Abstract
Organic acids are considered as potential substrates for biosynthesis of polyhydroxyalkaonates. The acids may also be the metabolic inhibitors at moderate concentration levels. In this study, Ralstonia eutropha was used to elucidate the protein regulations when the bacterial cells pre-cultivated on glucose were exposed to three representative short chain organic acids, acetic, propionic and levulinic acids. The research compared and examined the proteins that might participate in PHA metabolism, primary metabolism, and cell's defense systems. A number of proteins were found to be induced in R. eutropha by using 1D-PAGE and nano-liquid chromatography tandem MS/MS. With the proteins being up-regulated, a dramatic change occurred in the induction of PHA metabolism, including fatty acid biosynthesis for acetate, β-oxidation for propionate and both for levulinic acid. Acetate kinase was induced in response to the presence of acetate or levulinic acid. The organic acids induced several proteins involved in amino acid biosynthesis, purine and pyrimidine biosynthesis, and cofactor biosynthesis in R. eutropha, but the regulations had a great variation. R. eutropha might employ different regulation mechanisms to maintain cell growth and PHA formation when the cells are exposed to the organic acids as sole source of carbon and energy.
Keywords: Ralstonia eutropha, LC-MS, MS, organic acids, proteomics, biopolymer
1. Introduction
Ralstonia eutropha, an aerobic gram-negative bacterium, can use sugars, organic acids and alcohols to synthesize a family of polyesters, polyhydroxyalkanoates (PHAs) as carbon storage 1. PHAs are biodegradable, eco-friendly thermoplastics and have the similar material properties of petrochemical polymers such as polyethylene and polypropylene 2. Lignocellulosic biomass, after pretreatment, can be used as a renewable feedstock for microbial production of various bioproducts including ethanol and PHA bioplastics 3,4. Mineral acid-catalyzed thermal hydrolysis followed by enzymatic saccharification of the polysaccharides is widely used to convert biomass into fermentable sugars 5,6. Depending on the severity of processing conditions, short chain organic acids, such as formic, acetic, and levulinic acids are formed as the major hydrolytic byproducts 7,8.
Two problems pose the challenges to the microbial biosynthesis of PHAs from the hydrolytic sugars and organic acids. First, it is well known that the organic acids are toxic or inhibitive to microbial cells including R. eutropha, particularly at high concentration levels 9,10. The responses of the cells to the organic acids, such as regulation of proteins, can reveal the metabolic activities and mechanisms in detoxification and utilization of different organic acids by the microbial cells 10. The information is also useful in design and operation of PHA fermentation. Second, it is well known that R. eutropha will form different PHA biopolymers on different organic acids, involving different metabolic pathways and enzymes 1,2. More specifically, it produces a homopolymer, poly(3-hydroxybutyrate)(P3HB) on glucose and/or acetic acid, a copolymer, poly(3-hydroxybutyrateco-3-hydroxyvalerate)(P3HB3HV) on propionic acid or a mixture of acetic and propionic acids, and a terpolymer, poly(3-hydroxybutyrate-co-3-hydroxyvalerate-co-4-hydroxyvalerate)(P3HB3HV4HV) on levulinic acid 11-13. It is an interesting topic in production of bioplastics, because the PHA polymers show different material properties, from a brittle P3HB to a ductile P3HB3HV4HV with special applications 2. The information on proteins involved in different metabolic pathways, particularly the regulation of the enzymes in cells' responses to different organic acids, is invaluable to biosynthesis of PHA biopolymers.
Herein, we grew R. eutropha on glucose in a chemically defined mineral medium and exposed the cells to acetic, propionic and levulinic acids at a moderate concentration level. We examined the up- and down-regulations of the proteins in comparison with the cells grown on glucose. We further checked the possible roles of the proteins in PHA biosynthesis and general cell metabolism on different organic acids.
2. Materials and Methods
2.1 Strain and Cultivation
Ralstonia eutropha (a laboratory isolate) was maintained on nutrient slants containing 5 g/L of yeast extract, 5 g/L of peptone and 2.5 g/L of meat extract. The aerobic bacterium was cultivated in a mineral solution containing (per liter): 2 g NaH2PO4, 3.7 g K2HPO4.3H2O, 0.5 g NaHCO3, 0.5 g MgSO4.7H2O, 1 g NaCl, 0.01 g CaCl2.2H2O, 5 g (NH4)2SO4, and 5 mL of trace solution 10. The flask cultures were shaken at 200 rpm and 30 oC for 48 h. In the first 24 h, the cells were grown on glucose (2 g/L) and the dry cell mass (DCM) concentration reached about 1 g/L. The initial pH was controlled at 6.9 and the pH increased to 7.4 after the cultivation. Solutions (pH 6.5-7) of three organic acids were aseptically added into the cultures to a level of 5g/L. Glucose was also added into one flask for comparison. The medium pH was increased from initial 6.8 to 7.4 and the dry cell mass (DCM) concentrated reached about 1 g/L. The flask cultures were shaken in the same conditions for the second 24 hrs. The cells were harvested with centrifugation at 5,000 g for 20 min, and freeze-dried for later use.
2.2 One-dimensional SDS-polyacrylamide gel electrophoresis
One dimensional (1D) SDS-polyacrylamide gel was performed as described by Laemmli 14, 15. Samples of 20 μg were mixed with SDS-PAGE sample buffer and heated at 100 oC for 5 min. The denatured proteins were separated on 10-20% gradient polyacrylamide SDS gels and then stained by Coomassie dye (G-250). For determination of molecular weight, 10 μL of precision plus protein standards (Bio-Rad, California, USA) were applied on the gels. All protein bands were sliced from the gel, destained with 50% (v/v) acetonitrile in 50 mM NH4HCO3, and completely dried in a speed-vacuum centrifuge. Then 20 μL of sequencing-grade modified porcine trypsin (20 μg/μL in 50 mM NH4HCO3) was added to the dried gel slices that treated with DTT and iodoacetamide prior to addition of trypsin. The unabsorbed solution was removed before 20 μL of NH4HCO3 was added to the rehydrated slices. These samples were incubated at 37 oC overnight. Tryptic digestion was stopped by adding 5 μL of 2% trifluoroacetic acid (TFA). The digested peptides were extracted from each gel slice by sonication of 0.1% TFA and 50% acetonitrile/0.1% TFA for 45 min. Both supernatants were combined for LC-MS/MS analysis.
