Abstract
The human oral pathogen Porphyromonas gingivalis colonizes the gingival crevice and invades gingival epithelial cells. Multi-dimensional capillary HPLC coupled with tandem mass spectrometry and 2D gel electrophoresis were used to analyze the proteome of P. gingivalis as it adapts to a set of experimental conditions designed to reflect important features of an epithelial cell environment. 1014 proteins (46% of the total theoretical proteome) were identified in four independent analyses. 479 of these proteins showed evidence of differential expression after exposure of P. gingivalis to either conditioned epithelial cell growth medium or control conditions: i.e. they were only detected under one set of conditions. Moreover, 276 genes annotated as hypothetical were found to encode expressed proteins. Among the proteins upregulated in the presence of epithelial cell components were a homolog of the internalin proteins of Listeria monocytogenes and subunits of the ATP-dependent Clp protease complex. Insertional inactivation of clpP, encoding the Clp proteolytic subunit, resulted in an approximately 50% reduction in invasion of P. gingivalis. These results suggest that adaptation to an epithelial cell environment induces a major shift in the expressed proteome of the organism. Furthermore, ClpP, that is upregulated in this environment, is required for optimal invasive activity of P. gingivalis.
Keywords: Porphyromonas gingivalis, proteomics, gingival epithelial cells, invasion, protein expression, pathogenesis
1 Introduction
Porphyromonas gingivalis is a gram-negative anaerobe that resides within the subgingival crevice of the oral cavity. The organism is a predominant pathogen in severe and chronic manifestations of periodontitis, a disease characterized by destruction of the periodontal tissues and, ultimately, exfoliation of the teeth [1]. Periodontal diseases are the major cause of tooth loss in developed countries and in the USA these disease have been reported to afflict 14% of adults aged 45–54, and 23% of those aged 65–74 years (Oral Health in America, a report of the Surgeon General, U. S. Public Health Service, 2000). In addition to its pathogenic role in the mouth, P. gingivalis has been associated with systemic conditions such as coronary artery disease and preterm delivery of low birth weight infants [2]. P. gingivalis can invade and survive within both gingival epithelial cells and coronary artery endothelial cells, a property considered important in colonization, persistence and tissue destruction [1, 3, 4]. Invasion of gingival epithelial cells occurs rapidly (under 20 min), and intracellularly P. gingivalis cells remain viable and accumulate in high numbers in the perinuclear area. The epithelial cells undergo morphological changes after prolonged (24 h) association with intracellular P. gingivalis; however they do not undergo apoptosis and maintain physiologic integrity for extended periods [5, 6, 7, 8]. Thus, both bacteria and host cells adapt to their close association, leading to the hypothesis that P. gingivalis can regulate gene and protein expression according to the prevailing epithelial cell environment. The genome of P. gingivalis has been sequenced [9] and annotated (www.tigr.org; www.stdgen.lanl.gov/oragen) and DNA microarray approaches to P. gingivalis gene expression are now in progress. However, as cellular regulation can occur posttranscriptionally, and as proteins are, in the majority of cases, the effector molecules of cells, there is an increasing realization that protein expression data are required to fully comprehend biological processes and systems [10, 11, 12].
A variety of methodologies are currently extant to separate and identify bacterial proteins. The most commonly used separation method, 2D gel electrophoresis, has many limitations and cannot be relied upon to provide a complete inventory of expressed proteins, and is more useful for the initial detection of posttranslational modifications [10, 12]. Until recently, global protein based expression studies have been more difficult, less complete, less sensitive and have a lower throughput than RNA-based ones. The application of Multidimensional Protein Identification Technology (MudPIT) type approaches [13, 14] to proteomics is a significant advancement in this regard, and we have previously demonstrated that the MudPIT method can be modified for the identification of P. gingivalis proteins [15]. Protein separation by multi-dimensional HPLC can provide substantial coverage of the proteome, and reproducibility is enhanced by the detection of several peptide fragments of a protein. Reconstructed protein arrays are assembled computationally from genome annotations and experimental peptide sequence data.
