Abstract
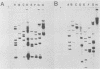
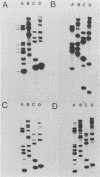
The Escherichia coli rRNA operon rrnB was used as a 32P-labeled hybridization probe in Southern blots of genomic DNAs from representative strains of the saccharolytic, gram-negative, obligate anaerobes of the genus Bacteroides. Control experiments with the B. fragilis type strain ATCC 25285 established that nearly identical rRNA fragment patterns were produced when either the E. coli rrnB gene probe or homologous rRNA isolated from B. fragilis was used as the probe. In addition, it was shown that a specific 16S or 23S rrnB gene probe also could be used to produce fragment patterns suitable for analysis. Thirty-one strains from 8 of the 10 recognized Bacteroides species were then examined. The resulting autoradiographs revealed specific fragment patterns for all but one (B. ovatus) of the species tested. Restriction fragment length polymorphisms were observed for many of the strains tested, but these differences did not hinder species classification. The five B. ovatus strains examined did not form a distinct group, and their rRNA fragment patterns displayed a marked heterogeneity. The same approach was applied to a unique set of enterotoxin-producing B. fragilis strains isolated from animals and humans with diarrhea. The results demonstrated that these strains were in fact B. fragilis and that they produce rRNA fragment patterns closely related to those of the type strain ATCC 25285. This set of strains did not appear to form a separate subgroup or genotype within the B. fragilis species, and there were no distinguishable restriction fragment length polymorphisms that could be used to specifically separate enterotoxin-producing strains from nonenterotoxigenic strains.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur M., Arbeit R. D., Kim C., Beltran P., Crowe H., Steinbach S., Campanelli C., Wilson R. A., Selander R. K., Goldstein R. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: pap-related sequences compared with rrn operons. Infect Immun. 1990 Feb;58(2):471–479. doi: 10.1128/iai.58.2.471-479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- De Buyser M. L., Morvan A., Grimont F., el Solh N. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1989 Apr;135(4):989–999. doi: 10.1099/00221287-135-4-989. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- Jacobs M. R., Spangler S. K., Appelbaum P. C. Beta-lactamase production, beta-lactam sensitivity and resistance to synergy with clavulanate of 737 Bacteroides fragilis group organisms from thirty-three US centres. J Antimicrob Chemother. 1990 Sep;26(3):361–370. doi: 10.1093/jac/26.3.361. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. E., Holdeman L. V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974 May;27(5):961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moureau P., Derclaye I., Gregoire D., Janssen M., Cornelis G. R. Campylobacter species identification based on polymorphism of DNA encoding rRNA. J Clin Microbiol. 1989 Jul;27(7):1514–1517. doi: 10.1128/jcm.27.7.1514-1517.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L. L., Shoop D. S. Antigenic characteristics of enterotoxigenic and nonenterotoxigenic isolates of Bacteroides fragilis. Am J Vet Res. 1987 Apr;48(4):643–645. [PubMed] [Google Scholar]
- Myers L. L., Shoop D. S., Collins J. E., Bradbury W. C. Diarrheal disease caused by enterotoxigenic Bacteroides fragilis in infant rabbits. J Clin Microbiol. 1989 Sep;27(9):2025–2030. doi: 10.1128/jcm.27.9.2025-2030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L. L., Shoop D. S., Stackhouse L. L., Newman F. S., Flaherty R. J., Letson G. W., Sack R. B. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J Clin Microbiol. 1987 Dec;25(12):2330–2333. doi: 10.1128/jcm.25.12.2330-2333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. J. Chromosomal DNA fingerprinting--a new method of species and strain identification applicable to microbial pathogens. J Med Microbiol. 1989 Oct;30(2):89–99. doi: 10.1099/00222615-30-2-89. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Smith C. J. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J Bacteriol. 1985 Mar;161(3):1069–1073. doi: 10.1128/jb.161.3.1069-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Spiegel H. Transposition of Tn4551 in Bacteroides fragilis: identification and properties of a new transposon from Bacteroides spp. J Bacteriol. 1987 Aug;169(8):3450–3457. doi: 10.1128/jb.169.8.3450-3457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Tally F. P., Cuchural G. J., Jacobus N. V., Gorbach S. L., Aldridge K. E., Cleary T. J., Finegold S. M., Hill G. B., Iannini P. B., McCloskey R. V. Susceptibility of the Bacteroides fragilis group in the United States in 1981. Antimicrob Agents Chemother. 1983 Apr;23(4):536–540. doi: 10.1128/aac.23.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer R. E. An improved method for detecting foreign DNA in plasmids of Escherichia coli. Anal Biochem. 1979 Sep 15;98(1):60–63. doi: 10.1016/0003-2697(79)90705-x. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A., Macrina F. L. Physical characterization of Bacteroides fragilis R plasmid pBF4. J Bacteriol. 1981 Feb;145(2):867–872. doi: 10.1128/jb.145.2.867-872.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst J., Wilkins T. D. Effect of clavulanic Acid on anaerobic bacteria resistant to Beta-lactam antibiotics. Antimicrob Agents Chemother. 1978 Jan;13(1):130–133. doi: 10.1128/aac.13.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]