Abstract
Congenital heart disease is the most common type of birth defect with an incidence of 1%. Previously, we described a point mutation in GATA4 that segregated with cardiac defects in a family with autosomal dominant disease. The mutation (G296S) exhibited biochemical deficits and disrupted a novel interaction between Gata4 and Tbx5. To determine if Gata4 and Tbx5 genetically interact in vivo, we generated mice heterozygous for both alleles. We found that nearly 100% of mice heterozygous for Gata4 and Tbx5 were embryonic or neonatal lethal and had complete atrioventricular (AV) septal defects with a single AV valve and myocardial thinning. Consistent with this phenotype, Gata4 and Tbx5 are co-expressed in the developing endocardial cushions and myocardium. In mutant embryos, cardiomyocyte proliferation deficits were identified compatible with the myocardial hypoplasia. Similar to Gata4, Gata6 and Tbx5 are co-expressed in the embryonic heart, and the transcription factors synergistically activate the atrial natiuretic factor promoter. We demonstrate a genetic interaction between Gata6 and Tbx5 with an incompletely penetrant phenotype of neonatal lethality and thin myocardium. Gene expression analyses were performed on both sets of compound heterozygotes and demonstrated downregulation of α-myosin heavy chain only in Gata4/Tbx5 heterozygotes. These findings highlight the unique genetic interactions of Gata4 and Gata6 with Tbx5 for normal cardiac morphogenesis in vivo.
Keywords: cardiac development, congenital heart defects, Gata4, Gata6, Tbx5, transcription factor
INTRODUCTION
Congenital heart disease (CHD) is the most common type of birth defect affecting nearly 1% of all live births and defects of cardiac septation are the most frequent form of CHD Hoffman, 2002). The majority of CHD has a multifactorial etiology but recent studies have identified monogenic etiologies for a subset of CHD (Garg, 2006; Ransom and Srivastava, 2007; Bruneau, 2008). The causative genes often result in disease in the setting of haploinsufficiency suggesting that gene dosage is critical for normal cardiac morphogenesis. CHD-causing genes have been well studied in murine heart development. Targeted deletion of these genes nearly always results in embryonic lethality and severe cardiac malformations while heterozygous mice are often unaffected. As is often the case, human cardiac disease-causing genes encode for proteins that are part of transcriptional complexes (Jimenez-Sanchez, 2001).
We previously discovered that mutations in GATA4 were associated with CHD, most commonly cardiac septal defects (CSD) and pulmonary valve stenosis (Garg et al, 2003). Subsequently, other investigators reported additional GATA4 mutations in individuals with CHD (Nemer et al, 2006; Rajagopal et al, 2007; Tomita-Mitchell et al, 2007). Gata4 belongs to a family of zinc finger transcription factors that includes six known family members, Gata 1–6, and Gata4 and Gata6 are important for cardiovascular development (Molkentin et al, 2000). Gata4 is expressed in multiple cell types that are critical for proper cardiac septation and these include endocardial, myocardial and cushion mesenchymal cells (Pu et al, 2004). Initial studies demonstrated that targeted deletion of Gata4 in mice results in early embryonic lethality and cardiac bifida likely secondary to an endodermal defect (Kuo et al, 1997; Molkentin et al, 1997). The dosage sensitivity and functions of Gata4 during later stages of cardiac morphogenesis have been demonstrated in numerous studies that have shown cardiac malformations in mice harboring a hypomorphic Gata4 allele and tissue-specific deletions of Gata4 (Crispino et al, 2001; Pu et al, 2004; Watt et al, 2004; Zeisberg et al, 2005; Rivera-Feliciano et al, 2006). Although Gata6 is expressed in the developing heart, targeted disruption of Gata6 in the mouse results in early embryonic lethality prior to cardiac development. Recent mouse studies have demonstrated an in vivo functional role for Gata6 in heart development as well (Koutsourakis et al, 1999; Morrissey et al, 1998; Xin et al, 2006; Lepore et al, 2006).
Tbx5 is a T-box containing transcription factor that is critical for proper limb and heart development. TBX5 mutations that result in haploinsufficiency cause Holt-Oram syndrome, which is characterized by congenital heart defects, conduction-system abnormalities and upper-limb deformities (Basson et al, 1997; Li et al, 1997). The structural cardiac manifestations of Holt-Oram syndrome range from atrial and ventricular septal defects to more severe defects such as hypoplastic left heart syndrome (Basson et al, 1994; Newbury-Ecob et al, 1996). The developmental expression pattern of the Tbx5 protein localizes to tissues affected by Holt-Oram syndrome (Bruneau et al, 1999; Hatcher et al, 2000). Tbx5 homozygous knockout mice are embryonic lethal due to arrested cardiac morphogenesis indicating a vital role for Tbx5 in early heart development. Mice heterozygous for Tbx5 develop heart and limb abnormalities and also express a unique gene expression pattern demonstrating the dosage sensitivity for this transcription factor during heart development (Bruneau et al, 2001; Mori et al, 2006).
Cardiac morphogenesis is a complex process that is regulated by numerous highly conserved transcription factors, which interact in a precise and coordinated manner (Olson 2006; Srivastava, 2006). We previously demonstrated that Gata4 and Tbx5 physically interact in co-immunoprecipitation assays and cooperatively activate a common luciferase reporter, atrial natiuretic factor (ANF), in transactivation assays (Garg et al., 2003). This novel interaction between Tbx5 and Gata4 was abolished by mutations in GATA4 or TBX5 that were identified in humans with CHD. These findings suggested that interaction between the Gata family of transcription factors and Tbx5 is necessary for proper cardiac morphogenesis.
Here, we show that Gata4, Gata6 and Tbx5 are co-expressed during cardiac morphogenesis and mice compound heterozygous for either Gata4 and Tbx5 or Gata6 and Tbx5 suffer embryonic or early neonatal lethality. The Gata4/Tbx5 compound heterozygotes had cardiovascular defects that included complete atrioventricular (AV) septal defects and thin myocardium. The cardiac phenotype of the Gata6/Tbx5 compound heterozygotes was less severe and involved only variable thinning of the myocardium. Cardiac gene expression analysis of Gata4+/−Tbx5+/− heterozygotesrevealed decreased mRNA levels of α-myosin heavy chain (or Myh6), a direct target of Gata4 and Tbx5 that has been implicated as a cause of human atrial septal defects (Molkentin et al, 1994; Huang et al, 1995; Ching et al, 2005). These findings provide insight into the genetic pathways cooperatively regulated by Gata4, Gata6 and Tbx5 that when altered lead to cardiac malformations.