P. gingivalis gene expression may vary during any stage during the association with epithelial cells including: bacterial localization, attachment, induction of uptake, and intracellular survival. A proteomic approach, however, is particularly suitable for the study of the initial adaptation of P. gingivalis to an epithelial cell environment without the confounding presence of high levels and numbers of epithelial cell proteins. Many bacteria are known to modulate protein expression in response to eukaryotic cell components released into culture media. For example, macrophage conditioned medium induces exposure of the DotH and DotO proteins on the surface of Legionella pneumophila [16]. In addition, Campylobacter jejuni secretes a distinct set of proteins when incubated in intestinal cell conditioned medium [17]. P. gingivalis cells were, therefore, reacted with epithelial cell culture supernatant (conditioned medium, cKGM) and protein expression compared with P. gingivalis cells in KGM growth medium that had not been used for cell culture. While this approach does not address regulation during the later stages of the P. gingivalis-GEC interaction, it has the additional advantage that as P. gingivalis cells do not replicate in epithelial cell culture medium, any changes in protein expression will not be the result of differential growth rate. We are presently engaged in defining the GEC-derived components of the cKGM medium, a task that may not be completed until expressed protein and (or) full length cDNA sequence databases based on the human genome are more complete than what we have access to at present.
The goal of this study was to determine on a global level, the differential protein expression that occurs in P. gingivalis as a result of contact with epithelial cell components. Mutation analysis was used to corroborate a role for one upregulated protein, ClpP, in the invasive process of P. gingivalis.
2 Materials and methods
2.1 Bacteria
P. gingivalis ATCC 33277 was cultured in an anaerobic chamber (85% N2, 10% H2 5% CO2) for 24 h at 37 °C in trypticase soy broth supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml). Bacteria were recovered from mid-log phase by centrifugation at 6000g and 4°C for 10 min.
2.2 Gingival epithelial cells (GEC) and conditioned KGM
Primary cultures of GEC were generated as described previously [6]. Briefly, healthy gingival tissue was obtained after oral surgery, and surface epithelium was separated by overnight incubation with 0.4% dispase. Cells were cultured as monolayers in serum-free keratinocyte growth medium (KGM) (Clonetics, San Diego, CA, USA) at 37°C in 5% CO2. Conditioned medium (cKGM) was collected from 48 h GEC cultures at 80% confluence.
2.3 Extraction and preparation of bacterial proteins
P. gingivalis cells (~1010) were incubated for 18 h in an anaerobic chamber (85% N2, 10% H2 5% CO2) in either cKGM (i.e. medium removed from a 48 h GEC culture) or reference state KGM (i.e. ordinary medium). As the P. gingivalis cells were incubated for 18 h in the media, incubation was anaerobic to avoid oxidative stress on the organism. Cells were pelleted, lysed and digested with Lys-C and trypsin sequentially as described [15]. Briefly, the cell pellet was resuspended and cells were lysed with 0.3% SDS at 100 °C for 3 min in the presence of DNAse and RNAse using a 10-fold dilution of the following stock solution: 1 mg/ml DNAse I, 500 μg/ml RNAse A, 50 mM MgCl2, 50 mM Tris-HCl at pH 7.0. Samples were then frozen in liquid N2 and lyophilized to dry powder. The powder was redissolved in 300 μl of resolubilization solution (7M urea, 200 mM NH4HCO3, 20 mM CaCl2). The proteins were reduced with 5 mM DTT at 37°C for 30 min and then alkylated with 10 mM iodoacetamide at 30°C for 30 min in the dark. Lys-C (5 μg, sequencing grade, Boehringer, USA) was added and incubated at 37°C for 20 h. The mixture was diluted to give a solution of 2M urea, 100 mM NH4HCO3, 5mM CaCl2; and 15 μg of trypsin (sequencing grade, Promega, Madison, WI, USA) was added and incubated at 37°C overnight. The sample was centrifuged at 14000 rpm using a Model 5415C bench top microcentrifuge (Eppendorf, Hamburg, Germany) and the supernatant was concentrated to 200 μl using a vacuum centrifuge (Jouan Model RCT 60, Nantes, France).
2.4 2D gel electrophoresis
18 cm IPG (immobilized pH gradient) strips and 18 × 18 cm × 1 mm second dimension SDS-PAGE gels were prepared and silver stained as described previously [15]. Proteins were identified conventionally by in situ digestion and tandem mass spectrometry followed by database searching.
