Abstract
Bipolar affective disorder (BPAD) is suspected to arise in part from malfunctions of the circadian system, a system which enables adaptation to a daily and seasonally cycling environment. Genetic variations altering functions of genes involved with the input to the circadian clock, in the molecular feedback loops constituting the circadian oscillatory mechanism itself, or in the regulatory output systems could influence BPAD as a result. Several human circadian system genes have been identified and localized recently, and a comparison with linkage hotspots for BPAD has revealed some correspondences.
We have assessed evidence for linkage and association involving polymorphisms in ten circadian clock genes (ARNTL, CLOCK, CRY2, CSNK1ε, DBP, GSK3β, NPAS2, PER1, PER2, and PER3) to BPAD. Linkage analysis in 52 affected families showed suggestive evidence for linkage to CSNK1ε. This finding was not substantiated in the association study. 52 SNPs in ten clock genes were genotyped in 185 parent proband triads. Single SNP TDT analyses showed no evidence for association to BPAD. However, more powerful haplotype analyses suggest two candidates deserving further studies. Haplotypes in ARNTL and PER3 were found to be significantly associated with BPAD via single-gene permutation tests (PG=0.025 and 0.008, respectively). The most suggestive haplotypes in PER3 showed a Bonferroni-corrected p-value of PGC=0.07. These two genes have previously been implicated in circadian rhythm sleep disorders and affective disorders.
With correction for the number of genes considered and tests conducted, these data do not provide statistically significant evidence for association. However, the trends for ARNTL and PER3 are suggestive of their involvement in bipolar disorder and warrant further study in a larger sample.
Keywords: manic-depressive illness, genetic linkage, genetic association, PER3, BMAL1
Introduction
Mood disorders cause about 1% of all deaths and are one of contemporary society’s most important causes of days lost to disability (Murray and Lopez, 1996). Affective illnesses, both unipolar major depressive disorder (MDD) and bipolar manic-depressive disorders (BPAD), have several features which have suggested a relationship to biologic clocks. Early authors commented on the periodicity of exacerbations and remissions of the illnesses, which in some patients are quite regular (e.g., Halberg, 1967; Gjessing, 1976). Others noted clinical features suggestive of circadian disorders, such as early awakening, short REM latency, and other sleep disturbances (Benca et al., 1992). A variety of not-very-consistent reports have described abnormalities of circadian phase and melatonin secretion in affective disorders. Evidence that the illness responds to circadian effects of light is quite strong for seasonal affective disorder (SAD) (Partonen and Magnusson, 2001), a phenotype of both MDD and BPAD, in which the circadian system may interact with photoperiodism. Nonseasonal depressions (both MDD and BPAD) likewise respond to light (Tuunainen et al., 2004), though the evidence for photoperiodic regulation of incidence is less extensive. These clinical features support the hypothesis that light treatment works through modifying circadian clock functions that might be closely related to the primary causes of the illness.
Both MDD and BPAD, as well as SAD appear to arise to some degree from partlyoverlapping genetic susceptibilities (Enoch and Goldman, 2001; Kelsoe, 2003; McGuffin et al., 2003). Given their relationships to circadian physiology, these disorders might be partly caused by genetic variations altering functions of the circadian clock (Kripke et al., 1978; Bunney and Bunney, 2000; Mitterauer, 2000; Gould and Manji, 2002). Thus in considering candidate genes with a plausible role in susceptibility to mood disorders, new knowledge of circadian system genetics is of great assistance.
In the past few years, the genetic and molecular bases of circadian clocks have been uncovered in a variety of organisms including Drosophila and mammals (see, e.g., the recent reviews of Dunlap, 1999; Albrecht and Eichele, 2003; Gachon et al., 2004b; Hirota and Fukada, 2004)). Affective disorders might arise from dysfunctions involved with input to the circadian oscillator (e.g., light synchronization), in the molecular feedback loops constituting the circadian oscillatory mechanism itself, or in the regulatory output systems. The primary mammalian circadian oscillator resides in the suprachiasmatic nucleus (SCN) and produces a nearly 24 h cycle through interacting positive/negative feedback loops. It is comprised of the basic helix-loop-helix-PAS transcription factors CLOCK and ARNTL (BMAL1), which act as positive regulators, and the negative regulators PER1, PER2, PER3, CRY1 and CRY2 (Hirota and Fukada, 2004). In addition, the basic helix-loop-helix transcription factors DEC1 and DEC2, the mammalian homolog of the Drosophila protein TIMELESS (TIM), the orphan nuclear receptor REV-ERBα and the basic leucine zipper transcription factors DBP and E4BP4 participate in the feedback loops. The stability and function of circadian system proteins is regulated by phosphorylation through CSNK1ε and MAPK (Hirota and Fukada, 2004). The feedback loops of the circadian clock regulate the output system, the expression of numerous clock-controlled genes, such as HLF and TEF which may also feed back on the clock (Gachon et al., 2004a), GSK3β, and others. GSK3β and perhaps GSK3α are circadian system genes of special interest as targets of the mood stabilizers lithium and valproate (Gould and Manji, 2002). It has been reported that a polymorphism in the GSK3β promoter is associated with the age of onset of bipolar disorder (Benedetti et al., 2004).
Circadian clock functions also exist outside the SCN and may be genetically distinct (Dudley et al., 2003). Indeed, the first evidence relating a circadian system gene to an affective disorder, an association of an NPAS2 allele with seasonal affective disorder (Johansson et al., 2003), highlights a circadian gene active mainly outside the SCN. In particular, NPAS2, a CLOCK paralogue in the forebrain, could be involved with circadian aspects of the sleep-wake cycle independent of the SCN.