MATERIALS AND METHODS
Breeding and Collection of Mouse Embryos
Mice were maintained on a 0600 to 1800h light-dark cycle, with noon of the day of observation of a vaginal plug defined as embryonic day (E) 0.5. Mice heterozygous for Gata4, Gata6, and Tbx5 were generated and genotyped as previously described (Molkentin et al, 1997; Bruneau et al, 2001; Xin et al, 2006). These lines have been maintained in mixed genetic backgrounds: Gata4/Gata6 are in 129/C57BL6 while Tbx5 are in Black Swiss/129. To generate the double heterozygote mice used in this study, mice heterozygous for Gata4 or Gata6 were mated to Tbx5 heterozygote mice and pregnant mothers or newborn litters were sacrificed at various embryonic and postnatal timepoints. Wildtype littermates were used as controls for histologic sections and gene expression studies.
Histologic and radioactive section in situ hybridization
To fully characterize the phenotype of the single and double heterozygote mice, histological analysis was performed. Pregnant mothers were sacrificed to obtain embryos, which were fixed in 4% paraformaldehyde for sectioning. Hematoxylin and eosin (H&E) staining was carried out on heart sections using standard methods to identify any defects. Histological 3-D reconstruction was performed using episcopic fluorescence image capture on E14.5 murine embryos to examine the cardiac phenotype (Rosenthal et al, 2004). Myocardial wall thickness was quantified by measuring the thickness of the compact myocardium in the right and left ventricles at the level of the tricuspid and mitral valve in coronal sections of embryonic hearts. A minimum of four measurements was obtained for each embryo and means and standard deviations were calculated. In situ hybridization was performed as described previously (Garg et al, 2001) using 35S-labeled antisense probes synthesized with T3, T7, or SP6 RNA polymerase (Maxiscript; Ambion Inc., Austin, TX) from mouse Myh6, Bmp2, Bmp4, and Notch1 cDNA.
Proliferation and Apoptosis Assays
Embryos were collected as described above. For these immunostaining studies, histologic sections were deparafinnized in xylene and rehydrated to phosphate buffered saline (PBS). Proliferation assays were performed using the phosphohistone H3 (PH3) antibody (Upstate Cell Signaling Solutions, Temecula, CA). The sections were permeabilized in 0.3% Triton X-100 in PBS. Sections were then blocked by 3.5% donkey serum in PBS followed by incubation with 1% rabbit anti-phosphohistone H3 antibody overnight at 4°C. Sections were then washed in PBS and Cy3 (1%) secondary antibodies (Vector Laboratories, Burlingame, CA) for 30 minutes. For cell proliferation studies, sister sections were stained for mouse heavy chain cardiac myosin using Cy3-conjugated antibody (Abcam, Cambridge, MA) to label the cardiomyocytes. The percentage of PH3-stained ventricular cardiomyocytes/total number of ventricular cardiomyocytes was calculated by analyzing a minimum of three embryos for each genotype. A minimum of four sections per embryo was analyzed, and the means and standard deviations are shown. Apoptosis (TUNEL) assays were performed using the In Situ Cell Death Detection Kit, Fluorescein (Roche) according to manufacturer instructions. Labeled ventricular cardiomyocytes were counted on a minimum of six sections of control and mutant embryonic hearts. Statistical analysis was performed using Student’s t-test.
Gene Expression Analysis
RNA was purified from embryonic hearts of Gata4+/− Tbx5+/− compound heterozygotes (E11.5 and E13.5); Gata 4+/− (E11.5 and E13.5), Tbx5+/− (E11.5 and E13.5), Gata6+/− (E13.5) single heterozygotes; and Gata6+/− Tbx5+/− (E13.5) double heterozygotes; and their respective wildtype littermates using Trizol (Invitrogen). Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using the Taqman Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA). 25 ng of total RNA was used for reverse transcription and amplification in each real-time PCR reaction using a Biorad iQ5 real-time PCR machine. Commercially available Taqman probes were utilized for the following genes: Gata4, Gata6, Nkx2.5, Hey1, Hey2, Myh7, Myh6, Bmp2, Bmp4, Tgfβ1, Tgfβ2, Nfatc1, and Notch1. For Tbx5, a previously published custom probe designed to amplify exon 3 was used (Mori et al, 2006). Mean relative gene expression was calculated from wild type and mutant hearts after normalization to 18s ribosomal RNA, minimum of n=3 per group. Statistical analysis was performed using Student’s t-test, and a P value of less than 0.05 was considered significant.
Luciferase Assays
HeLa cells were transfected using Fugene 6 (Roche) according to manufacturer’s instructions. The atrial natriuretic factor luciferase reporter plasmid (300ng) and CMV β-galactosidase expression plasmid (50ng) to control for transfection efficiency were transfected along with Gata6 and Tbx5 expression plasmids. Luciferase activity was measured 48h after transient transfection as previously described (Schluterman et al, 2007) and normalized to LacZ expression to generate relative luciferase activity. Three independent experiments were performed in duplicate. Statistical comparisons were performed using Student’s t-test.
RESULTS
Co-expression of Gata4 and Tbx5 in the developing heart
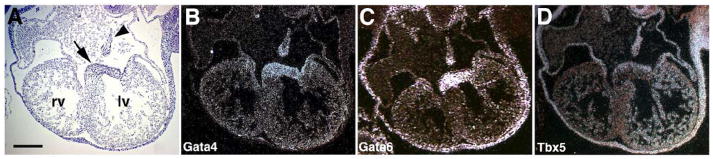
Gata4 and Tbx5 are known to be expressed in various cell lineages and at different stages of cardiac morphogenesis (Molkentin et al, 1997; Bruneau et al, 1999; Pu et al, 2004). To determine the cardiac cell lineages in which Gata4 and Tbx5 might genetically interact, we performed in situ hybridization in wildtype E11.5 mouse embryos. As previously reported, transcripts from both genes were present in the atrial and ventricular myocardium along with the developing endocardial cushion mesenchyme (Figure 1). Gata4 was predominantly expressed in the developing atrial septum and endocardial cushions while Tbx5 showed similar increased levels of expression in the atrial septum along with left ventricular myocardium and pericardium A low level of expression of Tbx5 mRNA was present in the right ventricular myocardium consistent with previous findings (Bruneau et al, 1999) (Figure 1D). The distinct but overlapping expression patterns of Gata4 and Tbx5 are consistent with these cardiac transcription factors having cooperative functions in vivo in later stages of cardiac morphogenesis.