2.5 Reverse transcriptase-PCR
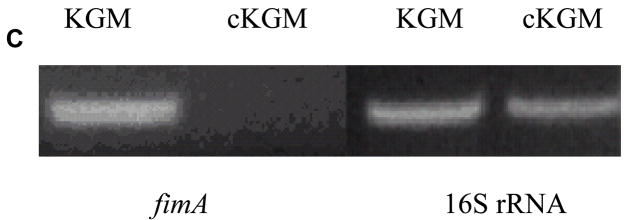
Total RNA was isolated from P. gingivalis exposed to cKGM or KGM for 18h using Totally RNA™ kit (Ambion, Austin, TX, USA) as described by the manufacturer. cDNA was synthesized from the same amount of total RNA isolated from two different conditions. PCR was performed under conditions of 1 min denaturation at 94°C, annealing at 55°C for 1 min followed by 1 min extension at 72°C for 30 cycles. The primers were: for fimA 5′-CGGGATCCCGTGGTATTGAAGACCAGCAAT-3′ and 5′-GGAATTCCAAGTAGCATTCTGACCAACGAG-3′; for 16S rRNA, 5′-TGGGTTTAAAGGGTGCGTAG-3′ and 5′-ACAACCATGCAGCACCTACA-3′;
2.6 Mutant construction
Insertional mutations of clpP and pg0350 were constructed as described previously [18]. Briefly, internal fragments of each gene were amplified by PCR with primers designed based on the DNA sequences in TIGR database, and cloned into pCRII®-TOPO. The cloned fragments were obtained by digesting with BamHI and XbaI, and were cloned into a suicide plasmid, pVA3000 carrying the erythromycin resistance gene cassette ermAM/ermF. These recombinant plasmids were introduced into E. coli S17-1 and conjugated into P. gingivalis. Transconjugants were selected on TSB-blood agar plates supplemented with erythromycin (20 μg/ml) and gentamicin (100 μg/ml). Mutagenesis was confirmed by Southern hybridization.
2.7 Invasion assay
Quantitation of viable P. gingivalis cells recovered intracellularly from GEC was determined by a conventional antibiotic protection assay, as modified for P. gingivalis [6]. P. gingivalis interaction with GEC was under conditions of 5% CO2 at 37°C.
2. 8 Adherence assay
Adherence of P. gingivalis strains to GEC was detected by ELISA. Briefly, GECs cultivated on 96-well plates were fixed with 5% buffered formalin, to prevent P. gingivalis invasion, and reacted with P. gingivalis strains at an MOI 100 in 5% CO2. After incubation for 30 min at 37°C, the cells were washed five times with PBS to removed non-adherent bacteria. Surface bacteria were then immunolabeled with P. gingivalis whole-cell antibodies (1:1000) [8] and horseradish peroxidase conjugated goat anti-rabbit IgG (ICN Biochemicals, Costa Mesa, CA, USA). Adherence of each strain was determined by a colorimetric reaction using 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system for ELISA (Sigma, USA). Wells with uninfected GECs were used as a negative control. Preliminary experiments showed that the ClpP mutant and parental strains exhibited an equivalent degree of reactivity with the antibodies raised to the 33277 parental strain.
2.9 Immunofluorescence microscopy
Immunofluorescence labeling and microscopy were performed essentially as described by Yilmaz et al. [8]. Briefly, GECs cultivated on 4-well chambered cover-glass slides (Nalge-Nunc, Rochester, NY, USA) were infected with P. gingivalis strains at an MOI 100 at 37°C in 5% CO2. for 30 min. The slides were washed four times with PBS containing 0.1% Tween 20 (PBS-T) to remove the non-adherent bacteria. Cells were fixed in 10% neutral buffered formalin, rinsed in PBS, and permeabilized for 15 min by 0.1% Triton X-100 in PBS at room temperature. Samples were incubated for 30 min in blocking solution (PBS-T supplemented with 5% goat serum) to mask non-specific binding sites prior to the fluorescence labeling. Visualization of intracellular bacteria was performed by indirect immunofluorescence microscopy using rabbit polyclonal antiserum to P. gingivalis 33277 (1:2500) and Oregon Green 488 goat anti-rabbit IgG (H+L) (Molecular Probes, Eugene, OR, USA) used at 1: 400. F-actin was also labeled with phalloidin-tetramethylrhodamine β-isothiocyanate (TRITC) (Sigma). The labeled samples were examined using an epifluorescence microscope (Zeiss Axioskope). Single exposure images were captured sequentially using a cooled CCD camera (Qimaging) and saved by Qcapture software v. 1394. Collected image layers were superimposed into a single image using Adobe Photoshop 6.0 software. At least 10 separate fields containing an average of 25 GECs were studied for each strain in triplicate assays.
