium (Vector Laboratories, Burlingame, CA, USA).
Analysis of Mouse Embryonic Fibroblasts
Mouse embryonic fibroblasts were generated from wild type and axJ embryos as previously described (Berthet et al., 2003). Briefly, primary wild type and axJ mouse embryonic fibroblasts were plated in triplicate at a density of 9 × 105 cells in a 6 cm dish. Cells were allowed to grow for 3 days, and were then harvested, counted and replated at the same density. The 3T9 assay was performed as previously described (Jones et al., 1996). To determine sensitivity to the DNA damaging agent Mitomycin C, mouse embryonic fibroblasts were plated in 6 well tissue culture plates and then, after adhering to the plates, were exposed to various concentrations of Mitomycin C for 24 h. To determine cell viability, cells were stained with Methylene blue as previously described (Bonora and Mares, 1982)
Cell and Tissue Imaging
Epifluorescence and differential interference contrast (DIC) images were acquired by using a Nikon Eclipse E800 microscope with a CoolSNAP hq monochrome camera and acquired by using MetaMorph v.4.6.7 software (Molecular Devices, Sunnyvale, CA, USA). Figure plates were edited using Adobe® Photoshop 5.5 (Adobe Systems, Inc., San Jose, CA, USA).
Quantitative PCR
Total RNA was isolated using RNA-STAT60 (Tel-Test, Friendswood, TX, USA) and 2 μg of the RNA was then reverse transcribed using the Applied Biosystems GeneAmp Gold RNA PCR Reagent Kit (Foster City, CA, USA). Real-time PCR reactions were set up in triplicate using TaqMan gene assays and amplified in an Applied Biosystems Step-One instrument. ΔΔCCT curves were generated using 18S TaqMan gene assays as internal standards. Quantitative PCR results are shown as the standard deviation of 3 different amplifications from RNA that was reverse transcribed from 3 different mice. Individual gene assay kits were purchased from Applied Biosystems for each of the RNAs analyzed. Paired t-tests were conducted on relative quantity (RQ) values for each group to determine their significance.
Results
The axJ males are infertile
To identify possible non-neuronal functions for Usp14, we first examined the reproductive capabilities of the axJ-Tg male and female mice, in which the movement disorders and early lethality of the axJ mice are restored by transgenically expressing Usp14 exclusively in neuronal tissues from the Thy1.2 promoter (Crimmins et al., 2006). To examine if the improved motor performance in the axJ-Tg mice corrects the reproductive deficiencies of the axJ mice, axJ-Tg mice were mated with wild type mice and monitored for their ability to produce offspring. All of the female axJ-Tg rescued mice exhibited normal fertility with litter sizes that were similar to those observed for the control wild type matings (Table 1). In contrast, none of the matings between the axJ-Tg males and wild type females were productive (Table 1), indicating that Usp14 expression is required for normal testes function.
Table 1.
Fertility of wild type (wt), axJ-Tg, and heterozygous Usp14rrk114 mice
| Genotypes of Breeding Pair | Fertile matings/Total attempted | Pups per litter n= 20 litters | |
|---|---|---|---|
| Female | Male | ||
| wt | wt | 10/10 | 7.3 ± 0.5 |
| axJ-Tg | wt | 10/10 | 7.1 ± 1.5 |
| wt | axJ-Tg | 0/10 | 0 |
| wt | Usp14rrk114/+ | 10/10 | 6.5 ± 1.7 |
| wt + axJ ovary | wt | 10/10 | 5.3 ± 0.8 |
Usp14 expression is reduced in the ovaries and testes of axJ and axJ-Tg mice
Our initial studies demonstrated that the axJ mutation results in hypomorphic expression of Usp14 in the axJ mice (Anderson et al., 2005). In order to compare the levels of Usp14 expressed in the axJ-Tg mice with the levels normally found in axJ and wild type mice, protein extracts from the brains, testes and ovaries of six-week old mice were immunoblotted for Usp14. Similar to our previous reports (Anderson et al., 2005), there was a 50 % reduction in the level of Usp14 in the testes and a 95 % reduction in the level of Usp14 in brain extracts of axJ mice as compared to wild type controls (Fig. 1). In the ovaries, there was greater than a 90 % reduction in Usp14 in the axJ ovaries as compared to controls (Fig. 1).
Figure 1.
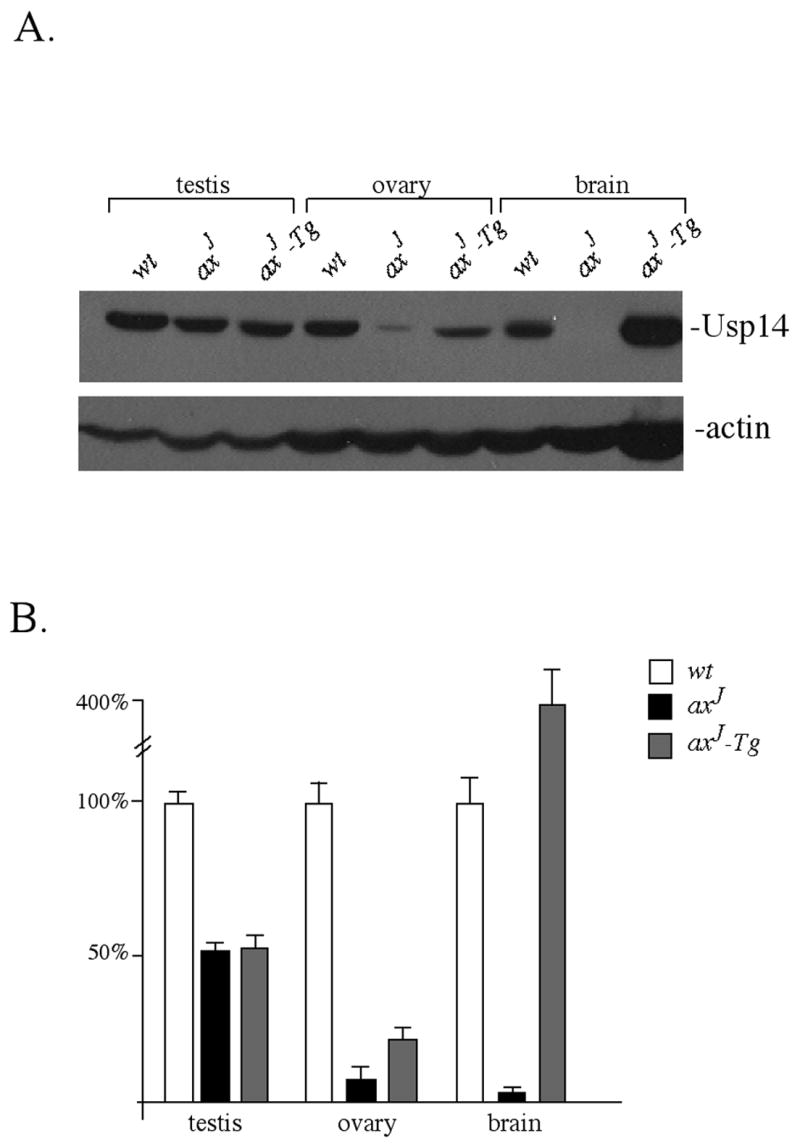
Expression of Usp14 in testes, ovaries, and brains of 6 week-old wild type (wt), axJ and axJ-Tg mice. (A) Immunoblots were probed with Usp14 antisera and with antibodies to β-actin as a loading control. (B) Quantitation of immunoblots with wild type levels of Usp14 set at 100%. Error bars indicate SE.