Circadian system genes have been associated with circadian rhythm sleep disorders, such as the PER2 gene in familial advanced sleep phase syndrome (Toh et al., 2001), and PER3 (Ebisawa et al., 2001; Archer et al., 2003; Pereira et al., 2005) and CSNKε (Takano et al., 2004) in delayed sleep phase syndrome (DSPS). Both of these disorders may be associated with affective symptoms (see Table I). However, evidence for a role of circadian system genes in non-seasonal affective disorders is sparse so far. Regarding BPAD, an abstract has appeared reporting an association with ARNTL polymorphisms (Mansour et al., 2003), and aspects of BPAD phenotypes have been associated with variations in the gene CLOCK (Benedetti et al., 2003). Another study reports association between a PER3 allele and response to antidepressant drugs (Lorenzi et al., 2003). If there is a genetic variation in the circadian system conferring susceptibility to BPAD, then it would likely occur in one or more of the circadian system genes.
TABLE I.
Summary table of 11 human circadian system genes, showing genetic position, functional implications, and analyses performed
| Gene (NCBI LocusID) | Location | Mutations associated with human traits/disorders |
Analyses performed |
|---|---|---|---|
|
ARNTL (Bmal1): aryl hydrocarbon receptor nuclear translocator-like (406) |
11p15 | association with bipolar disorder1 | linkage (2 STRs) association (5 SNPs) |
|
CLOCK: circadian locomotor output cycles kaput (9575) |
4q12 | diurnal preference 2; regulation of illness recurrence in bipolar disorder 3 |
linkage (2 STRs) association (5 SNPs) |
| CRY1: cryptochrome 1 (1407) | 12q23- q24.1 |
reported in Nievergelt et al. (2005) | |
| CRY2: cryptochrome 2 (1408) | 11p11.2 | linkage (2 STRs) association (5 SNPs) |
|
|
CSNK1 ε: casein kinase 1 epsilon (1454) |
22q13.1 | functional variant associated with circadian rhythm sleep disorders 4 |
linkage (2 STRs) association (6 SNPs) |
|
DBP: D site of albumin promoter binding protein (1628) |
19q13.3 | (mania and psychosis: microarray analysis 5) |
linkage (2 STRs) association (5 SNPs) |
|
GSK3 β: glycogen synthase kinase 3 beta (2932) |
3q13.3 | age of onset in bipolar disorder 6 (involvement in several CNS disorders, e.g., 7) |
association (9 SNPs) |
|
NPAS2: neuronal PAS domain protein 2 (4862) |
2q11.2 | seasonal affective disorder 8 | association (1 SNPs) |
| PER1: period 1 (5187) | 17p13.1- 17p12 |
(schizophrenia microarray analysis 9) | linkage (2 STRs) association (5 SNPs) |
| PER2: period 2 (8864) | 2q37.3 | advanced sleep phase syndrome 10; not associated with bipolar disorder 11 |
linkage (2 STRs) association (5 SNPs) |
| PER3: period 3 (8863) | 1p36.23 | delayed sleep phase syndrome 12, 13, 14; diurnal preference in seasonal affective disorder 8; bipolar disorder 15 |
linkage (2 STRs) association (6 SNPs) |
Key:
In this study, we focused on bipolar families, because BPAD is thought to have somewhat higher heritability and perhaps less genetic complexity compared to MDD. We examined linkage and association to BPAD in 11 circadian genes (Table I). CRY1 gene data were presented previously (Nievergelt et al., 2005). Because of the great complexity of input and output systems, we focused primarily on genes which are constituents of the complex feedback loops composing the molecular circadian clock. We gave preference to recognized function and proximity to reported linkage hot spots. For example, CSNK1ε is close to marker D22S278, which our group and others have found associated with BPAD (Kelsoe et al., 2001). In the present study we confirmed our previous finding of suggestive linkage to the region including CSNK1ε and found evidence of association of haplotypes in ARNTL and PER3 with BPAD. Replication studies in larger datasets are planned to confirm our initial findings and to study gene-gene interactions in this complex system.
Methods
Study subjects
Subjects were ascertained as part of two multi-site collaborations to collect families for linkage studies of bipolar disorder. Prior to participation, all subjects provided written informed consent through local IRB-approved procedures. The UCSD/UBC/UC family set (Set 1), was collected at UCSD, the University of British Columbia and the University of Cincinnati. Ascertainment and diagnostic methods have been described previously (Kelsoe et al., 2001). Briefly, all families were collected through a bipolar I or II proband and selected for the presence of at least two other mood disordered family members. The Structured Clinical Interview for DSM-III-R was used to directly interview subjects. Information from the interview, other family informants, and medical records were then reviewed by a panel of clinicians in order to make a best-estimate diagnosis. In addition, 106 families from the NIMH Genetics Initiative for Bipolar Disorder first-wave pedigree collection were used (Set 2). Ascertainment and diagnostic methods were similar to those described above. All families were ascertained through a bipolar I proband, and the Diagnostic Interview for Genetic Studies was employed.
We used 52 pedigrees from Set 1, consisting of 356 subjects with an average of 6.9 members/family (range 3-33) in the linkage study, and a sample of 159 families (564 individuals), each consisting of one or two affected (bipolar I or II) children and their parents (53 triads from Set 1, 106 triads from Set 2) in our association study.