2.3 Nano-electrospray LC-MS/MS analysis
LC-MS/MS analyses were carried out with UltimateTM system interfaced to a quadrople ion trap mass spectrometer (Bruker Dlatonics, Billerica, MA). The gradient was (A = 0.1% formic acid; B = 0.1% formic acid in acetonitrile) 5% B for 5 min, 60 % B in 88 min, 95% B in 10 min, 5% B in 15 min, 5% B for 20 min. Peptide spectra were recorded over a mass range of m/z 300-2500, MS/MS spectra were recorded in information dependent data acquisition over a mass range of m/z 50-1600. One peptide spectrum was recorded followed by two MS/MS spectra; the accumulation time was 1 sec for peptide spectra and 2 sec for MS/MS spectra. The collision energy was set automatically according to the mass and charge state of the peptides chosen for fragmentation. Doubly or triply charged ions were selected for product ion spectra. MS/MS spectra were interpreted by Mascot (Matrix Science Ltd, London, UK) via Biotools 2.2 software (Bruker Daltonics).
2.4 Analysis of peptide sequences
Peptide mass fingerprint (PMF) searches based on peptide masses measured were performed using the SWISSPROT database or MSDB database with the Mascot program. PMF used the assumption that peptides are monoisotopic, oxidized at methionine residues and carbamidomethylated at cysteine residues. Up to one missed trypsin cleavage was allowed, although most matches did not contain any missed cleavages. Mass tolerance of 1.0 Da was the window of error allowed for matching the peptide mass values. Probability based MOWSE scores were estimated by comparison of search results against estimated random match populations and were reported as: 10 x log10(p), where p is the absolute probability. Scores in Mascot greater than the score at p = 0.05 were considered significant, meaning that for scores higher than the score at p = 0.05 the probability of that match being a random event is lower than 0.05.
The algorithm used for determining the probability of a false-positive match with a given mass spectrum is described elsewhere 16.
3. Results and Discussions
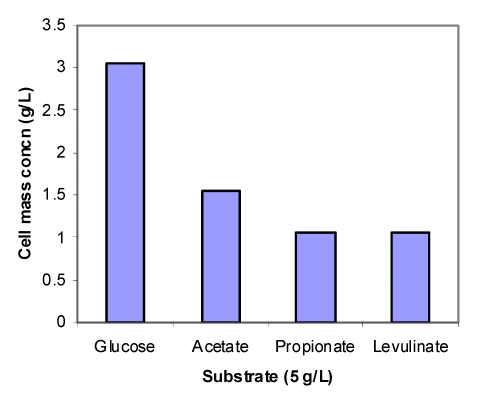
Fig. 1 shows the cell concentrations of R. eutropha grown on glucose and three organic acids in the mineral medium. The low cell mass concentration obtained on propionic and levulinic acids implies the inhibition of the acids on cell growth. For comparison, the cells grew very well on propionic and levulinic acids when extra nutrients in yeast extract and peptone are available (data not shown here).
Figure 1.
Cell mass concentrations of R. eutropha cultivated in a mineral solution (pH 6.8-7.5) at 30 oC. The cells were grew on glucose (2g/L) for 24 hrs and then exposed to glucose, acetate, propionate and levulinate of 5g/L for 24 hrs.
Electrospray tandem LC-MS/MS measurements of the samples prepared from 1D SDS-PAGE showed mass differences between R. eutropha cells fed with organic acids and glucose. Approximately, six hundred proteins from more than one thousand hits were identified by Mascot search and differently expressed proteins were used to elucidate organic acid metabolism and PHA production mechanisms employed in R. eutropha. The cells expressed differently with the diverse of organic acids (Tables 1-3) after they were exposed to organic acids for 24 h.
Table 1.
Up-regulated expression of proteins and their biological functions after acetic acid exposure to Ralstonia eutropha. Results are LC-MS/MS data processed with Mascot search engine and the homology alignments. Uniprot and TIGR classification were used to search cellular roles of identified proteins.
| Protein name | No. of matched peptides | Mascot Score (value P=0.05) | Accession number | Species | Biological function |
|---|---|---|---|---|---|
| Possible proteins involved in PHA production | |||||
| Acetoacetate metabolism regulatory protein | 3 | 32 (27) | Q06065 | Escherichia coli | Transcription |
| Acetoacetyl-CoA reductase | 1 | 78 (33) | RDALAE or P14697 | Ralstonia eutrophus | PHA metabolism |
| Putative acetyl-CoA:acetoacetyl CoA transferase | 3 | 33 (31) | Q8ZPR5_SALTY | Salmonella typhimurium | PHA metabolism |
| Myo-inositol catabolism iolD Probable malonic semialdehyde oxidative decarboxylase | 5 | 35 (21) | P42415 | Bacillus subtilis | Acetyl-CoA biosynthesis (propionyl-CoA metabolism) |
| Methylmalonyl-CoA mutase large subunit | 6 | 42 (25) | P11653 | Propionibacterium freudenreichii shermanii | Propionic acid fermentation |
| Carnitine O-acetyltransferase | 2 | 30 (29) | G90608 | Mycoplasma pulmonis | β-oxidation pathway |
| Probable acyl-CoA dehydrogenase FadE22b | 3 | 43 (26) | Q7TXC4_MYCBO | Mycobacterium bovis | β-oxidation pathway |