When we examined the level of Usp14 in the brains of axJ-Tg mice, we detected a 4-fold increase in expression of Usp14 compared to the level normally detected in wild type mice (Fig. 1), which is consistent with our previous reports (Crimmins et al., 2006) and with previous reports of gene expression from the Thy1.2 promoter (Caroni, 1997). In the testes, the level of Usp14 in the axJ-Tg mice was similar to that observed in the axJ mice, representing a 50 % reduction in Usp14 compared to the level observed in the testes of wild type mice (Fig. 1). In contrast, there was a 2-fold increase in Usp14 expression in the ovaries of the axJ-Tg mice as compared to the axJ mutant mice (Fig. 1). Although this slight increase was consistently observed, it still represented a 75% reduction in Usp14 compared to the levels observed in the ovaries of wild type mice.
To determine if the 2-fold increase in Usp14 expression in the axJ-Tg ovaries, relative to the level normally found in the axJ mice, was required for fertility, we examined the competence of the axJ ovaries by performing ovary transfer experiments. Ovaries from 4-week old axJ mice were transferred into wild type female mice and, following recovery, the recipient females were placed into breeding to determine fertility. All axJ ovaries were functional and resulted in the production of viable pups from the recipient hosts (Table 1), indicating that the 90% loss of Usp14 in the axJ mice is not detrimental to ovary function.
Usp14 expression is required for normal testicular function and sperm production
To determine if the fertility defect in the male axJ and axJ-Tg mice was due to a defect in testicular function, we initially compared the testes weights and sperm counts from the axJ, axJ-Tg, and wild type mice. At 6 weeks of age, there was a 50% reduction in the weights of the axJ and axJ-Tg testes as compared to wild type testes (Fig. 2A). In addition, there was a 100-fold reduction in epididymal sperm number in both the axJ and axJ-Tg mice compared to wild type mice (Fig. 2B). Many of the spermatozoa that were found in the axJ (Fig. 2C) and axJ-Tg mice (data not shown) exhibited abnormal morphology, with decapitated spermatozoa and two-tailed spermatozoa commonly observed.
Figure 2.
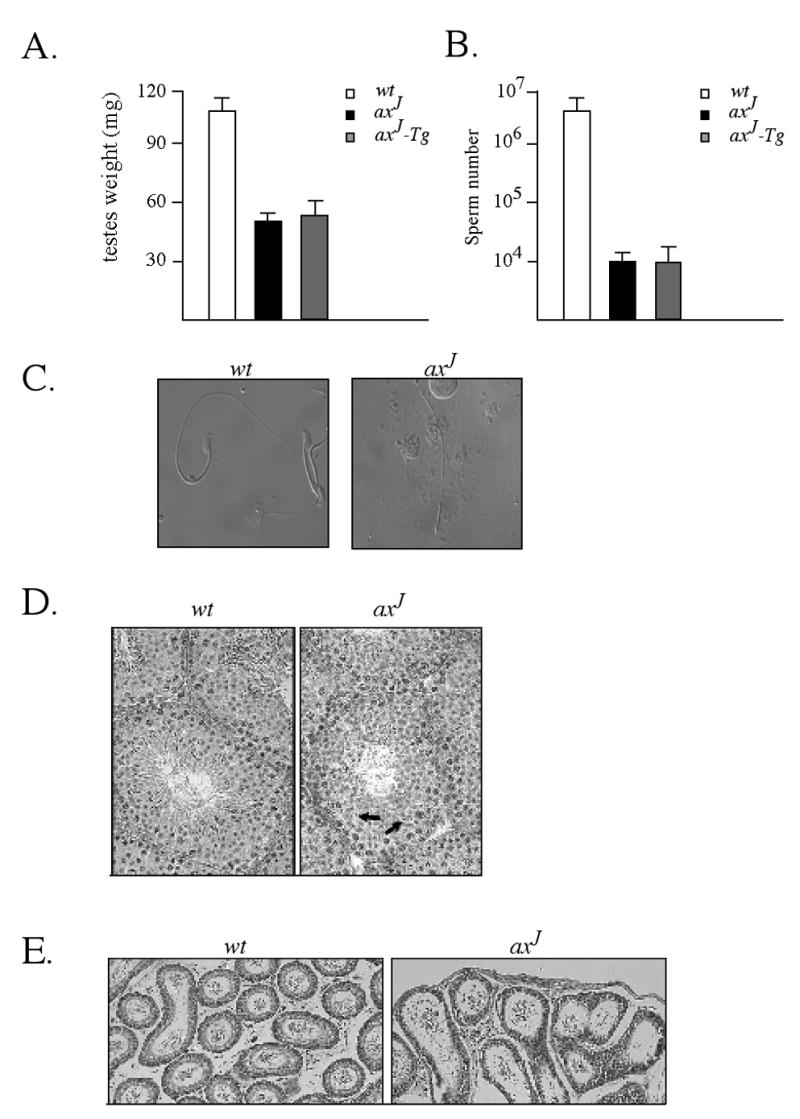
Reduced testis size and sperm production in axJ mice. (A) Testis weights were taken from 6 week-old wild type (wt), axJ and axJ-Tg mice (n=6). (B) Sperm counts were determined from the epididymis of 6 week-old wild type (wt), axJ and axJ-Tg mice (n=5). (C) Bright field microscopy of sperm isolated from the epididymis of 6 week-old wild type (wt) and axJ mice. (D) H&E staining of testis from 6 week-old wild type (wt) and axJ mice. Note few late step spermatids (arrows) in the axJ testis. (E) H&E staining of epididymis from 6 week-old wild type (wt) and axJ mice.
To investigate possible structural changes in the axJ testes, we examined testicular histology from 6-week old axJ and wild type mice. While we found normal numbers of spermatogonia, Sertoli cells and spermatocytes in the axJ mice, there were obvious defects in the spermatid elongation process, and multi-nucleated spermatids were commonly observed in the haploid phase of spermatogenesis (Fig. 2D). As a result, sperm release from the seminiferous tubules was minimal in the axJ mice, and few spermatozoa were found in the lumen of the epididymal tubule compared to wild type controls. Gross morphological defects were not observed in the epididymal epithelial cells of the axJ mice (Fig. 2E).
Differential effects of Usp14 expression on fertility
Our findings on the axJ-Tg mice indicated that a 50% loss of Usp14 expression in the testes is sufficient to cause male sterility in mice. However, since the axJ mutation has different affects on Usp14 expression in different tissues (Anderson et al., 2005), the effect of the intracisternal-A particle (IAP) may not be equivalent in all testicular cells. To determine if mice that had a uniform 50% loss of Usp14 in all tissues would also result in male sterility, we generated mice with a gene trap allele of Usp14. This allele of Usp14 (Usp14rrk114) was generated by the insertion of a gene-trap cassette containing a β-galactosidase reporter gene into intron 12 of Usp14 (Fig. 3A). To confirm allelism between the axJ mutation and the Usp14 rrk114 allele, we intercrossed heterozygous axJ mice with heterozygous gene trap mice. Analysis of the offspring of this mating revealed the presence of both normal and mutant animals, indicating that the Usp14 rrk114 and Usp14axJ alleles are non-complementing and are therefore allelic.