Figure 1. Gata4, Gata6, and Tbx5 are co-expressed in embryonic heart.
Coronal sections of E11.5 mouse hearts demonstrate that Gata4 mRNA(B), Gata6 mRNA (C) and Tbx5 mRNA (D) are co-localized to the atrial and ventricular myocardium by radioactive section in situ hybridization. All genes are co-expressed in the developing atrial septum (arrowhead) and endocardial cushion derivatives (arrow). Corresponding bright-field image is shown in (A). Scale bar represents 100 microns. rv, right ventricle; lv, left ventricle.
Embryonic lethality of Gata4+/− Tbx5 +/− mice
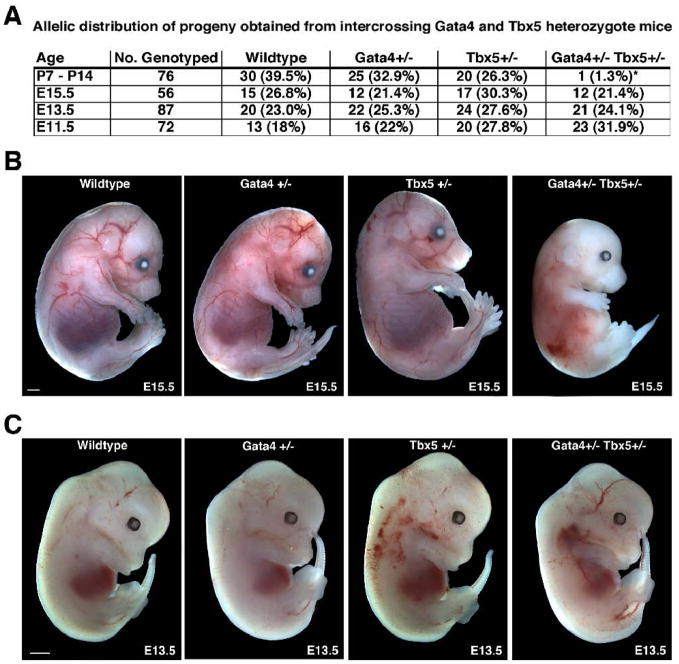
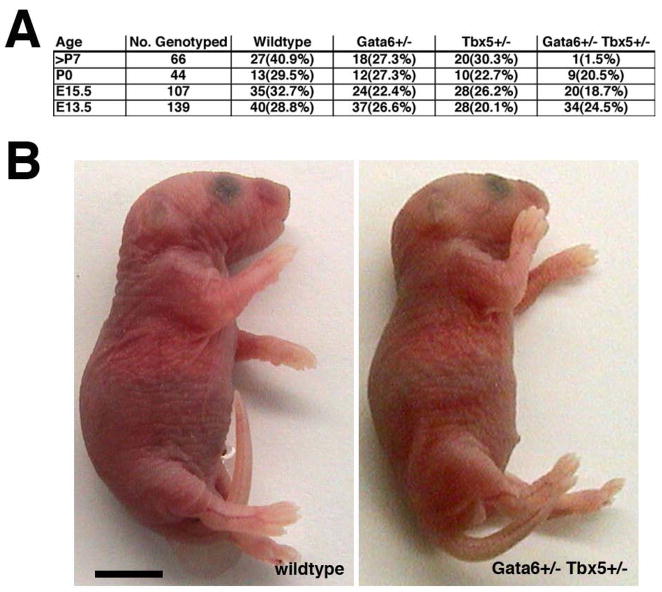
To investigate the functional significance of the in vivo interaction between Gata4 and Tbx5 during cardiac development, we generated mice heterozygous for both Gata4 and Tbx5 mutant alleles. According to Mendelian ratios, we expected equal numbers of wildtype, Gata4 heterozygote, Tbx5 heterozygote, and Gata4/Tbx5 double heterozygote pups to be present in utero and at birth, but only 60% of Tbx5 heterozygotes or Gata4/Tbx5 double heterozgygotes to be present at weaning (postnatal day 28) due to the reported perinatal lethality seen in Tbx5 heterozygote mice (Bruneau et al, 2001). Genotypic analysis of 36 embryonic and 14 post-natal litters demonstrated that mice heterozygous for both Gata4 and Tbx5 exhibited nearly 100% lethality by postnatal day 7 (Figure 2A). Although, normal Mendelian ratios were present at E15.5, nearly 50% of these embryos were growth retarded (Figures 2A and B). Analysis of E11.5 and E13.5 embryos showed expected Mendelian ratios with no evidence of growth retardation (Figure 2A and C; data not shown). Embryonic lethality of Gata4/Tbx5 double heterozygotes was observed in the C57BL6/129/Black Swiss mixed genetic background. Our analysis established that Gata4+/− Tbx5+/− embryos suffer lethality during the later stages of gestation.
Figure 2. Embryonic lethality and intrauterine growth retardation in Gata4+/− Tbx5+/− embryos.
(A) Frequency of genotypes obtained from intercrossing Gata4+/− mice with Tbx5+/− mice is shown and demonstrates embryonic lethality of Gata4+/− Tbx5+/− mice during late gestation. (B) Gata4+/− Tbx5+/− embryos are growth retarded at E15.5 when compared to wildtype and single heterozygote littermates while at E13.5 the mutant embryos (C) are of normal size when compared to littermates. *, p value < 0.05; scale bar represents 1 millimeter.