| 3-oxoacyl-acyl-carrier protein synthase (FabH) | 2 | 27 (26) | F69842 | Bacillus subtilis | Fatty acid biosynthesis |
| Enoyl-[acyl-carrier-protein] reductase (FabI) | 4 | 27 (23) | P54616 | Bacillus subtilis | Fatty acid biosynthesis |
| Fatty acid/phospholipid synthesis protein | 2 | 32 (23) | Q7NAZ1 | Mycoplasma gallisepticum | Fatty acid biosynthesis |
| Probable fatty acid-CoA ligase FadD30 | 4 | 29 (26) | Q7U226_MYCBO | Mycobacterium bovis | Fatty acid biosynthesis |
| Acetyl/propionyl-CoA carboxylase, beta subunit | 2 | 32 (27) | Q9L077_STRCO | Streptomyces coelicolor | Fatty acid biosynthesis |
| Energy metabolism | |||||
| Acetate kinase | 1 | 22 (22) | Q7NAZ6 | Mycoplasma gallisepticum | Acetyl-CoA biosynthesis |
| Enolase | 5 | 34 (18) | Q8DPS0 | Streptococcus pneumoniae | Energy metabolism |
| Hpr kinase/phosphrylase | 5 | 36 (31) | Q93FD2 | Lactobacillus delbrueckii subsp. bulgaritus | Regulation of carbon metabolism |
| Amino acid biosynthesis | |||||
| Ketol-acid reductoisomerase (Acetohydroxy-acid isomeroreductase) | 5 | 24 (16) | Q8UDV0 | Agrobacterium tumefasciens | Amino acid biosynthesis |
| D-isomer specific 2-hydroxyacid dehydrogenase | 11 | 33 (26) | Q5HW94_CAMJR | Campylobacter jejuni | Amino acid (L-serine) biosyntehsis |
| 5-methyl tetrahydropteroyltriglutamate--homocysteine methyltransferase | 4 | 28 (25) | Q8FQB2 | Streptomyces coelicolor | Methionine biosynthesis |
| Methionine synthase | 6 | 27 (22) | O33259 | Mycobacterium tuberclosis | Methionine biosynthesis |
| Glutamate 5-kinase | 3 | 35 (34) | Q7N7B2 | Photorhabdus luminescens subsp. laumondii | Proline biosynthesis |
| Putative cystathionine gamma-lyase | 2 | 29 (21) | Q59829 | Streptomyces coelicolor | Cysteine biosynthesis |
| Acetylornitine aminotransferase | 2 | 21 (17) | Q8UI71 | Agrobacterium tumefaciens | Arginine biosynthesis |
| Pyrimidine biosynthesis | |||||
| Quinolinate synthetase A | 2 | 24 (17) | Q9F364 | Streptomyces coelicolor | NAD biosynthesis |
| Bifunctional purine biosynthesis protein purH | 7 | 33 (19) | Q9JZM7 | Neisseria meningitidis | Nucleotide biosynthesis |
| Nicotinate phosphoribosyltransferase | 4 | 19 (16) | Q8UIS9 | Agrobacterium tumefaciens | NAD biosynthesis |
| Cofactor biosynthesis | |||||
| CinA-like protein | 3 | 33 (27) | Q67NW5 | Symbiobacterium thermophilum | Biosynthesis of molybdopterin cofactor |
| Coenzyme PQQ synthesis protein E | 3 | 35 (34) | Q01060 | Enterobacter agglomerans | Iron ion binding |
| Lipoyl synthase | 3 | 23 (16) | Q8UFG1 | Agrobacterium tumefaciens | Lipoate biosynthesis |
| Probable phosphoketolase | 4 | 25 (25) | Q5Z066 | Norcadia farcinica | Thiamine biosynthesis |
| Dethiobiotin synthetase | 1 | 19 (17) | Q9FCC1 | Streptomyces coelicolor | Cofactor biosynthesis |
| Stress response proteins | |||||
| Molecular chaperone GroEL | 1 | 71 (44) | Q75T66_BURPI | Burkhoderia picketii | Stabilize or protect disassembled polypeptides |
| Peroxidase/catalase | 3 | 21 (17) | Q9RJH9 | Streptomyces coelicolor | Response to oxidative stress |
| S-adenosylmethionine synthetase | 2 | 31 (25) | Q9X4Q2 | Streptomyces spectabilis | Methyl cycle and polyamine biosynthesis |
| Signal recognition particle protein (fifty-four homolog) | 4 | 25 (22) | P66844 | Mycobacterium tuberclosis | Signal transduction |
| Autoinducer synthesis protein solI | 2 | 35 (34) | P58584 | Ralstonia solanacearum | Signal transduction |
| Thioredoxin reductase | 2 | 22 (22) | P47348 | Mycoplasma gallisepticum | Oxidoreductase |
| Hyaluronate lyase precursor | 4 | 19 (18) | Q54873 | Streptococcus pneumoniae | invasive capacity of the pathogen |
| Toxic anion resistance protein homolog | 3 | 27 (26) | B69757 (MSDB) | Bacillus subtilis | Defense |
| Formate detoxification proteins | |||||
| FdhD protein homolog | 3 | 31 (17) | Q9ZBW0 | Streptomyces coelicolor | Formate dehydrogenase |
| Formate dehydrogenase homolog | 3 | 40 (26) | A27286 | Bacillus subtilis | Formate dehydrogenase |
| Probable Ni/Fe hydrogenase small chain | 4 | 28 (26) | G81284 | Campylobacter jejuni | Electron transport |
| Others | |||||
| Betaine aldehyde dehydrogenase | 5 | 17 (16) | Q8UH56 | Agrobacterium tumefaciens | Bataine biosynthesis Oxidoreductase |
| ComF operon protein | 6 | 29 (23) | P39145 | Bacillus subtilis | ATP binding |
| Phosphate import ATP-binding protein | 4 | 28 (23) | P75186 | Mycoplasma pneumoniae | Transporter |
Table 3.
Up-regulated expression of proteins and their biological functions after levulinic acid exposure to Ralstonia eutropha. Results are LC-MS/MS data processed with Mascot search engine and the homology alignments. Uniprot and TIGR classification were used to search cellular roles of identified proteins.