Figure 3.
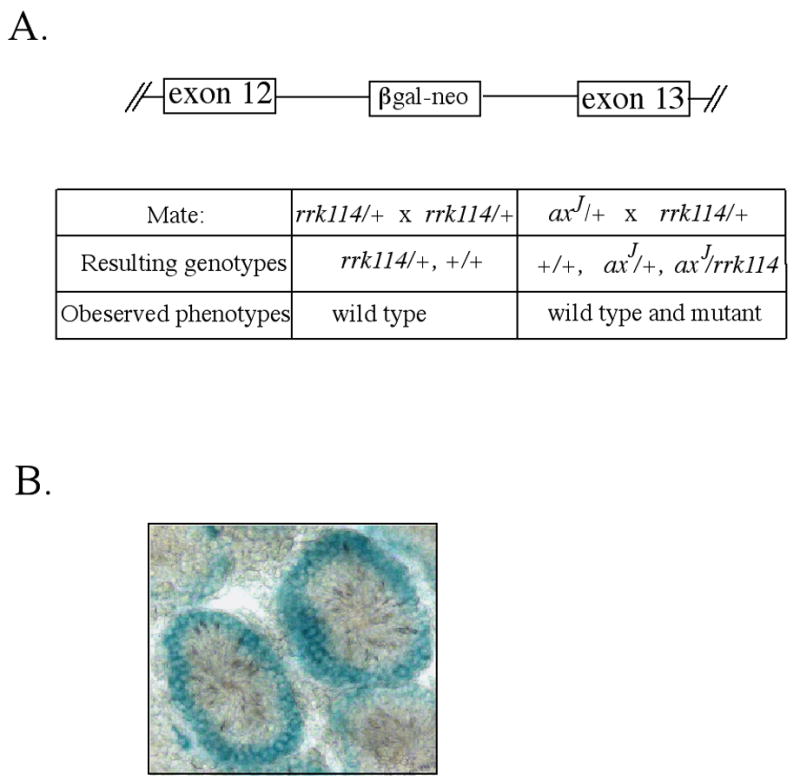
Structure and analysis of the Usp14 gene trap mice. (A) Schematic of the genomic structure of Usp14 containing the rrk114 gene trap insertion into intron 12. Breeding of heterozygous axJ/+ mice to heterozygous Usp14 gene trap (rrk114/+) mice demonstrated that the rrk114 gene trap is allelic to the axJ mutation. (B) X-gal staining of the testes from 6 week-old heterozygous gene trap mice (rrk114/+) demonstrating Usp14 expression in the periphery of the seminiferous tubules.
Immunoblot analysis of cell extracts from the testes, livers, and brains of heterozygous Usp14rrk114 mice demonstrated a 50 % reduction in the level of Usp14 compared to the levels normally found in wild type mice (Fig. 4A and B), which is consistent with the loss of expression of one allele of Usp14. Although the axJ-Tg mice also showed a 50% loss of Usp14 expression in the testes, there was a 95% reduction of Usp14 in the liver and a 4-fold increase in Usp14 expression in the brain relative to the levels normally observed in wild type mice (Fig. 4A and B). While mating of male axJ-Tg mice to wild type females did not result in the production of any offspring (Table 1), mating heterozygous Usp14 rrk114 males with wild type females generated normal litters (Table 1). These data therefore suggest that the axJ mutation has differential effects on Usp14 expression within testicular cells.
Figure 4.
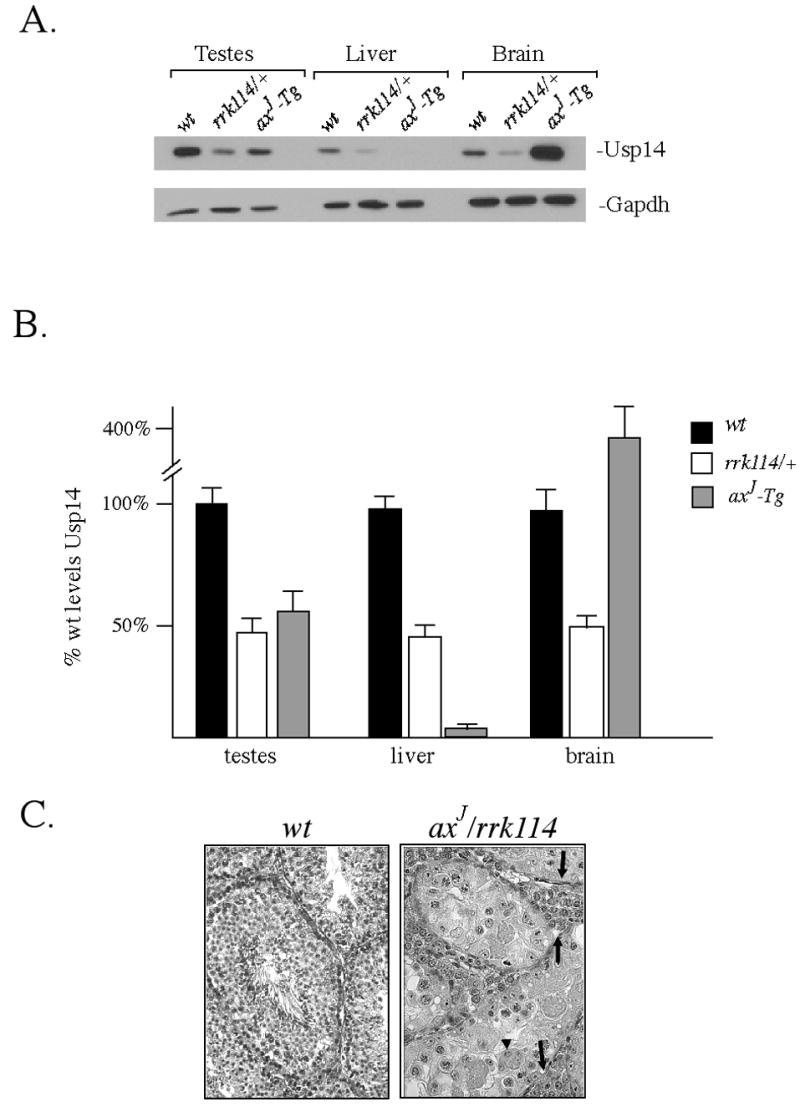
Analysis of Usp14rrk114 gene trap mice. (A) Immunoblot analysis of Usp14 expression in testes, livers, and brains of 6 week-old wild type (wt), heterozygous Usp14rrk114 gene trap (rrk114/+) and axJ-Tg mice. Blots were probed with antibodies for Usp14 and for GAPDH as a loading control. (B) Quantiation of Usp14 expression with wild type levels of Usp14 set at 100%. Error bars indicate SE. (C) Histological examination of H&E stained testis sections from 6 week-old wild type mice (wt) and mice that had both the Usp14rrk114 and axJ alleles (axJ /rrk114). Note multinucleated cells (arrowheads) and hypertrophied stroma (arrows) in the testis of the axJ /rrk114 mice.