Markers and genotyping
For the linkage study, two highly polymorphic, flanking microsatellite markers were chosen for genotyping for each of eight human circadian clock genes (see Table II in Results). Genotyping methods have been described in detail in (Nievergelt et al., 2005). Briefly, PCR reactions were run in a total volume of 20 μl, containing 100ng DNA, 0.5 μM of each primer, 0.25 mM dNTPs, 0.9 U AmpliTaq gold and 1x PCR buffer. Reactions were run with the following cycle parameters: 1 × 95°C, 10 min; followed by a touchdown protocol [starting at an annealing temperature of 70°C (D11S4116), or 65°C (D11S4170, D2S345, D19S867, D11S4109) and decreasing by 1°C every 2 cycles, followed by 10 cycles at 60°C, and 55°C, respectively] with denaturing at 94°C for 1 min., annealing for 1 min., extension at 72°C for 1 min., and a final extension at 72°C for 30 min. Markers D1S2663, D22S283, D1S1612, D4S1569, D22S423 (all multiplexed in one reaction), and markers D17S1829 and D11S1785 (both multiplexed in one reaction), and markers D4S3000, D17S1353, D2S2338, and D19S606 were run by a touchdown protocol starting at an annealing temperature of 65°C and decreasing by 1°C every 2 cycles, followed by 34 cycles at 55°C. PCR products were separated by electrophoresis and detected using an Applied Biosystems 377 automated sequencer with Genescan and Genotyper software. Markers were pooled for multiplex detection along with a molecular weight standard (TAMRA, PE Applied Biosystems). Consistency between different gels was assured by including a standard sample (CEPH GM7050).
TABLE II.
Linkage analysis for bipolar disorder and 16 microsatellite markers. Of the 6 different transmission models (narrow [BP1+BP2+SCZAFF]: nAD85, nAD50, nAR50; broad [BP1+BP2+SCZAFF+recurrent major depression]: bAD85, bAD50, bAR50), the model corresponding to the highest LOD score is selected for each marker at three different recombination fractions (θ)
| Gene | Marker | Model | θ = 0 | Model | θ = 0.05 | Model | θ = 0.1 | Physical dist. to gene (KB) |
deCODE position (cM) |
|---|---|---|---|---|---|---|---|---|---|
| CRY2 | D11S1785 | bAR50 | -6.46 | 6 | -3.8 | 6 | -2.26 | 3486 | 58.24 |
| D11S4109 | nAR50 | -6.54 | 3 | -3.24 | 3 | -1.63 | 1697 | 63.81 | |
| PER1 | D17S1828 | bAR50 | -11.69 | 6 | -7.55 | 6 | -4.97 | 4227 | 10.56 |
| D17S1353 | nAR50 | -4.24 | 2 | -0.68 | 2 | 0.13 | 426 | 20.88 | |
| PER2 | D2S345 | nAR50 | -3.81 | 3 | -1.98 | 3 | -0.97 | 1351 | 249.07 |
| D2S2338 | nAR50 | -10.16 | 3 | -6.19 | 3 | -3.91 | 303 | 251.82 | |
| PER3 | D1S2663 | nAR50 | -8.54 | 3 | -4.43 | 2 | -2.27 | 587 | 12.84 |
| D1S1612 | nAR50 | -7.34 | 3 | -3.84 | 3 | -2.05 | 213 | 13.82 | |
| CLOCK | D4S3000 | nAR50 | -4.56 | 2 | -1.7 | 2 | -0.63 | 835 | 71.58 |
| D4S1569 | nAD50 | -9.32 | 2 | -4.37 | 2 | -2.43 | 3236 | 74.52 | |
| ARNTL | D11S4116 | nAR50 | -6.63 | 3 | -3.49 | 3 | -1.93 | 349 | 22.05 |
| D11S4170 | nAR50 | -5.64 | 2 | -2.09 | 2 | -0.69 | 1027 | 22.90 | |
| CSNK1 ε | D22S283 | nAD50 | 0.07 | 2 | 1.92 | 2 | 2.22 | 1910 | 42.07 |
| D22S423 | bAR50 | -8.16 | 2 | -3.28 | 2 | -1.62 | 1668 | 49.14 | |
| DBP | D19S606 | bAR50 | -3.48 | 6 | -1.26 | 6 | -0.18 | 1160 | 75.60 |
| D19S867 | bAR50 | -6.67 | 6 | -3.8 | 3 | -2.14 | 1398 | 82.03 |
For the association study, we have identified 51 informative SNPs from public databases (dbSNP: http://www.ncbi.nlm.nih.gov/SNP/) and Celera (http://myscience.appliedbiosystems. com) (Supplemental Table). Preference was given to markers associated with disease phenotypes, nonsynonymous coding SNPs, and markers with minor allele frequencies > 0.1 in Caucasians. The Masscode System (Qiagen Genomics), using cleavable mass spectrometry tags in multiplex reactions (Kokoris et al., 2000) was employed to genotype 17 SNPs (rs1017168, rs10462024, rs1056560, rs1801260, rs2253820, rs228697, rs2304670, rs3789327, rs3809236, rs3809237, rs7684810, rs8192433, rs8192436, rs8192439, rs8192440, rs8192441, rs934945). All other SNPs and rs2253820 (repeated for quality control) were genotyped using TaqMan allele specific SNP genotyping assays (Applied Biosystems) using MGB probes according to the manufacturer’s protocols for the assays-on-demand and assays-by-design features. 5 μl reactions were run in 384 plates on an MJR cycler. End point reads were done on an ABI 7900. In addition, a 54 bp variable number of tandem repeat (VNTR) with 4 or 5 repeats in exon 18 of PER3 (Ebisawa et al., 2001) was genotyped with the method described above for microsatellite D11S4170 (PCR primers: F: AGGCAACAATGGCAGTGAG, R: AATGTCTGGCATTGGA GTTTG).
Statistical analysis
Deviation from Hardy-Weinberg equilibrium (HWE) of alleles at all 68 loci (16 microsatellites, 51 SNPs, and 1 VNTR) were tested among unrelated subjects using the computer program CERVUS 2.0 (http://helios.bto.ed.ac.uk/evolgen/cervus/cervus.html).