| Protein name | No. of matched peptides | Mascot Score (value P=0.05) | Accession number | Species | Biological function |
|---|---|---|---|---|---|
| Possible proteins involved in PHA production | |||||
| Acetoacetyl-CoA reductase | 1 | 88 (33) | RDALAE or P14697 | Ralstonia eutrophus | PHA metabolism |
| Probable trans-2-enoyl-CoA reductase | 3 | 28 (25) | Q6CBE4 | Yarrowia lipolytica | β-oxidation |
| 3-oxoacyl-(Acyl-carrier-protein) reductase | 2 | 53 (43) | Q8EDH3_SHEON | Shewanella oneidensis | β-oxidation |
| Putative fatty-acid-CoA ligase FadD11 | 5 | 23 (23) | Q10776 | Mycobacterium tuberclosis | Fatty acid biosynthesis |
| Energy metabolism | |||||
| Acetate kinase | 3 | 24 (23) | P37877 | Bacillus subtilis | Acetyl-CoA biosynthesis |
| Amino acid biosynthesis | |||||
| D-isomer specific 2-hydroxyacid dehydrogenase | 9 | 27 (26) | Q5HW94_CAMJR | Campylobacter jejuni | Amino acid (L-serine) biosyntehsis |
| Glutamate synthase | 3 | 30 (21) | P39812 | Bacillus subtilis | Amino acid biosynthesis |
| 5-methyl tetrahydropteroyltriglutamate--homocysteine methyltransferase | 4 | 28 (17) | Q93J59 | Streptomyces coelicolor | Methionine biosynthesis |
| Putative cystathionine gamma-lyase | 2 | 29 (21) | Q59829 | Streptomyces coelicolor | Cysteine biosynthesis |
| Chorismate synthase | 1 | 17 (17) | Q5HSF9 | Campylobacter jejuni | Aromatic amino acid biosynthesis |
| Ketol-acid reductoisomerase (Acetohydroxy-acid isomeroreductase) | 4 | 22 (17) | Q9PHN5 | Campylobacter jejuni | Amino acid biosynthesis |
| Histidinol-phosphate aminotransferase | 2 | 20 (17) | P16246 | Streptomyces coelicolor | |
| Purine and pyrimidine biosynthesis | |||||
| Dihydroorotate dehydrogease | 2 | 27 (25) | Q8NQC0 | Corynebacterium glutamicum | Nucleotide biosynthesis |
| Quinolinate synthetase A | 5 | 22 (17) | Q9F364 | Streptomyces coelicolor | NAD biosynthesis |
| Bifunctional purine biosynthesis protein purH | 4 | 30 (27) | Q8FB68 | Escherichia coli | Nucleotide biosynthesis |
| Nicotinate phosphoribosyltransferase | 2 | 36 (23) | Q5HWN2_CAMJR | Campylobacter jejuni | NAD biosynthesis |
| Cofactor biosynthesis | |||||
| Lipoyl synthase | 4 | 35 (30) | Q8ERL8 | Oceanobacillus iheyensis | Lipoate biosynthesis |
| Lipoyltransferase | 3 | 20 (17) | Q8CK04 | Streptomyces coelicolor | Lipoate metabolism |
| Biotin synthase | 10 | 50 (23) | A81117 | Neisseria meningitidis | Biotin biosynthesis |
| 3-octaprenyl-4-hydroxybenzoate carboxy-lyase | 3 | 20 (19) | Q9JT68 | Neisseria meningitidis | Ubiquinone biosynthesis |
| Stress response proteins | |||||
| 60 kDa chaperonin | 10 | 355 (34) | Q8Y1P8 | Ralstonia solanacearum | Stress |
| Transcription activator of acetoin dehydrogenase operon | 3 | 30 (25) | H69581 | Bacillus subtilis | Transcription |
| Signal recognition particle protein | 6 | 27 (26) | Q5HV72_CAMJR | Campylobacter jejuni | Signal transduction |
| Carboxylate-amine ligase | 2 | 16 (16) | Q9KY07 | Streptomyces coelicolor | Glutathione biosynthesis |
| Superoxide dismutase | 3 | 60 (43) | SODF_PSEPU | Pseudomonas putida | Defense |
| Peroxidase/catalase | 3 | 17 (17) | Q9RJH9 | Streptomyces coelicolor | Response to oxidative stress |
| Thioredoxin reductase | 3 | 31 (29) | O66790 | Aquifex aeolicus | Oxidoreductase |
| Thiol:disulfite interchange protein | 3 | 64 (43) | Q62MY5_BURMA | Burkholderia mallei | Oxidoreductase |
| Formate detoxification proteins | |||||
| Probable formate-tetrahydrofolate ligase | 7 | 29 (27) | Q9JVY8 | Neisseria meningitidis | Methyl cycle |
| Formate dehydrogenase, nitrate inducible | 3 | 29 (27) | P24183 | Escherichia coli | Formate dehydrogenase |
| Others | |||||
| Cation-transporting P-type ATPase B | 3 | 32 (22) | Q10877 | Mycobacterium tuberclosis | Transporter |
| Phosphate transport system protein | 2 | 23 (18) | P0A3Y7 | Streptococcus pneumoniae | Transporter |
| H+/K+-exchanging ATPase | 2 | 37 (25) | A81338 | Campylobacter jejuni | Transporter |
| Hyaluronate lyase precursor | 7 | 25 (19) | Q54873 | Streptococcus pneumoniae | invasive capacity of the pathogen |
| Formamidopyrimidine-DNA glycosylase | 1 | 36 (30) | P42371 | Lactococcus lactis subsp. cremoris | DNA repair |
| BSUB0010 | 12 | 77 (26) | CAD13602 | Bacillus subtilis | |
| SsrA-binding protein | 4 | 41 (25) | Q83N13 | Tropheryma whipplei | Protein biosynthesis |
| Foldase protein | 2 | 24 (21) | P24327 | Bacillus subtilis | Isomerase |
| Carnitine operon protein caiE | 3 | 32 (27) | Q8XA36 | Escherichia coli | Carnitine metabolism |
| Ornitine cabamoyltransferase | 2 | 24 (19) | Q9JTI4 | Neisseria meningitidis | |
| Radical SAM domain protein | 3 | 28 (26) | Q5HTL8_CAMJR | Campylobacter jejuni | |
| Hydrolase, alpha/beta hydrolase fold family | 4 | 26 (26) | Q7D8N4_MYCTU | Mycobacterium tuberclosis | |
3.1 Proteins induced by individual organic acids
Acetic acid causes the up-regulation acetoacetate metabolism regulatory protein, acetoacetyl-CoA reductase, putative acetyl-CoA:acetoacetyl CoA transferase, myo-inositol catabolism protein, methyl-malonyl-CoA mutase, carnitine o-acetyltransferase, probable acyl-CoA dehydrogenase, 3-oxo-acyl-carrier protein synthase, enoyl-[acyl-carrier-protein]reductase, fatty acid/phosphlipid synthesis protein, fatty acid-CoA ligase, and acetyl/propionyl-CoA carboxylase involved in PHA metabolism, β-oxidation, and fatty acid biosynthesis (Table 1). Propionic acid up-regulates acetoacetyl-CoA reductase, probable enoyl-CoA hydratase, probable acyl-CoA dehydrogenase, probable multi-domain beta keto-acyl synthase, and acetyl/propionyl-CoA carboxylase involved in PHA metabolism, β-oxidation, and fatty acid biosynthesis (Table 2). Levulinic acid results in the up-regulation acetoacetyl-CoA reductase, probable enoyl-CoA reductase, 3-oxo-acyl-carrier protein reductase, and fatty acid-CoA ligase in PHA metabolism, β-oxidation, and fatty acid biosynthesis (Table 3).
Table 2.