Localization of Usp14 in testes
Since the expression of Usp14 from the Usp14 rrk114 gene trap allele can be monitored in situ by the production of β-galactosidase, testicular sections from 8-week-old heterozygous Usp14 rrk114 mice were stained with X-gal. Analysis of these sections demonstrated that Usp14 is expressed predominately in the diploid germ cell types located in the periphery of the seminiferous tubules (Fig. 3B).
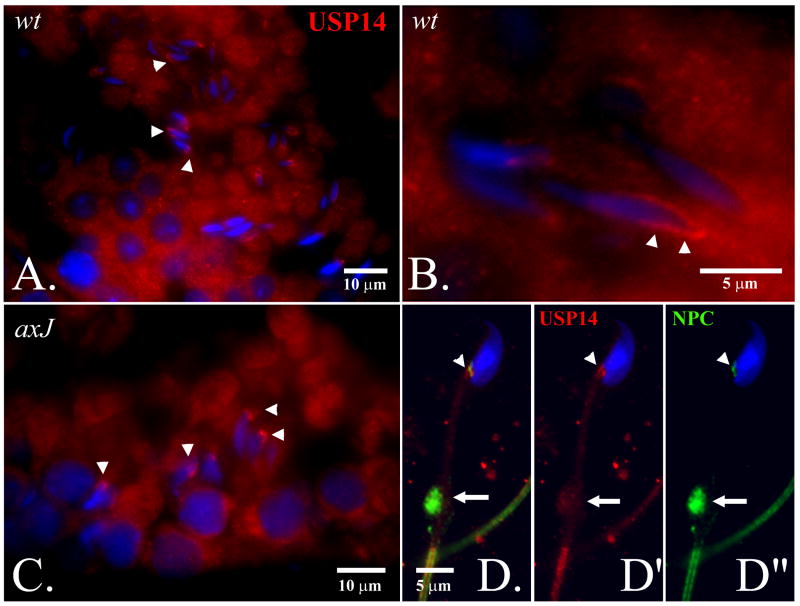
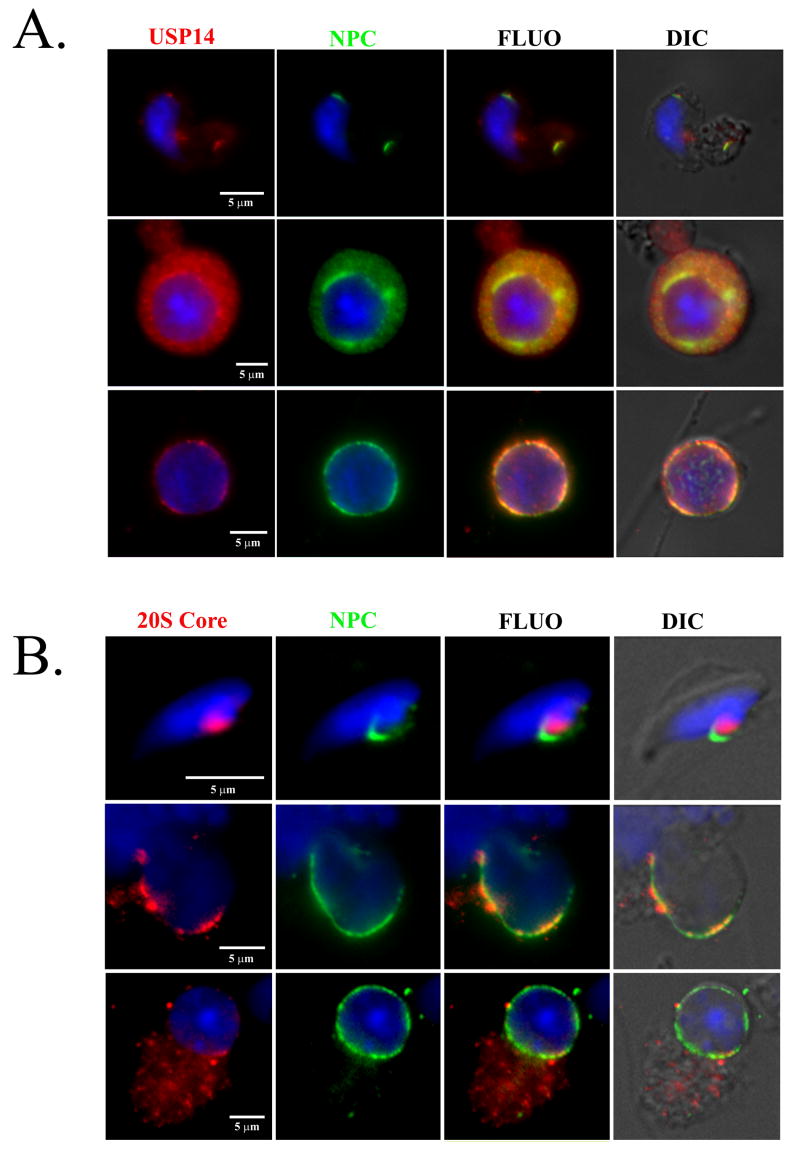
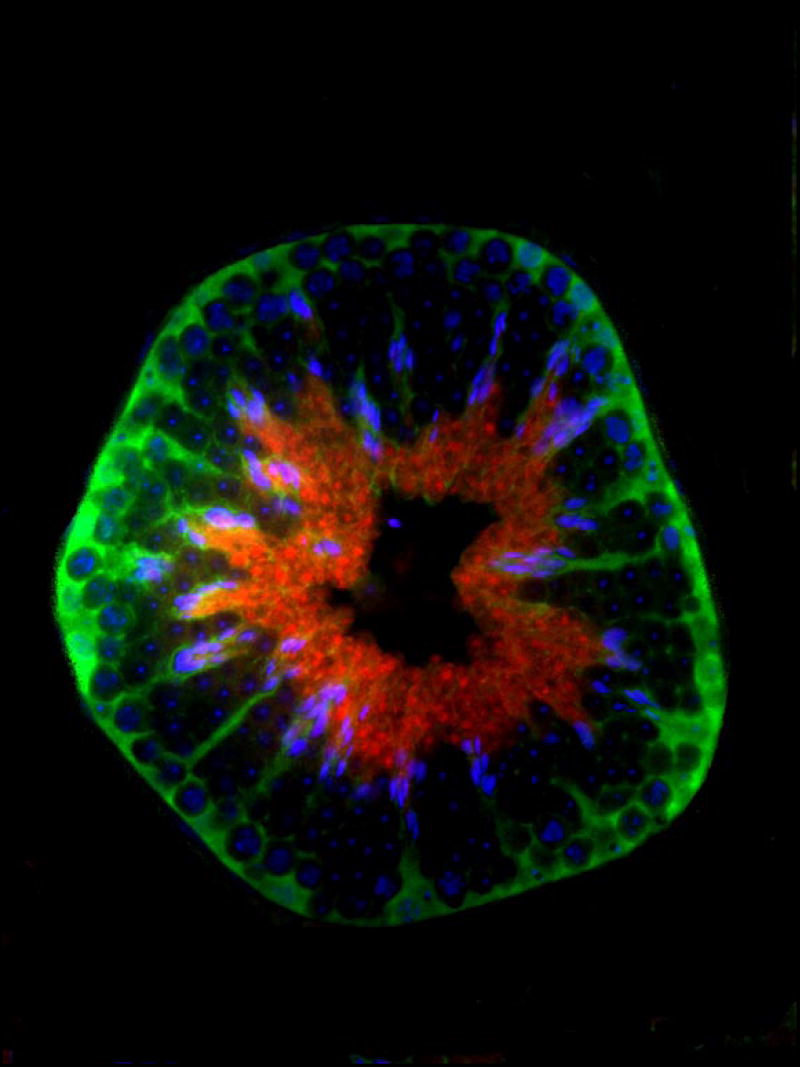
Due to notable defects of spermiogenesis in the axJ mutant males (Fig. 2), we investigated the distribution of Usp14 proteins in the testes of wild type and axJ mice, and in the spermatids and spermatozoa of wild type mice (Fig. 5). Usp14 was diffusely distributed in the cytoplasm of round and elongating spermatids and became associated with the postacrosomal segment of the spermatid nuclei in step 14-16 of wild type spermatids (Fig. 5A and B). This association was also observed whenever late step spermatids were present in the axJ testes (Fig. 5C). In step 16 spermatids, the area of Usp14 accumulation became smaller, coinciding with the localization of the redundant nuclear envelope (rNE) at the base of the spermatid nucleus. The appearance of rNE during spermatid elongation is a result of the progressive removal of nuclear pore complexes that parallels the hypercondensation of the spermatid nucleus (Sutovsky et al., 1999). Since the rNE is known to contain redundant nuclear pore complexes, we double-labeled isolated wild type spermatozoa (Fig. 5D-D”) and spermatids (Fig. 6A) to reveal colocalization of Usp14 with the redundant nuclear pore complexes. Consistent with Usp14 being a proteasome-associated deubiquitinating enzyme, the area of rNE also showed colocalization with Usp14 and proteasomes (Fig. 6).