For the linkage study, we used CRI-MAP 2.4 (Phil Green, http://compgen.rutgers.edu /multimap/crimap/index.html) to detect microsatellite genotyping errors (option ‘chrompic’). Parametric two-point linkage analyses were performed using LINKAGE (http://linkage.rockefeller.edu). We modeled BPAD in three ways: 1) as an autosomal dominant disease caused by a high penetrance allele (AD85), 2) as an autosomal dominant disease caused by alleles with medium penetrance (AD50), and 3) as an autosomal recessive disease caused by medium penetrance alleles (AR50) for both a broad (bipolar I + bipolar II + schizoaffective-bipolar type + recurrent major depression) and a narrow (bipolar I + bipolar II + schizoaffective-bipolar type) diagnostic model (Kelsoe et al., 2001). We assumed variable, age-dependant penetrance by using an age-of-onset curve with minimal risk below age 15, and with maximum penetrance at age 40. Penetrance and disease allele frequencies were adjusted for each genetic model to yield an approximate 5% phenocopy rate and disease prevalences of 2% for the broad and 1% for the narrow diagnostic model.
For the association study, SNP genotype quality was assured by the percentage of calls (>90%), the presence of Hardy-Weinberg equilibrium, and high cluster score values (samples which plotted more than 1/3 of the distance toward an adjacent cluster were considered not callable). In addition, marker rs2253820 was genotyped with both methods and showed a 98.6% concordance (i.e., the genotypes were scored identically). Mendelian inheritance as well as linkage disequilibrium (LD) structure were analyzed with HAPLOVIEW 3.1 (http://www.broad.mit.edu/mpg/haploview/index.php).
Because many of the 52 markers analyzed had alleles that were in LD, we calculated the “effective” number of independent markers using the method described by Nyholt (2004). The effective number of markers = 42.22 was then used to guide a Bonferroni correction for multiple comparisons in each single marker analysis as well as in power calculations. Power for the TDT analyses was calculated using Purcell et al.’s (2003) program for discrete traits assuming 42.22 effective markers for multiple comparisons corrected type I error rates, as well as perfect linkage disequilibrium (LD) between marker and disease alleles, and an additive allele model with a disease prevalence of 1%.
Associations between the 52 markers and BPAD were evaluated with a transmission disequilibrium test (TDT) (Spielman et al., 1993) as implemented in the module TDTPHASE of the UNPHASED v. 2.403 software package (Dudbridge, 2003). TDTPHASE analyzes single SNPs as well as haplotypes from unphased genotype data, and it performs permutation tests to assess significance by reassigning the transmitted and untransmitted labels. We used confirmed haplotypes only with frequencies >0.01 (rare haplotypes were dropped) and a robust permutation analysis involving 10 random restarts and 1000 random permutations. The analyses were performed with three different settings: Setting 1 included only phase-certain haplotypes; Setting 2 included phase-certain and uncertain haplotypes (i.e., the expectation-maximization (E-M) algorithm was used to account for haplotype ambiguities); and Setting 3 also included the estimation of missing parental genotypes (by looping over allele values which were consistent with the data) in addition to uncertain haplotypes. In single marker analyses, nominal P values (PN) were corrected with a Bonferroni correction assuming 42.22 effective markers (PC), e.g., only PN values <.0012 were considered statistically significant in the single marker analyses.
Haplotype-based tests were performed to increase the power of the association study for the nine genes for which multiple markers were available. We used the same three settings in TDTPHASE (S1 — S3) and a sliding window approach similar to that described by (Lin et al., 2004), whereby adjacent markers in the same gene were grouped into sliding windows of two, three, etc. up to a window including all markers available for the gene. To correct for multiple comparisons in haplotype analyses, we considered all settings with a nominal PN<0.05 and performed global permutation tests per gene to correct for multiple testing of haplotypes and markers. These global gene-wise significances (PG) were then corrected via a Bonferroni adjustment for the nine genes which could be analyzed in this way (PGC).
Results
Linkage analysis
16 microsatellite markers, with two in close proximity to each of eight circadian genes, were genotyped in 356 subjects from 52 bipolar families. All markers were highly polymorphic (i.e., heterozygosity >0.68). Observed allele distributions did not deviate from HWE for 13 markers, but markers D11S1785, D17S1353, and D11S4116 showed significant deviations, with slightly higher observed than expected heterozygosities (p<0.01).
With the exception of D22S283, no positive LOD scores were obtained under any of the six models for the overall marker vs. disease analysis (Table II). A maximum LOD of 2.22, which is suggestive for linkage, was obtained for D22S283 at θ = 0.1 for the narrow diagnostic and AD50 genetic model. Positive findings for linkage close to CSNK1ε have previously been reported in a subset of 20 families (Kelsoe et al., 2001).
Association analyses
Association of the 52 markers in 10 different circadian genes to bipolar disorder was analyzed with a linkage disequilibrium test among members of 159 affected families. The power of this study was calculated for 150 triads and 42.22 effective markers under an additive model at a disease prevalence of 1% in the general population and assuming that the candidate gene locus was the disease locus. In this setting, our study power was >80% to detect high-risk alleles at frequencies of 0.1 and 0.2 with genotype relative risks of 2.95 and 2.55, respectively.
Initially, single-marker TDT analyses were performed for 52 markers in 3 settings (data not shown). We did not find significant associations to bipolar disorder (PN>0.05 in all 156 tests; a corrected PC <.0012 would be considered significant).
A subsequent sliding-window haplotype analysis including 132 tests (combinations of SNPs analyzed together as haplotypes) and three different test settings (i.e., 396 total tests) showed 29 nominally significant associations (data not shown), of which eight were still significant within each gene when corrected for multiple testing of haplotypes and markers with the global permutation tests (PG) (Table III). However, none of these associations was significant when Bonferroni-corrections (PGC) were made for the nine genes included in these analyses.
TABLE III.