2.10 3D HPLC combined with data dependent tandem mass spectrometry
Details of our 2D capillary LC/MS instrumentation and protocol, and its deviations from the method as originally published [13] have been reported previously [15]. For the work reported here, approximately 0.5 mg of each whole-cell digest sample was separated into five fractions using a 2.1 mm i.d. reversed-phase HPLC column, and fractions were concentrated to 50 μl. A Magic 2002 HPLC (Michrom BioResources, Auburn CA, USA), an LCQ ion trap mass spectrometer (Thermo Finnigan, San Jose, CA, USA), an in-house built ESI interface and an in-house packed biphasic capillary column (75 μm i.d.) were used for each 2D capillary HPLC tandem MS analysis. 2 μl of each fraction were loaded pneumatically onto the biphasic capillary column. Taking into account an estimated loss of about 50% of the total cellular protein during the initial off-line fractionation, the actual amount of total protein injected onto the SCX resin of the biphasic capillary column for each MS run was on the order of 10 to 100 μg. The biphasic capillary column was not grossly overloaded due to the much larger sample capacity of the SCX resin, approximately a factor of 60, relative to the C18 material. Peptides were first partially eluted from the SCX packing, and were retained on the reverse phase material by ammonium acetate step gradients (0 mM, 10 mM, 25 mM, 50 mM, 100 mM, 250 mM, 500 mM). The peptides were eluted from the reverse phase packing with an acetonitrile gradient with 0.02% (v/v) HFBA, and were electrosprayed into the LCQ for semi-automated data dependent acquisition. The gradients programmed were 2–20% B in 1 min, hold 8 min, 20–45% B in 50 min, hold 4 min, 45-2% B in 2 min and equilibration for 10 min. For elution with 500 mM salt, the gradient continued from 45 to 90% B in 10 min and hold 30 min. The flow rate in the capillary column was 300 nl/min from 0–9 min, 150 nl/min from 10–60 min and 300 nl/min after 60 min. The MS scan range was 400–2000 m/z. After each main beam (MS1) scan, the four most intense signals were selected for collision-induced dissociation (CID, MS2) scans with 2 CID scans per parent ion scan. Default parameters under the Xcalibur 1.2 data acquisition software were used, with the exception of an isolation width of 3.0 m/z units. Dynamic exclusion was activated during all acquisitions.
2.11 Data reduction and bioinformatics
The results reported here are based on four complete analyses of the P. gingivalis proteome under both KGM and cKGM conditions using the capillary HPLC/tandem MS approach, and approximately 20 2D gels. The details regarding the purely qualitative computational reassembly in silico of the P. gingivalis proteins from their respective proteolytic fragments have been published [15]. SEQUEST search results [19] were processed into reconstructed protein arrays using a modified form of the P. gingivalis W83 ORF database from TIGR [9], that included additional sequences (from www.ncbi.nlm.nih.gov) known to be present in strain 33277. The use of a database based on strain W83 for our work with strain 33277 has not presented a significant problem [15, 20]. SEQUEST search results were summarized using DTASelect and Contrast [21] followed by display of the observed ORFs in a physical map of the genome using software developed in-house [15]. At the level of the batch processing of the *.dta files using SEQUEST, no enzyme specificity was indicated in the SEQUEST parameter file. However, at the DTASelect level, only full-tryptic and half-tryptic peptide sequences were retained. It has been our experience that in this type of experiment the high scoring non-tryptic fragments present an unacceptably high rate of false positive matches. A minimum of two unique peptides for a given ORF were required for qualitative identification. A peptide “hit” was retained in the dataset if its Xcorr value [19] was no less than 1.8 for singly charged precursors, 2.5 for doubly charged precursors, and 3.5 for triply charged precursors, its Delta Cn value was no less than 0.08, and it contained at least four amino acids. For results of particular importance with respect to P. gingivalis invasion, e.g. the down regulation observed for FimA in cKGM (see discussion), the peptide mass spectra were also inspected manually via local HTML links embedded in the fully processed dataset. For purposes of calculating semi-quantitative expression ratios for proteins, all MS1 signal intensities for peptides unique for a specific protein, as determined by their MS2 mass spectra, were summed and the total counts divided by the number of peptides used in the calculation. The resulting average signal intensity for a KGM protein was divided by that for the same protein from cKGM conditions in order to yield an estimate of the expression ratio. Such ratios were calculated only if duplicate determinations indicated the data were sufficiently reproducible to detect a two-fold change in expression. We routinely normalize the data employed for ratio calculations in our laboratory, in a manner similar to methods that are employed in the microarray field. Briefly, because a plot of the log transformed expression ratios is symmetric about zero, see Fig. S1 in the supplement, and the average signal intensities were similar under both growth conditions, we took this as evidence no normalization procedure was required for this particular study. Such normalization procedures are discussed more fully and referenced in the caption to Fig. S1. Intact cell optical density measurements and Bradford total protein assays, while less accurate, also supported the observation that we were looking at approximately the same amount of total protein and the same number of cells in each set of growth conditions. In certain cases processed data that were filtered out by the automated system, and hence failed to generate a numerical ratio, nonetheless showed reproducible evidence of differential expression as determined by manual inspection. Thus the final dataset (supplementary material, Table S1) was processed into three categories in terms of data quality: qualitative detection only, semi-quantitative with a calculated expression ratio, and semi-quantitative with a note indicating higher relative protein abundance under either KGM or cKGM conditions.