Figure 5.
Localization of Usp14 protein (red) in wild type (A,B) and mutant (C) testis, and in the wild type spermatozoon (D-D”). Usp14 associates with the nascent postacrosomal sheath of the step 13-15 spermatid nuclei (arrowheads in A-C). In fully differentiated spermatozoa, Usp14 (red; D’) associates with redundant nuclear pores complexes (D”, NPC, green) present within the area of redundant nuclear envelope at the base of the sperm head (arrowheads in D-D”) and in the sperm cytoplasmic droplet (arrows in D-D”). DNA was counterstained with DAPI (blue).
Figure 6.
Localization of Usp14 (A; red) and proteasomes (B; red; 20S core subunits are labeled) in the mouse round (bottom row in both panels), elongating (middle row) and elongated (top row) spermatids, respective to localization of nuclear pore complex (NPC, green in A & B). DNA was counterstained with DAPI (blue).
Effect of Usp14 gene dosage on testicular development
To further examine the effect of Usp14 on testicular development, we analyzed Usp14rrk114/Usp14axJ males to investigate Usp14 gene dosage effects on testicular development. Immunoblot analysis of Usp14 expression revealed a 75 % reduction in Usp14 levels in the Usp14 rrk114/Usp14axJ males relative to the levels normally observed in wild type mice (data not shown). Examination of the testes structure from fixed tissue sections of wild type and Usp14rrk114/Usp14axJ mice showed that the testes of the Usp14rrk114/Usp14axJ mice were severely abnormal and degenerating (Fig. 4C). The testicular pathology varied among individual Usp14rrk114/Usp14axJ animals and, in some cases, we observed a hypertrophy of testicular stroma and hyper-proliferation of Leydig cells, along with severely reduced numbers of germ cells, demonstrating that Usp14 is required for the normal development of the testes in mice. In particular, some of the males (Figure 4C, rt panel) showed conspicuous degeneration of round spermatids, while pachytene spermatocytes were still present and did not seem to show meiotic defects. In other males, normal numbers of spermatocytes and spermatids were present (see Figure 2D), but spermatid elongation seemed impaired.
Analysis of axJ mouse embryonic fibroblasts
Since mutations in several DNA damage repair genes result in fertility defects (Yang et al., 2001, Ng et al., 2002), and the Ubp6 orthologue of Usp14 has been shown to genetically interact with components of the DNA damage repair pathway (Tong et al., 2004; Chang et al., 2002), we examined if there are changes in DNA stability in the axJ mice that could contribute to their observed fertility defects. Four separate lines of mouse embryonic fibroblasts from wild type and axJ mice were analyzed for growth rates and sensitivity to Mitomycin C (Fig. 7). There was no significant difference in the proliferative rates, number of generations to senescence, or sensitivity to mitomycin C between wild type and axJ embryonic fibroblasts, indicating that loss of Usp14 may not influence genomic stability.
Figure 7.
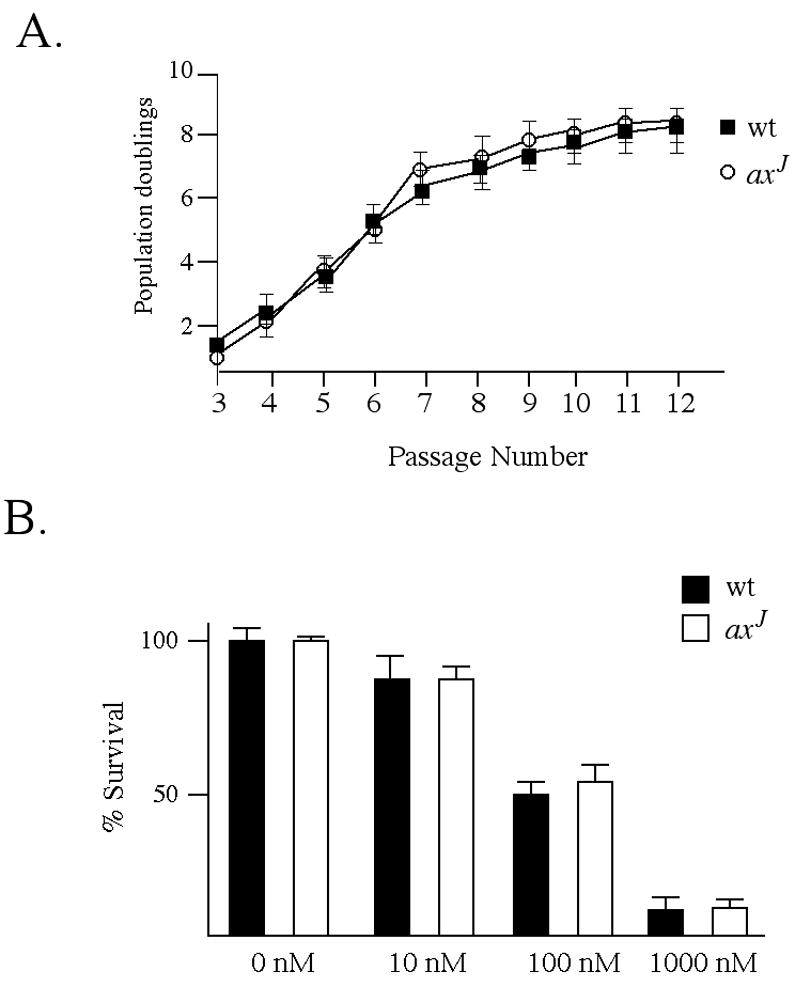
axJ mouse embryonic fibroblasts do not have altered growth rates or sensitivity to DNA damaging agents (A) 3T9 assay of mouse embryonic fibroblasts isolated from E14 wild type (wt) and axJ embryos. Cells were plated at a density of 9 × 105 cells per dish, allowed to grow for 3 days, harvested, counted and replated at the original density. Cell densities, indicated as population doublings, were recorded for the first 12 passages. (B) Sensitivities of mouse embryonic fibroblasts from E14 wild type (wt) and axJ embryos to the DNA damaging agent Mitomycin C. Cells were grown in the presence or absence of Mitomycin C for 24 h, and viability is presented as percent survival relative to wild type controls grown in the absence of Mitomycin C.