Haplotype association analysis for the most significant haplotypes for the two genes ARNTL and PER3
| Gene | W | Setting | SNPs | Haplotype | T | NT | %T | P haplo | LRS | PN | PG | PGC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARNTL | 2 | 2 | 4-5 | A.T | 8.29 | 30.3 | 21.49 | 0.0005464 | 13.03 | 0.00457 | 0.02597 | 0.234 |
| 2 | 3 | 4-5 | A.T | 11.67 | 38.29 | 23.36 | 0.001395 | 13.43 | 0.00379 | 0.02498 | 0.225 | |
| PER3 | 4 | 1 | 3-6 | C.C.A.4 | 25 | 52 | 32.47 | 0.002118 | 13.91 | 0.00759 | 0.03197 | 0.288 |
| C.C.G.4 | 21 | 9 | 70.00 | 0.031 | ||||||||
| 5 | 1 | 2-6 | T.C.C.A.4 | 20 | 43 | 31.75 | 0.003858 | 15.88 | 0.0144 | 0.04895 | 0.441 | |
| C.C.C.G.4 | 21 | 7 | 75.00 | 0.008638 | ||||||||
| 5 | 2 | 2-6 | T.C.C.A.4 | 58 | 84.02 | 40.84 | 0.01645 | 16.92 | 0.00958 | 0.03097 | 0.279 | |
| C.C.C.G.4 | 29 | 13 | 69.05 | 0.01037 | ||||||||
| C.T.C.A.5 | 28 | 13 | 68.29 | 0.01517 | ||||||||
| 6 | 1 | 1-6 | C.T.C.C.A.4 | 20 | 43 | 31.75 | 0.003858 | 16.9 | 0.00965 | 0.01898 | 0.171 | |
| C.C.C.C.G.4 | 21 | 7 | 75.00 | 0.008638 | ||||||||
| T.C.T.C.A.5 | 17 | 10 | 62.96 | 0.1755 | ||||||||
| 6 | 2 | 1-6 | C.T.C.C.A.4 | 57 | 84.03 | 40.42 | 0.01241 | 17.36 | 0.00804 | 0.00799 | 0.072 | |
| C.C.C.C.G.4 | 29 | 13 | 69.05 | 0.01036 | ||||||||
| T.C.T.C.A.5 | 28 | 13 | 68.29 | 0.01516 | ||||||||
| 6 | 3 | 1-6 | C.T.C.C.A.4 | 64.89 | 93.33 | 41.01 | 0.0148 | 14.9 | 0.02103 | 0.03596 | 0.324 | |
| C.C.C.C.G.4 | 30 | 15.78 | 65.53 | 0.03067 | ||||||||
| T.C.T.C.A.5 | 29 | 15 | 65.91 | 0.0293 |
w = window of SNPs, setting = TDT analysis method (see Methods), T = number of alleles transmitted, NT = number of alleles untransmitted, %T = percentage of alleles transmitted, Phaplo = p-value associated with the haplotype, PN = nominal p-value for the setting based on chi-square test, PG = global permutated p-value for each gene, PGC = Bonferroni corrected p-value for 9 genes. Note: fractional numbers of transmitted and untransmitted alleles in settings 2 and 3, respectively result from the inclusion of uncertain haplotypes (i.e., the expectation-maximization (E-M) algorithm was used to account for haplotype ambiguities).
Suggestive evidence for association was found in the gene ARNTL for an undertransmitted single haplotype comprised of the intronic markers 4 (rs3789327) and 5 (rs2278749) as part of the two-marker window analysis with settings 2 (PG = 0.02597) and 3 (PG = 0.02498), respectively. The gene and pairwise LD structure of the five SNPs is shown in Figure 1. No significant LD exists between SNPs 4 and 5 (D’ = 0.4). Interestingly, the only marker in this study that showed significant deviation from HWE was rs3789327 (observed heterozygosity (HO) = 0.646, expected heterozygosity (HE) = 0.499, p<0.001 in 144 affected probands). However, no deviation from HWE was found in 87 unaffected individuals (HO = 0.46, HE = 0.50, p>0.05).
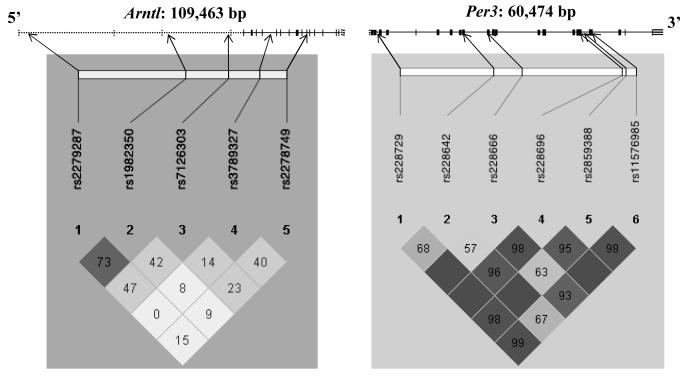
Fig. 1.
Map of the ARNTL and PER3 genes, indicating untranslated (dashed lines) and coding regions with exon and intron boundaries, and position and LD structure of the markers used in this study. Pair-wise D’ values and its confidence (i.e., LOD: dark shading = high LOD) are shown for all marker pairs.
The strongest evidence for association was found for the PER3 gene. We genotyped 6 markers in a 44315 bp region. Suggestive evidence for association was found in the 4, 5, and 6 marker window analyses in all three analysis settings, with one under-transmitted haplotype and one to two over-transmitted haplotypes (Table III). The strongest evidence was found on haplotypes including all six markers (PG = 0.00799) and was almost reaching Bonferronicorrected significance (PGC = 0.072). The 6-marker window includes four intronic markers (rs228729, rs228642, rs228666, and rs2859388), a non-synonymous SNP in exon 17 (rs228697) and the 54-bp length polymorphism in exon 18, all in strong LD with each other (Fig. 1).