Up-regulated expression of proteins and their biological functions after propionic acid exposure to Ralstonia eutropha. Results are LC-MS/MS data processed with Mascot search engine and the homology alignments. Uniprot and TIGR classification were used to search cellular roles of identified proteins.
| Protein name | No. of matched peptides | Mascot Score (value P=0.05) | Accession number | Species | Biological function |
|---|---|---|---|---|---|
| Possible proteins involved in PHA production | |||||
| Acetyl-CoA reductase | 1 | 120 (43) | RDALAE | Ralstonia eutropus | PHA metabolism |
| Probable enoyl-CoA hydratase | 4 | 27 (26) | B70695 | Mycobacterium tuberclosis | PHA metabolism |
| Probable acyl-CoA dehydrogenase FadE25 | 3 | 31 (23) | P63427 | Mycobacterium tuberclosis | β-oxidation |
| Probable multi-domain beta keto-acyl synthase | 4 | 37 (27) | T37056 | Streptomyces coelicolor | Fatty acid biosynthesis |
| Acetyl/propionyl-CoA carboxylase | 3 | 30 (26) | P46392 | Mycobacterium | Fatty acid biosynthesis |
| Energy metabolism | |||||
| Enolase 2 | 1 | 19 (17) | Q9F3P9 | Streptococcus coelicolor | Energy metabolism |
| Transaldolase B | 4 | 41 (23) | P66955 | Salmonella typhimurium | Pentose pathway |
| Amino acid biosynthesis | |||||
| Glutamine synthetase | 7 | 26 (23) | P0A590 | Mycobacterium tuberclosis | Amino acid biosynthesis |
| Putative cystathionine gamma-lyase | 1 | 19 (17) | Q59829 | Streptococcus coelicolor | Cysteine biosynthesis |
| Ketol-acid reductoisomerase 2 (Acetohydroxy-acid isomeroreductase) | 4 | 19 (17) | Q9FBR8 | Streptomyces coelicolor | Amino acid biosynthesis |
| 5-methyl tetrahydropteroyltriglutamate--homocysteine methyltransferase | 2 | 34 (29) | O67606 | Aquifex aeolicus | Methionine biosynthesis |
| Histidinol dehydrogenase | 4 | 17 (16) | Q9PM77 | Campylobacter jejuni | Amino acid biosynthesis |
| Carbamate kinase-like protein | 4 | 34 (26) | P77624 | Escherichia coli | Amino acid biosynthesis |
| Histidinol-phosphate aminotransferase | 3 | 29 (17) | Q8U9W3 | Agrobacterium tumefaciens | |
| Purine and pyrimidine biosynthesis | |||||
| Probable xanthine dehydrogenase | 4 | 25 (24) | O32144 | Bacillus subtilis | Purine catabolism |
| Probable inositol monophosphatase | 5 | 29 (27) | T35932 | Streptomyces coelicolor | Aromatic acid biosynthesis (chorismate biosynthesis) |
| Quinolinate synthetase A | 2 | 20 (16) | Q9F364 | Streptomyces coelicolor | NAD biosynthesis |
| NAD-dependent deacetylase (Regulatory protein Sir2) | 3 | 26 (17) | Q9JN05 | Campylobacter jejuni | Transcription |
| Cofactor biosynthesis | |||||
| Biotin synthase | 2 | 24 (23) | P12678 | Salmonella typhimurium | Biotin biosynthesis |
| Stress response proteins | |||||
| Glutathione biosynthesis bifunctional protein | 3 | 41 (31) | Q8DW15 | Streptococcus mutans | Glutathione biosynthesis |
| Sigma-70 | 3 | 32 (25) | F81375 | Campylobacter jejuni | Transcription |
| Signal recognition particle protein (sigma-54) | 2 | 22 (22) | Q01442 | Mycoplasma mycoides | Transcription |
| Alkyl hydroperoxide reductase | 2 | 72 (43) | Q7VTI5_BORPE | Bordetella pertussis | oxidoreductase |
| Gluconate operon transcription repressor | 2 | 24 (23) | P10585 | Bacillus subtilis | Transcription |
| Catalase | 4 | 28 (27) | Q50474 | Mycobacterium tuberclosis | Defense |
| Catalase 2 | 2 | 24 (23) | P42234 | Bacillus subtilis | Defense |
| Formate detoxification proteins | |||||
| Hydrogenase expression/formation protein | 3 | 33 (25) | Q5HVE5_CAMJR | Campylobacter jejuni | Transcription |
| FdhD protein | 3 | 24 (23) | P64118 | Mycobacterium coelicolor | Formate dehydrogenase |
| Transpoters | |||||
| H+-transporting two-sector ATPase | 2 | 29 (27) | Q97PT4_STRPN | Streptococcus pneumoniae | Transporter |
| Potassium-transporting ATPase B chain | 4 | 50 (29) | Q9R6X1 | Anabaena sp. (strain L31) | Transporter |
| H+/K+-exchanging ATPase | 3 | 30 (27) | T36652 | Streptomyces coelicolor | Transporter |
| p-hydroxybenzoic acid-efflux pump subunit | 3 | 26 (26) | Q8FD51 | Escherichia coli | Transporter |
| Others | |||||
| 2,3-dihydroxyphenylpropionate 1,2-dioxygenase | 2 | 30 (26) | P54711 | Escherichia coli | 3-hydroxyphenyl propionate metabolism |
| L-2,4-diaminobutyric acid acetyltransferase | 2 | 19 (17) | Q93RW2 | Streptococcus coelicolor | Polyamine biosynthesis |
| Ethanolamine ammonia-lyase | 1 | 24 (23) | Q8Z4U3 | Salmonella typhimurium | Ethnolamine utilization |
| Cyanate hydratase | 2 | 35 (30) | Q59948 | Synechococcus sp. | Cyanate metabolism |
| Hyaluronate lyase precursor | 5 | 20 (19) | Q54873 | Streptococcus pneumoniae | invasive capacity of the pathogen |
| Desaturase-related protein | 3 | 28 (27) | Q8VK28_MYCTU | Mycobacterium tuberclosis | Not known |
In the utilization of organic acids as energy sources, R. eutropha induces acetate kinase in acetate and levulinic acid-treated growth medium, whereas the bacteria do not change the protein in response to propionic utilization (Tables 1-3). Three organic acids refer to induce 5-methyl tetrahydropteroyltriglutamate-homocysteine methyltransferase simultaneously and presumably to synthesis methionine. In addition to these findings, R. eutropha induces several proteins that participated in amino acid biosynthesis, purine and pyrimidine biosynthesis and coenzyme and cofactor biosynthesis during the organic acid metabolism. Bifunctional purine biosynthesis protein is up-regulated within the medium containing acetate or levulinic acid (Table 1 and 3). Ketol-acid reductoisomerase or acetohydroxy-acid isomeroreductase is up-regulated in relation to the three tested organic acids (Table 1-3). Besides, stress responsible proteins such as catalase, peroxidase, superoxide dismutase and proteins involved in the glutathione biosynthesis are over-expressed to detoxify oxidative anion or hydrogen peroxide presumably produced in β-oxidation or other biochemical reactions (Table 1-3).