Genetic compensation of deubiquitinating enzymes in axJ testes
Since our previous data indicated that Usp14 functions to maintain ubiquitin levels (Anderson et al, 2005), we used quantitative PCR to examine the expression of other deubiquitinating enzymes involved in ubiquitin stability and determine if there is genetic compensation of deubiquitinating enzymes in the testes of the axJ mice. Consistent with our previous measurements of Usp14 protein levels (Fig. 1A), quantitative PCR analysis of Usp14 levels in the axJ testes demonstrated a 50% reduction in Usp14 mRNA compared to wild type controls (Fig. 8). When we examined the levels of the ubiquitin-carboxy-terminal hydrolases Uch-L1, Uch-L3 and Uch-L5, which are thought to play important roles in ubiquitin stability (Guterman and Glickman, 2002; Osaka et al., 2003), there was a 2-fold increase in all three of these transcripts in the axJ testes (Fig. 8). Usp5, a deubiquitinating enzyme that is important for processing linear ubiquitin chains, was also increased 1.4-fold in the axJ testes as compared to wild type controls (Fig. 8).
Figure 8.
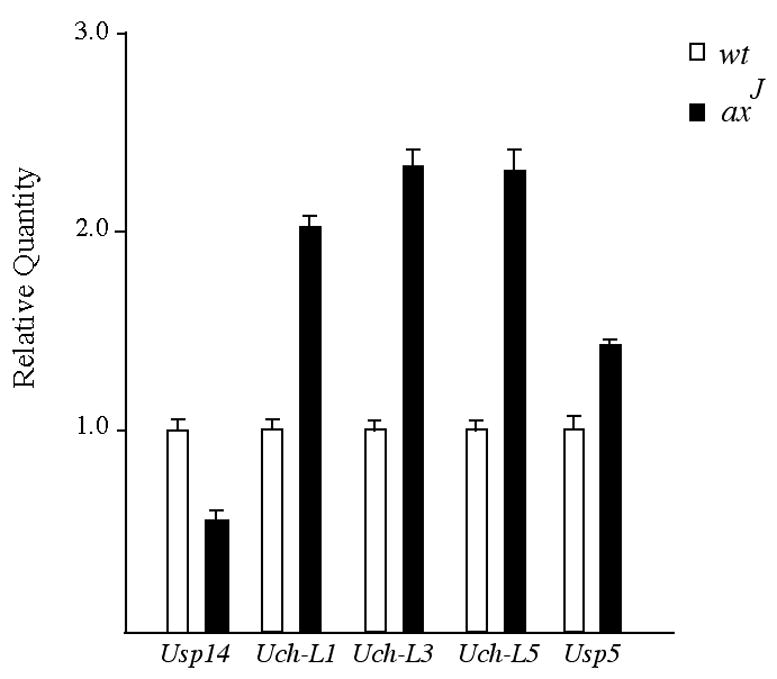
Genetic compensation of deubiquitinating enzymes in the axJ testes. Quantitative PCR was performed on RNA isolated from the testes of 6 week-old wild type and axJ mice. Gene assay primers were used to amplify individual deubiquitinating enzymes and ΔΔCT were generated by comparison to the internal GAPDH control.
Since the spermiogenesis defects were still observed in the axJ testes despite the presumed compensatory over-expression of the Uch-L1, Uch-L3, Uch-L5 and Usp5 deubiquitinating enzymes, we examined whether testis-expressed DUBs are spatially isolated from the rNE area in which Usp14 accumulates in spermatids (Fig. 9). Usp15 accumulated in the developing acrosomal cap of round spermatids in the testes of both wild type and axJ mice (Fig. 9A and B). As expected based on previous studies (Kon et al., 1999), the ubiquitin-C-terminal hydrolase Uch-L1 accumulated in the spermatogonia and Sertoli cells, but did not accumulate in spermatids from the wild type and axJ testes (Fig. 9C and D). In contrast, Uch-L3 accumulated in the cytoplasmic lobes of the elongating spermatid, as reported previously in boar (Yi et al., 2007), but was not found in the rNE of late step spermatids in the testes of wild type mice. Due to abnormal spermatid elongation, few elongating spermatids with cytoplasmic lobes containing Uch-L3 were found in the axJ testes (Fig. 9D). There was a striking accumulation of the ubiquitin activating enzyme E1 in the nuclei of step 10-15 elongating spermatids in the testes of both wild type and axJ mice (Fig. 9E and F). The 19S proteasomal regulatory complex subunit PSMC1 showed localization in the nascent postacrosomal sheath of the spermatid nucleus in late step spermatids (Fig 9G and H), which was similar to the localization of Usp14 in the wild type and axJ testes. None of these localization patterns were observed with non-immune rabbit sera in negative control slides (Fig. 9I and J). The deubiquitinating enzymes that are expressed in the male germ line therefore show a spatially compartmentalized and temporally-regulated expression pattern, indicating that they may have unique, non-redundant, non-compensating functions during spermatogenesis, and spermiogenesis in particular.
Figure 9.