Gata4+/− Tbx5 +/− embryos display atrioventricular septation defects
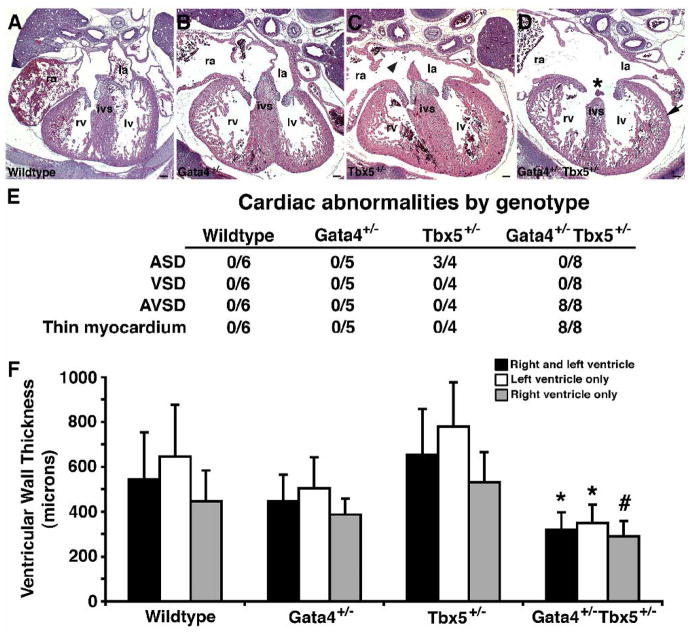
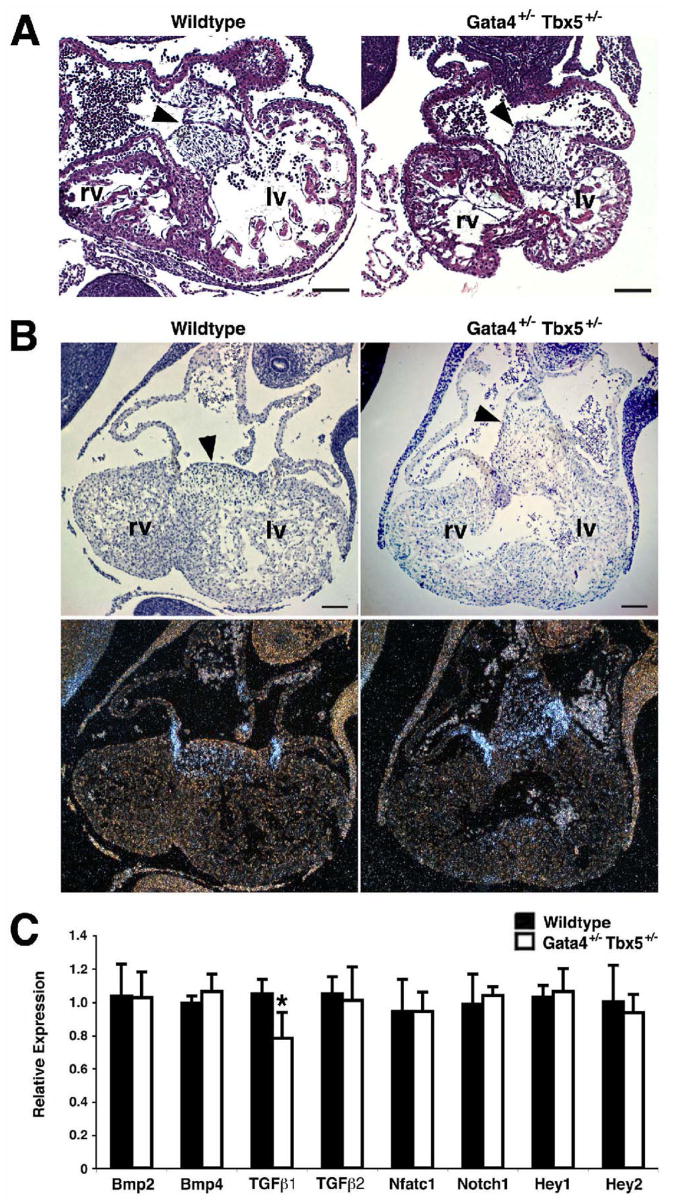
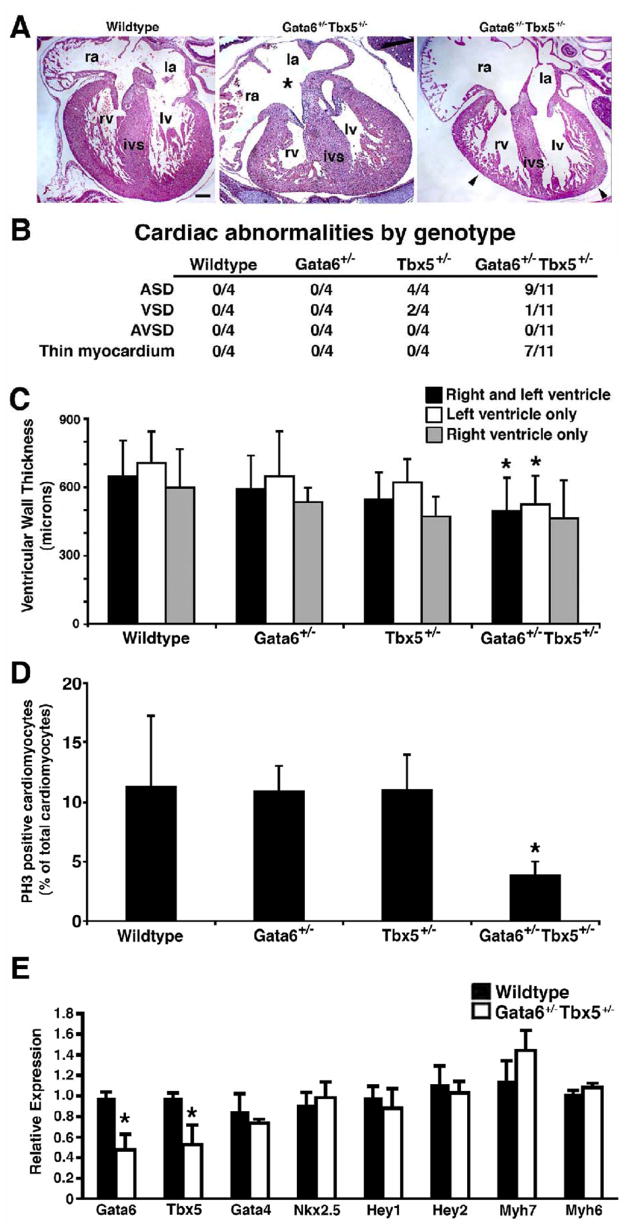
In order to determine the cause of this early lethality, histological analysis was performed on E15.5 Gata4+/− Tbx5+/− embryonic hearts and demonstrated a defect of atrioventricular septation with a single common atrioventricular valve and associated atrial and ventricular septal defects in all embryos examined (n=7) (Figure 3). This phenotype was also demonstrated by histologic 3-D reconstruction of the heart of an E14.5 embryo (see Supplementary movie). At E15.5, wildtype, Gata4 heterozygote or Tbx5 heterozygote littermates did not demonstrate atrioventricular septal defects and had two separate atrioventricular valves with a normal myocardium (Figures 3A–C). Isolated atrial septal defects were seen in a portion of Tbx5 heterozygotes, as has been reported, however none were similar to the Gata4+/−Tbx5+/− intracardiac defects (Figures 3C and D). The embryologic basis of complete atrioventricular septal defects is thought to be abnormal development of the endocardial cushions. Consistent with this, the atrioventricular septum, which is derived from endocardial cushion cells, was almost non-existent in the crux of the Gata4+/− Tbx5+/− E15.5 hearts (Figure 3D, asterisk). The process of atrioventricular septation begins at E9.5, and examination of Gata4+/− Tbx5+/− embryos at E10.5 demonstrated normal cellularized endocardial cushions when compared to wildtype, Gata4 heterozygote and Tbx5 heterozygote littermates (Figure 4A and data not shown). The endocardial cushions of the mutant embryos had normal expression of Bmp2, Bmp4, and Notch1 by radioactive in situ hybridization (Figure 4B and data not shown). The endocardial cushions are remodeled into separate tricuspid and mitral valves by E13.5, and the atrioventricular septation defect is present at this timepoint in the compound heterozygotes. In a similar fashion, gene expression studies using real time qRT-PCR of E11.5 mutant hearts demonstrated no significant alteration in RNA levels of genes implicated in endocardial cushion formation except for a mild decrease in Tgfβ1 to 80% of wildtype levels (Figure 4C). Additional qRT-PCR experiments performed using isolated atrioventricular cushions harvested from E11.5 embryonic hearts did not confirm downregulation of Tgfβ1 (data not shown). These findings suggested epithelial to mesenchymal transformation (EMT) was normal in these mutant hearts and that the defect was due to improper endocardial cushion maturation and remodeling.