Discussion
This study confirmed evidence for linkage of BPAD to the region of CSNK1ε (LOD of 2.22), consistent with a previous report of our group (Kelsoe et al., 2001). However, this linkage region on 22q spans 32 cM and includes hundreds of identified genes, and in our current study showed no evidence for association of six CSNK1ε markers to BPAD. CSNK1ε has roles in both the degradation of PER proteins through phosphorylation and in their transport to the nucleus. It also autoinhibits its kinase activity by autophosphorylation of its carboxyl-terminal extensions. A mutation in hamsters that decreases kinase activity causes a short-period tau phenotype (Lowrey and Takahashi, 2000). A very recent study found a human functional genetic variant to be highly associated with circadian rhythm sleep disorders (Takano et al., 2004). The marker coverage for CSNK1ε in the present association study (i.e., six intronic SNPs with mostly low LD structure) was clearly insufficient to exclude a role of this gene in BPAD. Our group is currently investigating the recently detected functional mutation and other markers in this region.
No additional support for linkage of BPAD families to the other seven circadian clock genes was found. As reported previously (Shaw et al., 2003), these 52 pedigrees have 74% and 61% power to detect with a lod score >3 (under a dominant and recessive model, respectively, and assuming 25% heterogeneity), and we can therefore not exclude a modest effect of the genes reported here and in a previous study including CRY1 (Nievergelt et al., 2005).
Of the 52 polymorphisms studied in 10 circadian clock genes, none was individually associated with BPAD. We calculated that our sample had reasonable power to detect a common risk variant, but would be insufficient to detect a susceptibility locus exerting a minor effect.
To make optimal use of our data, we also subjected haplotypes involving sliding windows of adjacent loci to TDTs. However, adjacent windows are correlated and have to be corrected for multiple comparisons. Therefore, we used permutation tests to adjust the nominal p-values to correct for multiple testing of haplotypes and markers for each gene (Purcell et al., 2003). We then used a Bonferroni correction that adjusted p-values for the nine genes analyzed. Similar methods to deal with these issues have recently been described by (Lin et al., 2004) and require further investigation.
Haplotype analyses suggested two candidate genes deserved further studies. One haplotype in the ARNTL gene and several haplotypes in PER3 were significantly associated from the permutation tests (PG=0.025 and 0.008, respectively), and the most suggestive haplotypes in PER3 showed borderline significance of PGC=0.07 when Bonferroni adjusted.
ARNTL is a key element of the positive feedback loop of the molecular circadian oscillator. It heterodimerizes with CLOCK and NPAS2 and may bind in larger complexes with the PER and CRY proteins, the latter inhibiting the stimulation of gene transcription which occurs when ARNTL binds to E-boxes in the promoters of several circadian genes (Hosoda et al., 2004). We found suggestive evidence for association of a haplotype comprised of two intronic SNPs (rs3789327 and rs2278749, D’=0.4), 12.56 KB apart and surrounding a region of six exons. This haplotype is preferentially non-transmitted and, if it is real, may represent a protective factor. A potential role of rs3789327 (or more likely a functional mutation elsewhere in this region that is in LD with this marker) was further implicated by significant deviation from HWE in affected probands, but not in unaffected individuals. Association of BPAD to a region 5′ to our haplotype has previously been reported (Mansour et al., 2003). We included two of the SNPs previously reported to be associated (rs2279287 and rs1982350) in our study, but could not reproduce preliminary findings of Mansour et al. (2003).
PER3, a negative regulator of the feedback loop, has been found to be associated with DSPS (Ebisawa et al., 2001; Archer et al., 2003; Pereira et al., 2005), and may be implicated in phenotypic aspects of bipolar disorder, such as antidepressant response (e.g., report of Ploia et al., 2004). In our study, haplotypes including windows of 4 to 6 adjacent SNPs that are in strong LD with each other showed suggestive association to BPAD. These haplotypes covered most of the 21 exons spanning the >60 KB gene. They included the interesting region of amino acid similarity to the CSNK1ε binding domain of PER1 and PER2, in which a mutation in PER2 has been found to be associated with familial advanced sleep phase syndrome (Toh et al., 2001), a non-synonymous mutation in exon 15 (V639G, rs10462020) implicated in the pathogenesis of DSPS in Japan (Ebisawa et al., 2001) and diurnal preference (Johansson et al., 2003), and a 54-bp length polymorphism in exon 18 with evidence for association to diurnal preference (Archer et al., 2003; Pereira et al., 2005) and breast cancer (Zhu et al., 2005). The structure of the PER proteins is complex and includes casein-kinase binding sites, phosphorylation sites, PAS domains, etc. (Ebisawa et al., 2001; Travnickova-Bendova et al., 2002) and the molecular basis for the clock-relevant functions is not yet fully understood (but see (Lee et al., 2004)).
Further studies are necessary to identify the functional mutation/s in PER3 associated with circadian and affective phenotypes. There is likewise growing evidence that depression (whether unipolar or bipolar or seasonal) may sometimes be characterized by circadian phase delay, suggesting possible overlap of susceptibility factors between BPAD and DSPS (e.g., Drennan et al., 1991; Chelminski et al., 1999; Johansson et al., 2003).
Although a much-studied SNP in CLOCK (rs1801260) has not been found to be associated with the occurrence of unipolar or bipolar illness (Desan et al., 2000; Serretti et al., 2003a) and in our data, this polymorphism may be related to the presence of insomnia in affective phenotypes (Serretti et al., 2003a), and to bipolar illness recurrence rate and treatment response (Benedetti et al., 2003; Serretti et al., 2003b). Thus, further study of CLOCK polymorphisms and bipolar phenotypes is indicated.