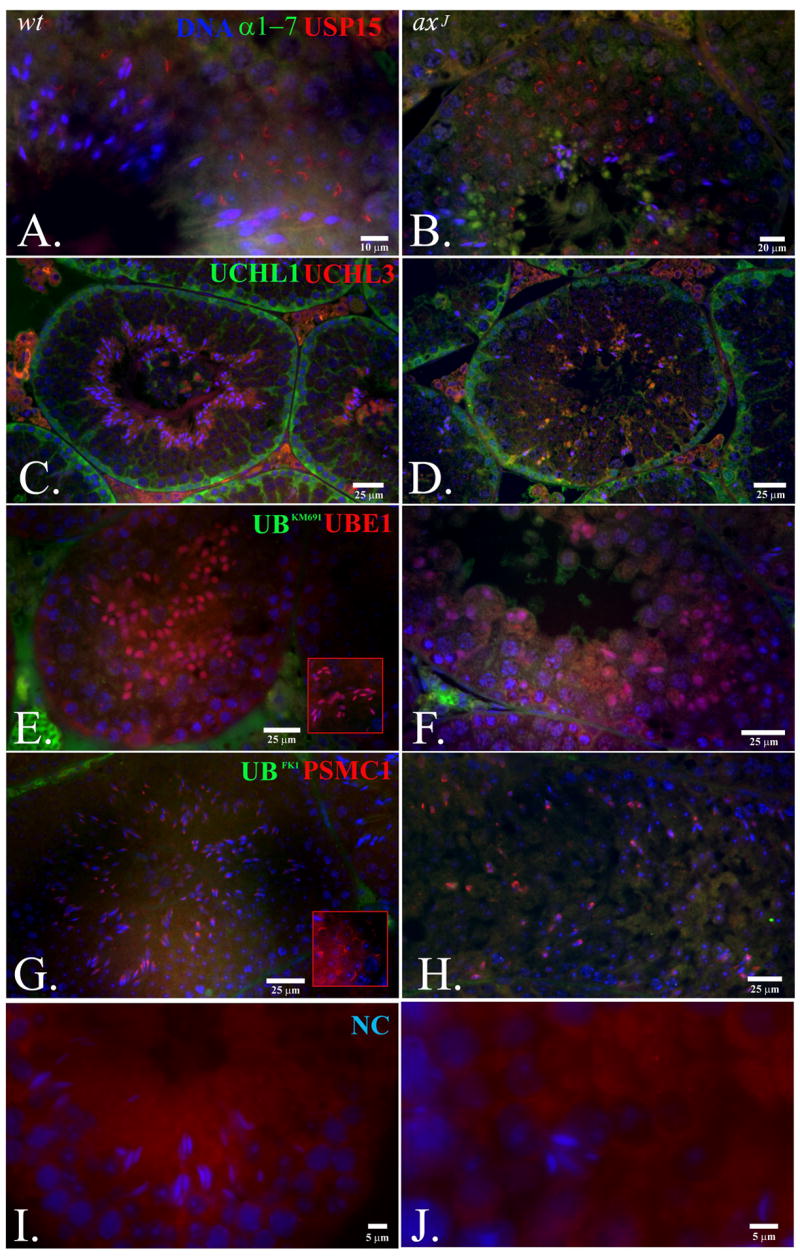
Distribution of deubiqutinating enzymes and other components of the ubiquitin proteasome system in wild type (wt, left column) and axJ (right column) testis. Usp15 (red staining in A and B) accumulated in the developing acrosomal cap of round spermatids in the testis of both wt and axJ mice. The 20S alpha subunits (α1-7; green staining in A and B) accumulated in the cytoplasmic lobes of spermatids from the axJ mice. The Ubiquitin-C-terminal hydrolase Uch-L1 (green staining in C and D) accumulated in the spermatogonia adjacent to the basement membrane of the seminiferous tubules. Uch-L3 (red staining in C and D) accumulated in the cytoplasmic lobes of the elongating spermatids. The Ubiquitin activating enzyme E1 (UBE1; red staining in E and F) accumulated in the nuclei of elongating spermatids. Ubiquitin is labeled green with antibody KM691 (E and F). Labeling of the 19S proteasomal regulatory complex subunit PSMC1 (red staining in G and H) showed the accumulation of proteasomes in the postacrosomal sheath of the late step elongating spermatids. Testis were also labeled with the anti-ubiquitin antibody FK1 (green staining in G and H). Cells were stained with non-immune rabbit serum as a negative control (I and J).
Discussion
Research on the ubiquitin proteasome system has demonstrated that proteins involved in ubiquitin-dependent signaling and proteolysis are critically important for a variety of cellular functions. Central to all ubiquitin signaling events is the maintenance of cellular pools of monomeric ubiquitin. Mutations in Usp14 and Uch-L1, two different deubiquitinating enzymes that have been implicated in ubiquitin stability, have resulted in both nervous system and testicular dysfunction (Kwon et al., 2005; Anderson et al., 2005; this study). However, only the Usp14 mutation (this study) caused complete male infertility. By stabilizing the mono-ubiquitin pool, Uch-L1 and Usp14 are believed to provide sufficient levels of ubiquitin to allow for ubiquitin-mediated proteolysis, receptor endocytosis and lysosomal trafficking. The extensive remodeling of cellular constituents that occurs during spermatogenesis is dependent on the ubiquitin proteasome system for efficient protein turnover, which is evident by the identification of fertility defects in animals with mutations in the ubiquitin proteasome system (Kwon et al., 2005, Sun et al., 1999).
Usp14 is a ubiquitin-specific protease that associates with the proteasome and is expressed in all tissue types. Loss of Usp14 expression leads to an early onset neurological disorder that results in death by 8 weeks of age. To identify a role for Usp14 in reproductive function, we utilized axJ mice containing a Thy1-Usp14 transgene that corrects the premature death, muscle wasting, and tremor observed in the axJ mice. While both male and female axJ-Tg mice have normal neuromuscular function and life spans, loss of Usp14 only affected male reproduction. Furthermore, our results indicate that the absolute level of Usp14 expression is not necessarily an indicator of fertility. Male Usp14rrk114 heterozygous gene trap mice exhibit a 50% decrease in testicular Usp14 levels compared to wild type mice and are fertile, without any obvious testes or neurological phenotypes. The axJ and axJ-Tg mice, on the other hand, also show a 50% decrease in the expression of Usp14 in the testes but are infertile.
We hypothesize that the expression pattern of Usp14 from the axJ mutation is responsible for the fertility defects observed in the axJ mice. The axJ mutation results from the insertion of an intra-cisternal A-particle (IAP) into intron 5 of Usp14 and exerts its effect on Usp14 expression by the presence of cryptic splice acceptor and donor sites (Wilson et al., 2002). Splicing of the IAP into the Usp14 mRNA would therefore produce an unstable truncated Usp14 protein. As a result, one possibility for the differential fertility observed between the heterozygous Usp14rrk114 gene trap mice and the axJ mice may be that the splicing of the IAP is not uniform and that the Usp14 IAP is not efficiently removed in a specific population of testicular cells, such as the germ cells, resulting in a differential loss of Usp14 in the axJ testes. In contrast, the heterozygous Usp14rrk114 gene trap mice would exhibit a uniform 50% decrease in Usp14 in all testicular cells in which it is expressed.
Usp14 is expressed in several cell types within the testes. Usp14rrk114 gene trap mice have demonstrated that Usp14 mRNA is expressed in diploid germ cells (spermatogonia and spermatocytes) and in Sertoli cells of the seminiferous tubules. At the protein level, Usp14 was most abundant in the haploid germ cells and round and elongating spermatids, suggesting that some of the mRNA that is produced during the diploid phase of spermatogenesis is stored and translated during the haploid phase. In addition, immunostaining revealed that Usp14 accumulation can be detected in the rNE and cytoplasmic lobe of elongating spermatids, and in the rNE and cytoplasmic droplet of fully differentiated spermatozoa, which are all sites of ubiquitin-dependent proteolysis. The male-specific sterility observed in the axJ mice may therefore be due to altered regeneration of monoubiquitin and/or altered proteasomal deubiquitinating activity.