Figure 3. Cardiac defects in Gata4+/− Tbx5+/− embryos.
Coronal sections through E15.5 embryonic hearts (A–D). In Gata4+/− Tbx5+/− embryo, an atrioventricular septation defect (*) and thin myocardium (arrow) is demonstrated (D). Normal cardiac septation and myocardium are seen in wildtype (A) and Gata4+/− (B) littermates. An atrial septal defect (arrowhead) is found in hearts of Tbx5+/− heterozygotes (C). Scale bar represents 100 microns; ra, right atrium; la, left atrium; ivs, interventricular septum; rv, right ventricle; lv, left ventricle. (E) Table representing the frequency of cardiac abnormalities identified in each genotype at E14.5 and E15.5. ASD, atrial septal defect; VSD, ventricular septal defect; AVSD, atrioventricular septal defect. (F) Decreased ventricular wall thickness in Gata4+/− Tbx5+/− double heterozgotes at E15.5 compared to wildtype, Gata4+/− single heterozygote and Tbx5+/− single heterozygote littermates. Mean thickness of compact myocardial layer of ventricular wall is shown in microns. *, p value < 0.05; #, p value < 0.06.
Figure 4. Endocardial cushion development in Gata4+/− Tbx5+/− embryos.
(A) Coronal sections through wildtype and Gata4+/− Tbx5+/− E10.5 embryos demonstrate normal endocardial cushion formation (arrowhead). (B) Radioactive section in situ hybridization shows normal expression of Bmp2 mRNA in the mesenchyme of the endocardial cushions (arrowheads) at E10.5. (C) No statistically significant alterations in the expression of Bmp2, Bmp4, Tgfβ2, Nfatc1, Notch1, Hey1 and Hey2 in Gata4+/− Tbx5+/− E11.5 embryonic hearts are seen by real time qRT-PCR. Tgfβ1 is present at ~80% of wildtype levels. Scale bar represents 100 microns. rv, right ventricle; lv, left ventricle. *, p value < 0.05.
Gata4+/− Tbx5 +/− embryos have abnormal myocardium
The presence of a complete atrioventricular septal defect should not lead to prenatal lethality and therefore we examined the Gata4+/− Tbx5+/− embryos to determine the possible etiology for the embryonic lethality. On histologic examination, the E15.5 Gata4+/− Tbx5+/− embryos also displayed a thin myocardium that appeared to affect the right and left ventricle. (Figure 3D and Supplementary movie). Examination of the compound mutants at E11.5 showed no obvious myocardial hypoplasia (data not shown) but thinning was observed by E13.5. We quantified the myocardial wall thickness in wild type, Gata4+/−, Tbx5+/−, and Gata4+/− Tbx5+/− heterozygote E15.5 hearts and the Gata4+/− Tbx5+/− embryos had significantly thinner ventricular walls as compared to wildtype and single heterozygote littermates (Figure 3F). Our data suggests that the mutant embryos were possibly dying after E15.5 from heart failure secondary to the abnormal ventricular myocardium.
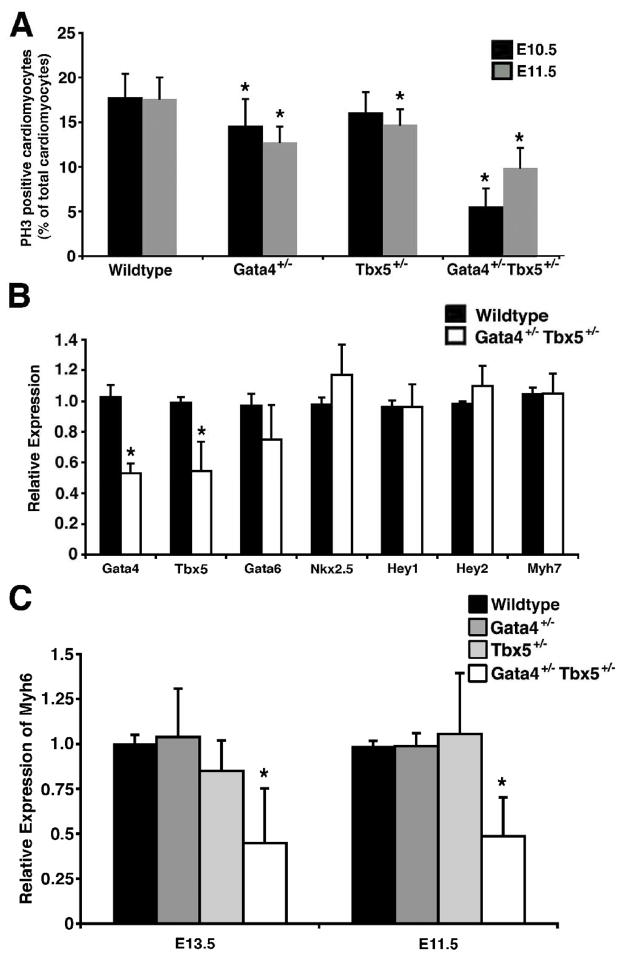
In order to determine the etiology of the thin ventricular myocardium in Gata4+/− Tbx5+/− embryos, we examined mutants at earlier embryonic timepoints for defects in cardiomyocyte proliferation and apoptosis. In the Gata4+/− Tbx5+/− embryos, we did not identify any changes in apoptosis in the ventricular myocardium at E10.5, 11.5 and E13.5 (data not shown). We did observe decreased cellular proliferation by phosphohistone H3 staining at E10.5 and E11.5 in the compound heterozygotes that was more severe than the single heterozygotes (Figure 5A). These data suggest that the dosage of Gata4 and Tbx5 is critical for normal cardiomyocyte proliferation during the middle stages of cardiac development.