It is likely that BPAD is influenced by several susceptibility factors, no one of which accounts entirely for the disorder even in a single family. This heterogeneity may be one reason why many reports of loci linked to BPAD have been difficult to replicate. Also, since many of the circadian proteins heterodimerize or combine in larger complexes, and paralogues have overlapping functions, gene-gene interactions are likely.
No findings in our data met conservative statistical criteria that account for multiple testing. However, given the inter-marker distance and the SNP-SNP LD, it is likely that an insufficient number of SNPs was used to interrogate these genes. We believe that further exploration of association of BPAD with polymorphisms in circadian genes would be promising, but will require both larger samples and examination of more polymorphisms to adequately test for association. Of the circadian system genes examined so far, PER3, ARNTL, CLOCK and CSNK1ε appear the most likely to yield positive results. In addition, it is possible that BPAD association will be found with circadian system genes which have not yet been explored.
Supplementary Material
Acknowledgements
We are grateful to the family members who participated in this study, without whom it would not have been possible. CMN has been supported by a grant from the Swiss National Science Foundation (823A-061200) and HL071123, DFK by grants from NIH (HL071123, AG15763, AG12364, and HL61280), NJS by grants from NIH (HL054998-09, HL064777-06, HL069758-03, HLMH065571-02, HL074730-02, MH059567-05A2, MH068503-01A1, HL070137-01A1, and AG023122-01), TBB by MH067959, and JRK by grants from NIH (MH47612, MH59567, MH68503, DA13769), the Department of Veterans Affairs and by the UCSD General Clinical Research Center (M01 RR00827). JRK is a founder and holds equity in Psynomics, Inc. We would like to thank Tatyana Shekhtman, Zofi Mroczkowski-Parker and Meghan Alexander for technical assistance, and Sarah Shaw for statistical assistance. Data and biomaterials were collected in four projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1991-98, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, UO1 MH46282, John Nurnberger, M.D., Ph.D., Marvin Miller, M.D., and Elizabeth Bowman, M.D.; Washington University, St. Louis, MO, UO1 MH46280, Theodore Reich, M.D., Allison Goate, Ph.D., and John Rice, Ph.D.; Johns Hopkins University, Baltimore, MD UO1 MH46274, J. Raymond DePaulo, Jr., M.D., Sylvia Simpson, M.D., MPH, and Cohn Stine, Ph.D.; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, Elliot Gershon, M.D., Diane Kazuba, B.A., and Elizabeth Maxwell, M.S.W.
References
- Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev. 2003;13:271–277. doi: 10.1016/s0959-437x(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, von Schantz M. A length polymorphism in the circadian clock gene PER3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- Benca RM, Obermeyer WH, Thisted RA, Gillin JC. Sleep and psychiatric disorders. A meta-analysis. Arch Gen Psychiatry. 1992;49:651–668. doi: 10.1001/archpsyc.1992.01820080059010.discussion 669-670.
- Benedetti F, Bernasconi A, Lorenzi C, Pontiggia A, Serretti A, Colombo C, Smeraldi E. A single nucleotide polymorphism in glycogen synthase kinase 3-beta promoter gene influences onset of illness in patients affected by bipolar disorder. Neurosci Lett. 2004;355:37–40. doi: 10.1016/j.neulet.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, Smeraldi E. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet. 2003;123B:23–26. doi: 10.1002/ajmg.b.20038. [DOI] [PubMed] [Google Scholar]
- Bhat RV, Budd SL. GSK3beta signalling: casting a wide net in Alzheimer’s disease. Neurosignals. 2002;11:251–261. doi: 10.1159/000067423. [DOI] [PubMed] [Google Scholar]
- Bunney WE, Bunney BG. Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression. Neuropsychopharmacology. 2000;22:335–345. doi: 10.1016/S0893-133X(99)00145-1. [DOI] [PubMed] [Google Scholar]
- Chelminski I, Ferraro FR, Petros TV, Plaud JJ. An analysis of the “eveningnessmorningness” dimension in “depressive” college students. J Affect Disord. 1999;52:19–29. doi: 10.1016/s0165-0327(98)00051-2. [DOI] [PubMed] [Google Scholar]
- Desan PH, Oren DA, Malison R, Price LH, Rosenbaum J, Smoller J, Charney DS, Gelernter J. Genetic polymorphism at the CLOCK gene locus and major depression. Am J Med Genet. 2000;96:418–421. doi: 10.1002/1096-8628(20000612)96:3<418::aid-ajmg34>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Drennan MD, Klauber MR, Kripke DF, Goyette LM. The effects of depression and age on the Horne-Ostberg morningness-eveningness score. J Affect Disord. 1991;23:93–98. doi: 10.1016/0165-0327(91)90096-b. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Uchiyama M, Kajimura N, Mishima K, Kamei Y, Katoh M, Watanabe T, Sekimoto M, Shibui K, Kim K, Kudo Y, Ozeki Y, Sugishita M, Toyoshima R, Inoue Y, Yamada N, Nagase T, Ozaki N, Ohara O, Ishida N, Okawa M, Takahashi K, Yamauchi T. Association of structural polymorphisms in the human PERIOD3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A, Goldman D. Genetic influences. In: Partonen T, Magnusson A, editors. Seasonal Affective Disorder: Practice and Research. Oxford University Press; Oxford: 2001. pp. 259–266. [Google Scholar]
- Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, Schibler U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004a;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004b;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gjessing R. Contribution to the Somatology of Periodic Catatonia. Pergamon; Oxford: 1976. [Google Scholar]
- Gould TD, Manji HK. The Wnt signaling pathway in bipolar disorder. Neuroscientist. 2002;8:497–511. doi: 10.1177/107385802237176. [DOI] [PubMed] [Google Scholar]