Although Usp14 is required to maintain ubiquitin levels, our previous studies have shown that there is not a significant reduction in the levels of free ubiquitin in the axJ testes (Anderson et al., 2005). While the total ubiquitin pool may not be altered, our data now suggest that the axJ mutation may have differential effects on Usp14 expression, and therefore ubiquitin levels, in a subset of testicular cells. By decreasing the mono-ubiquitin pool, substrate ubiquitination and ubiquitin-dependent proteolysis would not be able to proceed, resulting in a block to spermatogenesis. Alternatively, loss of Usp14 in the axJ mice may have directly affected the rate of protein degradation by the proteasome. It has previously been shown that inhibition of deubiquitination can stimulate proteasomal proteolysis (Guterman and Glickman, 2004). In agreement with that observation, it has recently been shown that loss of Ubp6, the yeast orthologue of Usp14, increases the rate of proteasomal proteolysis in vitro and in vivo (Hanna et al., 2006). As a result, loss of Usp14 may result in the incorrect degradation of critical factors required for sperm differentiation. Since oocytes do not undergo the extensive cellular remodeling that is seen during spermatogenesis, egg maturation may be less sensitive to changes in ubiquitin levels than spermato/spermiogenesis.
In this study, we have also found that loss of Usp14 results in up-regulation of several other deubiquitinating enzymes thought to be important in ubiquitin stability. Like Usp14, Uch-L5 is a proteasome-associated deubiquitinating enzyme that acts to edit ubiquitin side chains at the proteasome (Lam et al., 1997). The up-regulation in Uch-L5 expression may be a cellular response to deal with decreased ubiquitin recycling at the proteasome due to lose of Usp14, indicating that Uch-L5 may play an important role in ubiquitin stability as well as editing of ubiquitin side chains. In addition, Uch-L1 and Uch-L3 were increased 2-fold in the axJ testes. While Uch-L1 and Uch-L3 have not been shown to associate with the proteasome, Uch-L1 is thought to stabilize ubiquitin in a catalytically-independent manner (Osaka et al., 2003). Due to the high expression levels of these deubiquitinating enzymes, direct binding and sequestration of ubiquitin by Uch-L1 and Uch-L3 may be important aspects of their ubiquitin-stabilizing function. These results offer a possible explanation for our previous findings that demonstrated the absence of ubiquitin depletion in the axJ testis. Ubiquitin recycling at the proteasome could be enhanced by up-regulating expression of Uch-L1, Uch-L3 and Uch-L5, resulting in the stabilization of free ubiquitin pools. Changes in gene expression induced by the loss of Usp14 may also underlie the absence of cellular proliferation and chemical sensitivity in the axJ mouse embryonic fibroblasts. If the induction of other deubiquitinating enzymes facilitates ubiquitin stabilization similar to what is seen in the testes, then it is not surprising that wild type and Usp14-deficient mouse embryonic fibroblasts show similar growth properties and sensitivities to Mitomycin C.
The data presented in this study demonstrate a requirement for Usp14 and the ubiquitin proteasome system in male reproductive function. While other studies have shown that mutations in components of the ubiquitin proteasome system that are necessary for ubiquitin-substrate conjugation can cause sterility, the present data show that loss of a proteasome-associated deubiquitinating activity specifically results in male infertility. Further studies will be required to determine what protein substrates require proteasomal degradation to ensure proper spermiogenesis.
Acknowledgments
We thank Kathy Craighead for clerical assistance and the staff of the histology core of the University of Missouri for tissue processing and sectioning. Funding to the laboratory of P.S. was provided by the Food for the 21st Century Program of the University of Missouri-Columbia, and by the National Research Initiative Competitive Grant No. 2007-01319 from USDA CSREES. This work was supported by NIH Neuroscience Blueprint Core Grant NS57098 to the University of Alabama at Birmingham and NIH/NINDS Grant NS047533 to S.M.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci U S A. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Bonora A, Mares D. A simple colorimetric method for detecting cell viability in cultures of eukaryotic microorganisms. Curr Microbiology. 1982;7:217–221. [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J Neurosci Methods. 1997;71:3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA, Wilson SM. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–11431. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato CJ, Hicks SP. Neuropathologic alterations in the ataxia (paralytic) mouse. Arch Path. 1965;80:604–612. [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH. Deubiquitinating enzymes are IN/(trinsic to proteasome function) Curr Protein Pept Sci. 2004;5:201–211. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;6:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Sands AT, Hancock AR, Vogel H, Donehower LA, Linke SP, Wahl GM, Bradley A. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc Natl Acad Sci U S A. 1996;93:14106–14111. doi: 10.1073/pnas.93.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon Y, Endoh D, Iwanaga T. Expression of protein gene product 9.5, a neuronal ubiquitin C-terminal hydrolase, and its developing change in sertoli cells of mouse testis. Mol Reprod Dev. 1999;54:333–341. doi: 10.1002/(SICI)1098-2795(199912)54:4<333::AID-MRD3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Kwon J, Mochida K, Wang YL, Sekiguchi S, Sankai T, Aoki S, Ogura A, Yoshikawa Y, Wada K. Ubiquitin C-terminal hydrolase L-1 is essential for the early apoptotic wave of germinal cells and for sperm quality control during spermatogenesis. Biol Reprod. 2005;73:29–35. doi: 10.1095/biolreprod.104.037077. [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Lyon MF. A new recessive mutant of the house mouse. J Hered. 1955;46:77–80. [Google Scholar]
- Ng JM, Vrieling H, Sugasawa K, Ooms MP, Grootegoed JA, Vreeburg JT, Visser P, Beems RB, Gorgels TG, Hanaoka F, Hoeijmakers JH, van der Horst GT. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol Cell Biol. 2002;22:1233–12345. doi: 10.1128/MCB.22.4.1233-1245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, Harada T, Hara Y, Kimura I, Chiba S, Namikawa K, Kiyama H, Noda M, Aoki S, Wada K. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- Papa FR, Amerik AY, Hochstrasser M. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connenction. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R, Page DC. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Sutovsky P. Visualization of sperm accessory structures in the mammalian spermatids, spermatozoa, and zygotes by immunofluorescence, confocal, and immunoelectron microscopy. Methods Mol Biol. 2004;253:59–77. doi: 10.1385/1-59259-744-0:059. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Ramalho-Santos J, Moreno RD, Oko R, Hewitson L, Schatten G. On-stage selection of single round spermatids using a vital, mitochondrion-specific fluorescent probe MitoTracker(TM) and high resolution differential interference contrast microscopy. Hum Reprod. 1999;14:2301–2312. doi: 10.1093/humrep/14.9.2301. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–425. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kuang Y, Montes De Oca R, Hays T, Moreau L, Lu N, Seed B, D’Andrea AD. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001;1:3435–3440. doi: 10.1182/blood.v98.12.3435. [DOI] [PubMed] [Google Scholar]
- Yi YJ, Manandhar G, Sutovsky M, Li R, Jonakova V, Oko R, Park CS, Prather RS, Sutovsky P. Ubiquitin C-terminal hydrolase-activity is involved in sperm acrosomal function and anti-polyspermy defense during porcine fertilization. Biol Reprod. 2007;77:780–793. doi: 10.1095/biolreprod.107.061275. [DOI] [PubMed] [Google Scholar]