Figure 5. Cardiomyocyte defects in Gata4+/− Tbx5+/− embryos.
(A) Decreased cardiomyocyte proliferation as determined by phosphohistone H3 (PH3) staining is quantified in Gata4+/− Tbx5+/− embryonic hearts when compared to wildtype, Gata4+/− single heterozygote and Tbx5+/− single heterozygote littermates at E10.5 and E11.5. *, p value < 0.05. (B) Real-time qRT-PCR demonstrates no significant changes in Gata6, Nkx2.5, Hey1, Hey2 and Myh7 expression in the embryonic ventricles. Gata4 and Tbx5 mRNA levels are decreased by approximately 50% as predicted by heterozygosity of Gata4 and Tbx5. (C) Decreased expression of Myh6 mRNA is found in E13.5 and E11.5 embryonic ventricles by real-time qRT-PCR while expression is unchanged in the Gata4+/− and Tbx5+/− single heterozygotes. *, p value < 0.05 when compared to wildtype, Gata4+/− heterozygote or Tbx5+/− heterozygote littermates.
To determine if cardiac gene expression was altered in the Gata4+/− Tbx5 +/− mice, we performed real time qRT-PCR using RNA harvested from ventricles of E13.5 embryos. We found the expression levels of the cardiac transcription factors, Gata6, Nkx2.5, Hey1, and Hey2 in E13.5 mutant ventricles were unchanged when compared to wildtype ventricles (Figure 5B). As expected, Gata4 and Tbx5 were downregulated to ~50% of wildtype levels. We did identify downregulation of Myh6 (α-myosin heavy chain) but not Myh7 (β-myosin heavy chain) in these ventricles (Figures 5B and 5C). Real-time qRT-PCR demonstrated that Myh6 mRNA was expressed at lower levels in the compound heterozygotes when compared to Gata4 and Tbx5 single heterozygotes and was also decreased at an earlier timepoint (E11.5) (Figure 5C). Radioactive section in situ hybridization suggested that the downregulation was more pronounced in the ventricular myocardium, but downregulation of Myh6 was also seen by semi-quantitative RT-PCR when mRNA from whole hearts was analyzed (Supplementary Figure 1).
Interaction of Gata6 and Tbx5
Gata6 has been shown to be important for cardiovascular development and has been postulated to be functionally redundant with Gata4. Similar to Gata4, Gata6 was co-expressed with Tbx5 in the developing heart (Figure 1) and was able to synergistically activate an ANF-luciferase reporter in transactivation assays (Supplementary Figure 2). To determine if Gata6 exhibited a similar in vivo genetic interaction with Tbx5 as Gata4, we generated Gata6+/− Tbx5 +/− mice. The Gata6+/− Tbx5 +/− mice displayed an early neonatal lethality but exhibited no growth retardation at E18.5 and post-natal day 1 (Figure 6 and data not shown). Histologic analysis at E15.5 demonstrated normal cardiac septation but myocardial thinning that predominantly involved the left ventricle was found in a subset of compound mutants (Figure 7A, 7B and 7C). The etiology for the nearly 100% lethality is unclear and maybe due to a role for Gata6 and Tbx5 in the conduction system or to abnormal contractile properties of the myocardium (Davis et al, 2001; Zhu et al, 2008). No significant amount of apoptosis was found in the myocardium of E13.5 mutant embryos when compared to wildtype littermates (data not shown), but a deficit in myocardial proliferation was demonstrated at E13.5 in these Gata6/Tbx5 compound heterozygotes (Figure 7D). Interestingly, the myocardial defect was less severe as compared to the Gata4+/− Tbx5 +/− embryos highlighting the unique and functional importance of Gata4 at these stages. By real time qRT-PCR, there was normal expression of several cardiac transcription factors except for the expected downregulation of Gata6 and Tbx5. Interestingly unlike the Gata4+/− Tbx5 +/− embryos, there was also normal expression of α-myosin heavy chain in Gata6+/−Tbx5+/− double heterozygote hearts (Figure 7E).
Figure 6. Neonatal lethality and no intrauterine growth retardation in Gata6+/− Tbx5+/− embryos.
(A) Frequency of genotypes obtained from intercrossing Gata6+/− mice with Tbx5+/− mice. Neonatal lethality of Gata6+/− Tbx5+/− mice is demonstrated. (B) No growth retardation of Gata6+/− Tbx5+/− embryos is seen at postnatal day 1. Scale bar represents 5 mm.
Figure 7. Cardiac defects in Gata6+/− Tbx5+/− embryos.