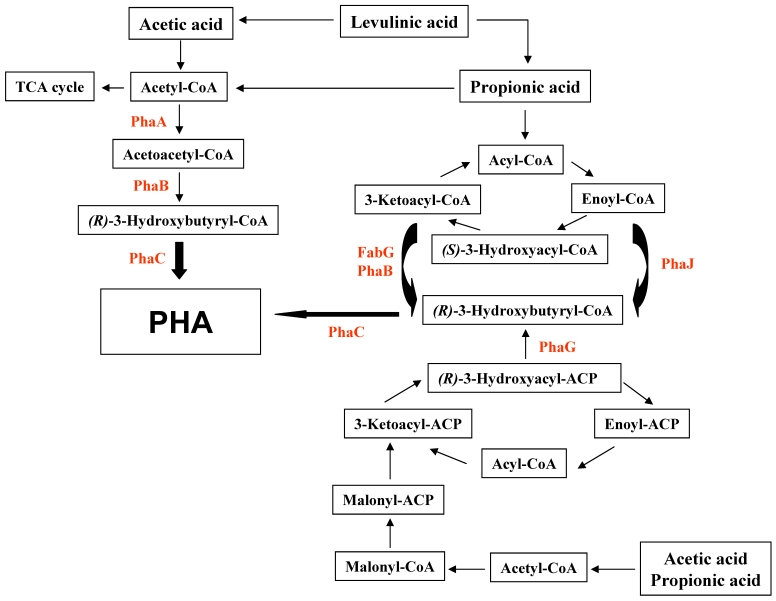
There are at least three different mechanisms to synthesize PHAs from organic acids including acetate, propionic acid, and levulinic acid (Fig. 1). The first pathway is the conversion of substrates to acetyl-CoA, leading to form PHAs via acetoacetyl-CoA and 3-hydroxybutyryl-CoA. This pathway is compact and has been known to be employed in R. eutropha if glucose and other sugars are used as substrates. The second pathway is the β-oxidation of fatty acids. Finally, fatty acid biosynthesis is the avenue of PHA biosynthesis when the acetyl-CoA carboxylase catalyzes the conversion of acetyl-CoA to malonyl-CoA.
3.2 Induction of acetoacetyl-CoA reductase in R. eutropha
By R. eutropha, two acetyl-CoAs are left to be condensed to form acetoacetyl-CoA with the activity of β-ketothiolase and the resultant acetoacetyl-CoA turns to be R-β-hydroxybutyryl-CoA by an NADPH-dependent acetoacetyl-CoA reductase. The final step of the PHA biosynthesis is the reaction of the PHB synthase that convert the moiety of R-β-hydroxybutyryl-CoA to the hydroxyl groups of carboxyl end of a pre-existing PHB molecule. With all of the three substrates such as acetate, propionate, and levulinic acid, R. eutropha up-regulated acetoacetyl-CoA reductase when compared to the control (Table 1-3). With these findings, a putative acetyl-CoA:acetoacetyl CoA transferase was induced in the growth medium containing acetate for R. eutropha. However, this type of protein was not found in the medium containing the other two substrates. The protein has been assumed to act as acetoacetyl-CoA thiolase during the PHA formation. Therefore, the addition of three different substrates into the growth medium of R. eutropha may induce similar routes of the production of PHAs, except for the induction of a putative acetyl-CoA:acetoacetyl CoA transferase with acetate addition.
3.3 Induction of β-oxidation in R. eutropha
Most naturally occurring fatty acids have an even number of carbon atoms. The pathway for catabolism of fatty acids is referred to as the β-oxidation pathway, because oxidation occurs at the β-carbon (C3).
Carnitine o-acetyl transferases catalyze transfer of a fatty acid between the thiol of CoA and the hydroxyl on carnitine. R. eutropha grown in acetate in the growth medium increased an expression of a carnitine o-acetyl transferase, whereas it did not change the protein after propionate and levulinic acid treatments (Table 1). A probable acyl-CoA dehydrogenase was also induced after the treatments of acetate and propionate into the growth medium, whereas the addition of levulinic acid did not induce it.
Feeding propionate only increased the level of enoyl-CoA hydratase, whereas treatments with acetate and levulinic acid did not over-express the protein in R. eutropha. The enzyme catalyzes enoyl-CoA into hydroxyacyl-CoA during β-oxidation of fatty acids. The reaction catalyzed by enoyl-CoA hydratase may be a key process for the formation of PHAs via β-oxidation. Therefore, R. eutropha may prefer propionate as a substrate for generating PHAs via β-oxidation rather than acetate and levulinic acid.
3.4 Induction of fatty acid biosynthesis in R. eutropha
The fatty acid-CoA synthetases ligate CoA to a free fatty acid. This step requires ATP and magnesium, as well as the CoASH. It is interesting that R. eutropha grown in the addition of acetate and levulinic acid in the growth medium up-regulated a probable fatty acid-CoA synthetase (or probable fatty acid-CoA ligase), whereas the bacteria did not change the protein after propionate treatment (Table 1). However, we found the induction of a probable multi-domain beta keto-acyl synthase presumed as a fatty acid-CoA synthetase in the propionate treatment of the growth medium. Therefore, R. eutrohpa may induce fatty acid biosynthesis when the three tested substrates introduced into the growth medium.
Malonyl-CoA, which is a precursor of fatty acid biosynthesis, is produced from acetyl-CoA by the enzyme acetyl-CoA carboxylase. In R. eutropha, acetyl/propionyl-CoA carboxylase was over-expressed after the bacteria fed with the acetate and propionate. Thus, the two substrates might be the inducers of acetyl-CoA carboxylase during fatty acid biosynthesis. However, we did not find the induction of the protein with the levulinic acid treatment. In fatty acid biosynthesis, 3-oxo-acyl-carrier protein synthase (FabH) and enoyl-[acyl-carrier-protein] reductase (FabI) were over-expressed after acetate was treated, whereas the enzyme was not changed after propionate and levulinic acid were treated into the growth medium. In the case of 3-oxo-acyl-carrier protein reductase, R. eutropha increased it when levulinic acid was added into the growth medium. Therefore, R. eutropha may prefer acetate as a substrate via fatty acid biosynthesis as one of PHA-generating routes, even though the bacteria also induced the fatty acid biosynthesis after the treatments of propionate and levulinic acid.