- Halberg F. Physiologic considerations underlying rhythmometry, with special reference to emotional illness. Paper presented at Symposium on Biological Cycles and Psychiatry Symposium Bell-Air III; Geneva. 1967. [Google Scholar]
- Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci. 2004;21:359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- Hosoda H, Motohashi J, Kato H, Masushige S, Kida S. A BMAL1 mutant with arginine 91 substituted with alanine acts as a dominant negative inhibitor. Gene. 2004;338:235–241. doi: 10.1016/j.gene.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppa T, Lichtermann D, Praschak-Rieder N, Neumeister A, Nilsson LG, Kasper S, Peltonen L, Adolfsson R, Schalling M, Partonen T. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology. 2003;28:734–739. doi: 10.1038/sj.npp.1300121. [DOI] [PubMed] [Google Scholar]
- Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, Mignot E. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- Kelsoe JR. Arguments for the genetic basis of the bipolar spectrum. J Affect Disord. 2003;73:183–197. doi: 10.1016/s0165-0327(02)00323-3. [DOI] [PubMed] [Google Scholar]
- Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H. A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci U S A. 2001;98:585–590. doi: 10.1073/pnas.011358498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoris M, Dix K, Moynihan K, Mathis J, Erwin B, Grass P, Hines B, Duesterhoeft A. High-throughput SNP genotyping with the Masscode system. Mol Diagn. 2000;5:329–340. doi: 10.1007/BF03262094. [DOI] [PubMed] [Google Scholar]
- Kripke DF, Mullaney DJ, Atkinson M, Wolf S. Circadian rhythm disorders in manic-depressives. Biol Psychiatry. 1978;13:335–351. [PubMed] [Google Scholar]
- Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- Lorenzi C, Serretti A, Benedetti F, Tubazio V, Ploia C, Colombo C, De Ronchi D, Smeraldi E. Human period 3 gene influences SSRIs antidepressant activity in mood disorders. Am J Med Genet. 2003;122B:124. [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system: Photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu Rev Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- Mansour H, Wood J, Devlin B, Logue T, Chowdari K, Kupfer D. Family based and case-control association analysis of circadian gene polymorphisms in bipolar I disorder. Am.J.Hum.Genet. 2003;73:504. [Google Scholar]
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- Mitterauer B. CLOCK genes, feedback loops and their possible role in the etiology of bipolar disorders: an integrative model. Med Hypotheses. 2000;55:155–159. doi: 10.1054/mehy.1999.1039. [DOI] [PubMed] [Google Scholar]
- Murray C, Lopez A. The Global Burden of Disease. Harvard University Press; Cambridge: 1996. [Google Scholar]
- Niculescu AB, 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Kripke DF, Remick RA, Sadovnick AD, McElroy SL, Keck PE, Jr., Kelsoe JR. Examination of the clock gene Cryptochrome 1 in bipolar disorder: Mutational analysis and absence of evidence for linkage or association. Psychiatric Genetics. 2005;15:45–52. doi: 10.1097/00041444-200503000-00008. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partonen T, Magnusson A. Seasonal Affective Disorder. Oxford University Press; Oxford: 2001. [Google Scholar]
- Pereira DS, Tufik S, Louzada FM, Benedito-Silva AA, Lopez AR, Lemos NA, Korczak AL, D’Almeida V, Pedrazzoli M. Association of the length polymorphism in the human PER3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- Ploia C, Serretti A, Benedetti F, Fontana V, Colombo C, Catalano M, Smaraldi E. Mood disorders and antidepressants treatment: haplotype analysis of human period 3 gene. Am J Med Genet. 2004;130B:34. [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Serretti A, Benedetti F, Mandelli L, Lorenzi C, Pirovano A, Colombo C, Smeraldi E. Genetic dissection of psychopathological symptoms: insomnia in mood disorders and CLOCK gene polymorphism. Am J Med Genet. 2003a;121B:35–38. doi: 10.1002/ajmg.b.20053. [DOI] [PubMed] [Google Scholar]
- Serretti A, Cusin C, Lorenzi C, Benedetti F, Pirovano A, Arnoldi A, Zanardi R, Colombo C, Smeraldi E. Genetic predictors of antidepressants efficacy: the state of the art. Am J Med Genet. 2003b;122B:36. [Google Scholar]
- Shaw SH, Mroczkowski-Parker Z, Shekhtman T, Alexander M, Remick RA, Sadovnick AD, McElroy SL, Keck PE, Kelsoe JR. Linkage of a bipolar disorder susceptibility locus to human chromosome 13q32 in a new pedigree series. Mol Psychiatry. 2003;8:558–564. doi: 10.1038/sj.mp.4001267. [DOI] [PubMed] [Google Scholar]
- Shiino Y, Nakajima S, Ozeki Y, Isono T, Yamada N. Mutation screening of the human period 2 gene in bipolar disorder. Neurosci Lett. 2003;338:82–84. doi: 10.1016/s0304-3940(02)01290-9. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Takano A, Uchiyama M, Kajimura N, Mishima K, Inoue Y, Kamei Y, Kitajima T, Shibui K, Katoh M, Watanabe T, Hashimotodani Y, Nakajima T, Ozeki Y, Hori T, Yamada N, Toyoshima R, Ozaki N, Okawa M, Nagai K, Takahashi K, Isojima Y, Yamauchi T, Ebisawa T. A Missense Variation in Human Casein Kinase I Epsilon Gene that Induces Functional Alteration and Shows an Inverse Association with Circadian Rhythm Sleep Disorders. Neuropsychopharmacology. 2004;29(10):1901–1909. doi: 10.1038/sj.npp.1300503. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPER2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U.S.A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuunainen A, Kripke DF, Endo T. Light therapy for non-seasonal depression. Cochrane Database Syst Rev:CD004050. 2004 doi: 10.1002/14651858.CD004050.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. PERIOD3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–270. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.