(A) Variable myocardial thinning (arrowhead) is seen in a subset of Gata6+/− Tbx5+/− E15.5 embryos in coronal section of hearts when compared to wildtype. Atrial septal defect (asterisk) is found in a portion of double heterozygotes. Scale bar represents 100 microns; ra, right atrium; la, left atrium; ivs, interventricular septum; rv, right ventricle; lv, left ventricle. (B) Table representing the frequency of cardiac abnormalities identified in each genotype at E15.5. ASD, atrial septal defect; VSD, ventricular septal defect; and AVSD, atrioventricular septal defect. (C) Decreased thickness of compact myocardium in Gata6+/− Tbx5+/− double heterozgotes at E15.5 compared to wild type, Gata6+/− single heterozygote and Tbx5+/− single heterozygote littermates. Mean thickness as measured in microns is shown for both ventricles, left ventricle, and right ventricle. *, p value < 0.05. (D) Decreased cardiomyocyte proliferation as determined by phosphohistone H3 (PH3) staining is quantified in Gata6+/− Tbx5+/− embryonic hearts when compared to wildtype, Gata6+/− and Tbx5+/− single heterozygote littermates at E13.5. *, p value < 0.05. (E) Real-time qRT-PCR demonstrates no significant change in expression levels of Gata4, Nkx2.5, Hey1, Hey2, Myh7 and Myh6 in E13.5 Gata6+/− Tbx5+/− embryonic hearts when compared to wildtype littermates. Gata6 and Tbx5 mRNA levels are decreased by approximately 50% as predicted by heterozygosity of Gata6 and Tbx5. *, p value < 0.05.
DISCUSSION
The transcription factors, Gata4, Gata6 and Tbx5, are known to play essential roles in cardiac development. Here, we have demonstrated that combined haploinsufficiency of Gata4 and Tbx5 results in early neonatal lethality and cardiovascular defects that are not seen in mice heterozygous for null mutations of either gene. Furthermore, these studies suggest that Gata4 and Tbx5 function in a cooperative manner during formation of the atrioventricular septum and maturation of the myocardium. In addition, we describe a genetic interaction between Gata6 and Tbx5. The cardiac phenotype in the Gata6+/− Tbx5+/− double heterozygotes is less severe than the Gata4+/− Tbx5+/− double heterozygotes suggesting unique in vivo roles for Gata4 and Gata6. In addition, we identified that a common transcriptional target of Gata4 and Tbx5, Myh6, is specifically downregulated in Gata4+/− Tbx5+/− embryos. These findings highlight the dosage sensitivity of cardiac morphogenesis for appropriate levels of these critical transcription factors.
Cooperative functions of Gata4, Gata6 and Tbx5 during myocardial development
Cardiac development is regulated by a highly conserved group of cardiac transcription factors (Olson, 2006). Two of these, Gata4 and Tbx5, have been well studied and play critical roles in the early stages of cardiac morphogenesis. We previously reported the discovery of a novel biochemical interaction between Gata4 and Tbx5 and demonstrated their ability to cooperatively activate common luciferase reporters in an in vitro system. Here we extended these findings to show that Myh6, a proposed in vitro target of Gata4 and Tbx5, was downregulated in the hearts of Gata4+/− Tbx5+/− embryos. Myh6 is critical for normal cardiac development and Myh6-null mice die between E11.0 and E12.0 of severe heart defects (Jones et al., 1996). In vitro studies demonstrated that the cooperative activation of common downstream targets required the independent binding of both Gata4 and Tbx5 (Garg et al, 2003). Our in vivo studies demonstrate that haploinsufficiency of Gata4 and Tbx5 results in altered gene expression of a common downstream target, but it is not clear if this is the result of decreased physical interaction Gata4 and Tbx5 or simply a dosage effect in activation of target genes by Gata4 and Tbx5.
Our work demonstrates the importance of transcription factor interactions in cardiac morphogenesis as we show that Gata4 and Gata6 can genetically interact with Tbx5. Interestingly, haploinsufficiency of both Gata4 and Tbx5 led to defects in two cardiac morphogenetic processes, atrioventricular septum formation and myocardial development, while haploinsufficiency of Gata6 and Tbx5 resulted in only mild defects in myocardial development. Similar to Tbx5, Gata4 is known to play an important role in myocardial development as multiple mouse models of Gata4 deficiency result in myocardial hypoplasia (Crispino et al, 2001; Zeisberg et al, 2005; Watt et al, 2004; Pu et al, 2004) In addition, Gata6 has been shown to be critical for cardiac differentiation and cardiomyocyte proliferation in vivo such that haploinsufficiency of both genes causes lethality at E13.5 (Xin et al, 2006; Zhao et al, 2008). Our studies support a combination of unique and redundant roles for Gata4 and Gata6 in myocardial development. It is likely other cardiac transcription factors such as Nkx2.5, which has been shown to interact with Gata4 and Tbx5, may also play a role in regulating these later developmental processes. Of note, we have not found an increased lethality in mice that are heterozygous for all three transcription factors (unpublished data, V.G.).
Atrioventricular septation defects in Gata4+/− Tbx5+/− mice
The cardiovascular defects identified in mice heterozygous for Gata4 and Tbx5 were limited to atrioventricular septation and cardiomyocyte development. An atrioventricular septal defect has traditionally been proposed to be the result of abnormal endocardial cushion development. The process of endocardial cushion formation is complex and starts at E9.5 in the mouse. At that stage, epithelial-mesenchymal transformation (EMT) is critical for formation of the endocardial cushion cells. In the Gata4/Tbx5 compound heterozygotes, this initial process appears normal. This phenotype is similar to what has been reported in several mouse models of atrioventricular septal defect including myocardial-deletion of Bmp4; deletion of the Bmp type I receptor, Alk3, in the cardiomyocytes of the atrioventricular canal; and combined deficiency of Connexin 40 or 43 (Jiao et al, 2003; Gaussin et al, 2002; Kirchhoff et al, 2000). In these mice, a normal mesenchymal mass of endocardial cushion cells develops by E10.5 but later remodeling does not occur resulting in an atrioventricular septation defect. It has been proposed that this is due to continued requirements of reciprocal signaling between the endocardium and myocardium for these later stages of endocardial cushion remodeling. In our Gata4/Tbx5 double heterozygote embryos, it is likely that a similar phenomenon occurs. Gata4 and Tbx5 are co-expressed in multiple cardiac cell lineages during cardiac morphogenesis, and further studies are needed to identify the cell lineage that requires the interaction of Gata4 and Tbx5 to result in this unique phenotype. Ultimately, the elucidation of common downstream target genes in these cells will be critical for understanding the mechanistic basis for the atrioventricular septal defects in these mice.