Interestingly, two proteins involved in the propionyl-CoA metabolism such as a methylmalonyl-CoA mutase large subunit and a probable malonic semialdehyde oxidative decarboxylase were over-expressed in the growth medium including acetate only. They were not found in the growth medium containing propionate and levulinic acid. The methylmalonyl-CoA mutase catalyzes the conversion of methylmalonyl-CoA into succinyl-CoA during synthesis of propionate from TCA cycle. The probable malonic semialdehyde oxidative decarboxylase may convert malonic semialdehyde with Coenzyme A to acetyl-CoA. Those two proteins are the key enzymes to produce succinyl-CoA and acetyl-CoA from propionyl-CoA.
3.5 Induction of energy metabolism in R. eutropha
Acetate and propionate has been known to be used as carbon and energy sources for procaryotes, where acetate and propionate are the most abundant organic acids 17. By utilizing acetate and propionte, they have to be catabolized or activated into their corresponding acyl-CoA forms (Fig. 2). Acetyl-CoA enters directly into the TCA cycle, whereas propionyl-CoA can be catabolized via a number of different pathways that convert it into pyruvate, acetate and succinyl-CoA which they can enter the TCA cycle 18. Acetate is activated into acetyl-CoA via either one of two pathways. The first pathway requires the involvement of the acetate kinase (AckA) and phosphotranacetylase (Pta) enzymes. In the enteric bacteria, AckA and Pta are responsible for the synthesis of acetyl-CoA when acetate is present in high concentrations in the environment (> 30 mM acetate). The second pathway for the activation of acetate requires the activity of the ATP-dependent acete:CoA ligase (or acetyl-CoA synthetase). Acetyl-CoA synthetase (Acs) is required when the concentration of acetate in the environment is low (<10mM acetate) 19,20. Therefore, the induction of AckA is related to the concentration of acetate in the growth medium and the protein may be involved in the conversion of acetate into the TCA cycle in R. eutropha. Interestingly, AckA was over-expressed in the growth medium containing levulinic acid and it demonstrated that leuvulinic acid could be metabolized to acetate in the cells by β-oxidation. Thus, levulinic acid may be similar to acetate as substrate for R. eutropha.
Figure 2.
Proposed schematic representation of the polyalkanoate (PHA) production of R. eutropha exposed to organic aicds. PhaA, β-ketothiolase; PhaB, NADPH-dependent acetoacetyl-CoA reductase; PhaC, polyalkanoate synthase; PhaG, 3-hydroxyacyl-ACP-CoA transferase; PhaJ, (R)-specific enoyl-CoA hydratase; FabG, 3-ketoacyl-CoA reductase.
The two substrates, acetate and levulinic acid, also enhanced acetyl-CoA synthetase protein and it might be participated in the activation of acetate into the TCA cycle at low concenration of acetate. By the addition of acetate and levulinic acid, R. eutropha induced those two pathways to use acetate at low or high concentrations in the growth medium. By the addition of propionate, R. eutropha induced propionyl-CoA synthetase to form propionyl-CoA from propionate with a high-affinity 18.
Recently, acetate and propionate activation by acyl-CoA synthetase are related to Sir2 protein which has a NAD+-dependent histone deacetylase activity 21. In our findings, Sir2 protein was induced in R. eutropha fed with the addition of propionate. Therefore, propionic acid may induce the Sir2 protein for the activation of acyl-CoA synthetase in R. eutropha.
3.6 Induction of proteins in primary metabolism
With the tested organic acids as carbon and energy sources, R. eutropha enhanced a variety of primary metabolism with the induction of several key proteins. By addition of acetate, R. eutropha cells up-regulated several enzymes involved in the production of amino acids such as arginine, proline, serine, methionine and cysteine. However, R. eutropha cells grown in propionate and levulinic acid showed the up-regulation of histidinol-phosphate aminotransferase which converts phenylpyruvate to phenylalanine. In the R. eutropha cells grown in the propionate treated medium, histidinol dehydrogenase was up-regulated and it might play an important role in histidine. Biosynthesis of three aromatic amino acids, phenylalanine, tyrosine, and tryptophan uses a shared starting compound as phosphoenolpyruvate (PEP). The condensation of PEP with D-erythrose-4-phosphate forms 7-phosphate-2-dehydro-3-deoxy-D-arabino-hepnoate and the further reactions produce an important intermediate, chorismate. Chorismate can be further metabolized in two divergent paths; one leading to tryptophan and the other to phenylalanine and tyrosine. In our study, chorismate synthase was up-regulated by levulinic acid treatment, leading to induction of the biosynthesis of the three amino acids (Table 1).
By utilizing three organic substrates, R. eutropha enhanced several enzymes participated in pyrimidine and purine biosynthesis and cofactor biosynthesis. The induction pattern was similar between the three organic acid treatments.
3.7 Induction of defense systems in R. eutropha
During β-oxidation, acyl-CoA oxidase introduces a double bond between the α and β-carbons of the acyl-CoA and passes the electrons to oxygen molecule, leading to generate hydrogen peroxide. Catalase converts the potentially toxic hydrogen peroxide produced by acyl-CoA oxidase to water and oxygen molecule. By the addition of propionate, R. eutropha induced catalase 1 and 2 forms, whereas R. eutropha increased the level of thioredoxin, peroxide/catalase, and toxic anion resistance protein with the treatment of acetate, and of superoxide dismutase, peroxide/catalase, and thioredoxin reductase with the treatment of levulinic acide (Tables 1-3). Therefore, we may assume that the oxidative stress occurs during PHA formation or acquisition of organic acid substrates for carbon and energy sources in R. eutropha via β-oxidation at least.
On the other hand, R. eutropha cells grown in the acetate-treated medium enhanced glutathione biosynthesis by up-regulation of S-adenosylmethionine (SAM) synthetase catalyzing SAM formation from methionine and ATP (Table 1). SAM is an important methyl donor for transmethylation and polyamine biosynthesis. SAM is also a key substrate of certain methylases for the regeneration of glutathione. Thus, the acetate-treated R. eutropha cells may allow SAM as an important substrate for glutathione production in response to acetic acid exposure. Cytstathione β-lyase, a key enzyme catalyzing cystathione to cysteine for glutathione biosynthesis was also induced in the acetate treatment. However, R. eutropha grown in the propionate-treated medium showed a simple employment of glutathione biosytnehsis bifuntional protein for glutathione biosynthesis. Finally, R. eutropha cells up-regulated a carboxylate-amine ligase in the levulinic acid treatment. The enzyme is considered as γ-glutamylcysteine ligase to produce the final product, glutathione 22. Therefore, R. eutropha may vary the induction pathway of glutathione biosynthesis via a different kind of routes.