GATA4, GATA6 and TBX5 in human congenital heart disease
Heterozygous mutations in GATA4 and TBX5 have been linked to congenital heart defects in humans and likely result in haploinsufficiency leading to cardiac malformations (Garg et al, 2003; Basson et al, 1997; Li et al, 1997). The CHD associated with mutations of GATA4 and TBX5 are predominantly ostium secundum type atrial septal defects although atrioventricular septal defects have been reported in children with either GATA4 mutations or Holt-Oram syndrome (Garg et al, 2003; Pierpont et al, 2000). From an embryologic standpoint, there is no clear link between ostium secundum atrial septal defects and atrioventricular septal defects as they are thought to have different cellular bases. Our findings provide a plausible explanation for these clinical observations since it is possible that children with atrioventricular septal defects who have mutations in either GATA4 or TBX5 may harbor additional mutations in other cardiac developmental genes.
To date, no mutations in GATA6 have been reported in children with CHD but our data, along with recent publications, suggest that GATA6 plays an important role in cardiac morphogenesis and is an excellent candidate gene for causing CHD (Xin et al, 2006; Lepore et al, 2006). In addition, our studies suggest that downstream targets of GATA4 and TBX5 such as MYH6, which has already been implicated as a cause of human atrial septal defects, are candidate genes for CHD (Ching et al, 2005). Future studies that elucidate the molecular pathways regulated by these transcription factors will lead to an increased knowledge of the genetic basis of CHD.
Supplementary Material
Acknowledgments
We thank members of the Molecular Pathology Core for histology and radioactive section in situ hybridization; A. Krysiak for technical assistance; B.G. Bruneau for providing the Tbx5 mutant mice; E. N. Olson for providing the Gata4 and Gata6 mutant mice; and B.G. Bruneau and E.N. Olson for critical review of the manuscript. This work was supported by grants from NHLBI/NIH, March of Dimes Birth Defects Foundation, and American Heart Association to D.S.; NHLBI/NIH, March of Dimes Birth Defects Foundation, and the American Heart Association-Texas Affiliate to V.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basson CT, Cowley GS, Solomon SD, Weissman B, Poznanski AK, Traill TA, Seidman JG, Seidman CE. The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome) N Engl J Med. 1994;330:885–91. doi: 10.1056/NEJM199403313301302. [DOI] [PubMed] [Google Scholar]
- 2.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–5. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 3.Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–8. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 4.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–21. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 5.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 6.Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37:423–8. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 7.Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, Orkin SH. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001;15:839–44. doi: 10.1101/gad.875201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis DL, Edwards AV, Juraszek AL, Phelps A, Wessels A, Burch JB. A GATA-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–19. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- 9.Garg V, Yamagishi C, Hu T, Kathiriya IS, Yamagishi H, Srivastava D. Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol. 2001;235:62–73. doi: 10.1006/dbio.2001.0283. [DOI] [PubMed] [Google Scholar]
- 10.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 11.Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63:1141–8. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaussin V, Van de Putte T, Mishina Y, Hanks MC, Zwijsen A, Huylebroeck D, Behringer RR, Schneider MD. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc Natl Acad Sci U S A. 2002;99:2878–83. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatcher CJ, Goldstein MM, Mah CS, Delia CS, Basson CT. Identification and localization of TBX5 transcription factor during human cardiac morphogenesis. Dev Dyn. 2000;219:90–5. doi: 10.1002/1097-0177(200009)219:1<90::AID-DVDY1033>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 15.Huang WY, Cukerman E, Liew CC. Identification of a GATA motif in the cardiac alpha-myosin heavy-chain-encoding gene and isolation of a human GATA-4 cDNA. Gene. 1995;155:219–23. doi: 10.1016/0378-1119(94)00893-w. [DOI] [PubMed] [Google Scholar]
- 16.Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–7. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature. 2001;409:853–5. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 18.Jones WK, Grupp IL, Doetschman T, Grupp G, Osinska H, Hewett TE, Boivin G, Gulick J, Ng WA, Robbins J. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J Clin Invest. 1996;98:1906–17. doi: 10.1172/JCI118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchhoff S, Kim JS, Hagendorff A, Thonnissen E, Kruger O, Lamers WH, Willecke K. Abnormal cardiac conduction and morphogenesis in connexin40 and connexin43 double-deficient mice. Circ Res. 2000;87:399–405. doi: 10.1161/01.res.87.5.399. [DOI] [PubMed] [Google Scholar]
- 20.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–32. [PubMed] [Google Scholar]
- 21.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–60. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 22.Lepore JJ, Mericko PA, Cheng L, Lu MM, Morrisey EE, Parmacek MS. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J Clin Invest. 2006;116:929–39. doi: 10.1172/JCI27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–9. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin JD, Kalvakolanu DV, Markham BE. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the alpha-myosin heavy-chain gene. Mol Cell Biol. 1994;14:4947–57. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–72. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 26.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–52. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 27.Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, Henkelman RM, Bruneau BG. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297:566–86. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–90. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat. 2006;27:293–4. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 30.Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID. Holt-Oram syndrome: a clinical genetic study. J Med Genet. 1996;33:300–7. doi: 10.1136/jmg.33.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierpont ME, Markwald RR, Lin AE. Genetic aspects of atrioventricular septal defects. Am J Med Genet. 2000;97:289–96. [PubMed] [Google Scholar]
- 33.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–44. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, Goldmuntz E, Broman KW, Benson DW, Smoot LB, Pu WT. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43:677–85. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ransom J, Srivastava D. The genetics of cardiac birth defects. Semin Cell Dev Biol. 2007;18:132–9. doi: 10.1016/j.semcdb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–18. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenthal J, Mangal V, Walker D, Bennett M, Mohun TJ, Lo CW. Rapid high resolution three dimensional reconstruction of embryos with episcopic fluorescence image capture. Birth Defects Res C Embryo Today. 2004;72:213–23. doi: 10.1002/bdrc.20023. [DOI] [PubMed] [Google Scholar]
- 38.Schluterman MK, Krysiak AE, Kathiriya IS, Abate N, Chandalia M, Srivastava D, Garg V. Screening and biochemical analysis of GATA4 sequence variations identified in patients with congenital heart disease. Am J Med Genet A. 2007;143A:817–23. doi: 10.1002/ajmg.a.31652. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–48. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet. 2007;44:779–83. doi: 10.1136/jmg.2007.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–8. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, Olson EN. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci U S A. 2006;103:11189–94. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest. 2005;115:1522–31. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–9. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y, Gramolini AO, Walsh MA, Zhou YQ, Slorach C, Friedberg MK, Takeuchi JK, Sun H, Henkelman RM, Backx PH, Redington AN, Maclennan DH, Bruneau BG. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Natl Acad Sci U S A. 2008;105:5519–24. doi: 10.1073/pnas.0801779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.