4. Concluding remark
The proteomic examination reveals that R. eutropha up-regulated expression of proteins when the bacterium utilized acetate, propionate and levulinic acid as carbon and energy sources. According to the TIGR protein classification, most of the over-expressed proteins in relation to PHA formation were involved in fatty acid biosynthesis for acetate, β-oxidation for propionate, and both for levulinic acid. R. eutropha also enhanced detoxifying proteins to suppress oxidative stress caused by β-oxidation. Glutathione biosynthesis mechanism was also differently up-regulated via different proteins in R. eutropha. Biosynthesis for pyridines and pyrimidines, amino acids, cofactors was up-regulated in the cells grown on three organic acids. Therefore, R. eutropha may utilize acetic acid, propionic acid and levulinic acid in different metabolisms to produce PHAs, amino acids, purine and pyrimidine, and other primary intermediates. These findings are similar to the previous report which R. eutropha up-regulated PHA forming-enzyme systems with glutathione production as a defense system to formic acid toxicity 23. Further studies on changes in gene levels in R. eutropha will be necessary to validate roles of differently expressed proteins in response to the three tested organic acid metabolism and production PHAs.
Acknowledgments
This work was supplied in part with grants (QXL) from Hawaii State Civil Defense and Hawaii Department of Agriculture Pesticides Branch; grants (JY) from the Consortium of Plant Biotechnology Research Inc. and US Department of Energy.
References
- 1.Reinecke F, Steinbuchel A. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J Mol Microbiol Biotechnol. 2009;16:91–108. doi: 10.1159/000142897. [DOI] [PubMed] [Google Scholar]
- 2.Sudesh K, Abe H, Doi Y. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog Polym Sci. 2000;25:1503–1555. [Google Scholar]
- 3.Rubin E.M. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 4.Somleva M.N, Snell K.D, Beaulieu J.J, Peoples O.P, Garrison B.R, Patterson N.A. Production of polyhydroxybutyrate in switchgrass, a value-added co-product in an important lignocellulosic biomass crop. Plant Biotechnol J. 2008;6:663–678. doi: 10.1111/j.1467-7652.2008.00350.x. [DOI] [PubMed] [Google Scholar]
- 5.Grohmann K, Bothast R.J. Saccharification of corn fiber by combined treatment with dilute sulphuric acid and enzymes. Proc Biochem. 1997;32:405–415. [Google Scholar]
- 6.Yu J, Zhang J, He J, Liu Z, Yu Z. Combinations of mild physical or chemical pretreatment with biological pretreatment for enzymatic hydrolysis of rice hull. Bioresour Technol. 2009;100:903–908. doi: 10.1016/j.biortech.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 7.Torget R, Walter P.J, Himmel M.E, Grohmann K. Dilute-acid pretreatment of. corn residues and short-rotation woody crops. Appl Biochem Biotechnol. 1991;28:75–86. [Google Scholar]
- 8.Larsson S, Palmqvist E, Hahn-Hagerdal B et al. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enz Microb Technol. 1999;24:151–159. [Google Scholar]
- 9.Yu J, Wang J. Metabolic flux modeling of detoxification of acetic acid by Ralstonia eutropha at slightly alkaline pH levels. Biotech Bioeng. 2001;73:458–464. doi: 10.1002/bit.1080. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Stahl H. Microbial utilization and biopolyester synthesis of bagasse hydrolysates. Bioresour Technol. 2008;99:8042–8048. doi: 10.1016/j.biortech.2008.03.071. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Si Y. Metabolic carbon fluxes and biosynthesis of polyhydroxyalkanoates by Ralstonia eutropha on short chain fatty acids. Biotechnol Prog. 2004;20:1015–1024. doi: 10.1021/bp034380e. [DOI] [PubMed] [Google Scholar]
- 12.Du G, Si Y, Yu J. Inhibition by medium-chain-length fatty acids of formation of polyhydroxyalkanoates from volatile fatty acids by Ralstonia eutropha. Biotechnol Lett. 2001;23:1613–1617. [Google Scholar]
- 13.Gorenflo V, Schmack G, Vogel R. Steinbuchel, A, Development of a process for the biotechnological large-scale production of 4-hydroxyvalerate-containing polyesters and characterization of their physical and mechanical properties. Biomacromolecules. 2001;2:45–57. doi: 10.1021/bm0000992. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.E, Seo J.S, Keum Y.S, Lee K.J, Li Q.X. Fluoranthene metabolism and associated proteins in Mycobacterium sp. JS14. Proteomics. 2007;7:2059–2069. doi: 10.1002/pmic.200600489. [DOI] [PubMed] [Google Scholar]
- 16.Berndt P, Hobohm U, Largen H. Reliable automatic protein identification from matrix-assisted laser desorption/ionization mass spectrometric peptide fingerprints. Eletrophoresis. 1999;20:3521–3526. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3521::AID-ELPS3521>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Buckel W. Anaerobic energy metabolism. In: Chlegel HG, editor. Biology of the Procaryotes. Germany: Thieme, Stuttgart; 1999. pp. 278–326. [Google Scholar]
- 18.Horswill A.R, Escalante-Semerena J.C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horswill A.R, Escalante-Semerena J.C. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology. 1999;145:1381–1388. doi: 10.1099/13500872-145-6-1381. [DOI] [PubMed] [Google Scholar]
- 20.Horswill A.R, Escalante-Semerena J.C. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J Bacteriol. 1999;181:5615–5623. doi: 10.1128/jb.181.18.5615-5623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starai V.J, Takahashi H, Boeke J.D, Escalante-Semerena J.C. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics. 2003;163:545–555. doi: 10.1093/genetics/163.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann C, Doseeva V, Pullalarevu S et al. YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays. Proteins. 2004;56:376–383. doi: 10.1002/prot.20103. [DOI] [PubMed] [Google Scholar]
- 23.Lee S.E, Li Q.X, Yu J. Proteomic responses to formic acid on Ralstonia eutropha. Proteomics. 2006;6:4259–4268. doi: 10.1002/pmic.200500824. [DOI] [PubMed] [Google Scholar]