3 Results and Discussion
3.1 General considerations
In this study we have adapted the 2D capillary HPLC tandem mass spectrometry method to examine global differential protein expression in P. gingivalis in response to released epithelial cell components. Protein sequence data were derived from HPLC separation and of whole proteome enzymatic digests followed by tandem mass spectrometry and database searching of ~250,000 product ion (MS2) spectra, with relative abundance estimates based on observed signal intensities of the parent ions in MS1, as explained in detail under Materials and methods.
3.2 MudPIT analysis of differential expression of proteins in KGM and cKGM
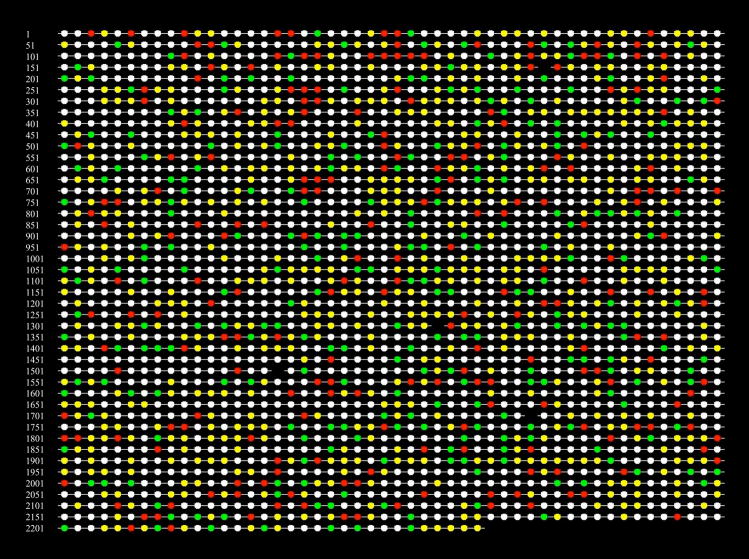
As shown graphically in Fig. 1, peptide mass spectra were mapped to 258 ORFs unique to KGM, 221 unique to cKGM, and 535 common to both conditions. This represents about 46% of the 2224 ORFs in the TIGR database. A complete list of the observed proteins, with expression ratios where it was possible to calculate them (see below), can be found in the supplementary material as Table S1. The total number of ORFs that express protein under any particular set of conditions in P. gingivalis is unknown, but estimates based on the duty cycle limitations of our mass spectrometer suggest a value of around 60% of the ORF database. This level of expression is slightly higher than that reported for Pseudomonas aeruginosa, which has been reported to transcribe approximately 40% of its much larger genome [22].
Figure 1.
Qualitative reconstructed protein array map of protein expression in P. gingivalis 33277 when exposed to cKGM or to KGM conditions (see text). Each spot represents an ORF in the P. gingivalis database. The order of the entries on the map follows the physical map of the genome. ORFs uniquely expressed under cKGM conditions are coded red, those associated only with KGM conditions are green, those common to both conditions are yellow. A portion of those entries coded yellow showed differences in abundance between the two sets of conditions. White indicates an entry in the database for which there was no evidence for expression. Black (gaps) indicates an unused number or an ORF that has been judged not to contain valid sequence information. By ordering the entries according to a physical map of the P. gingivalis genome, it is also possible to use the image to look for regulatory relationships among genes. A complete listing of observed P. gingivalis proteins shown in the reconstructed array image is given in the supplementary material as Table S1. Within our own laboratory, such images are also used as a graphical user interface to the various levels of raw and processed data collected for each ORF.
With the exception of hypothetical proteins, disrupted reading frames, other categories and unknown function; the identified proteins generally account for over 50% of the total predicted proteins in a functional class. The percentage is higher (over 70%) for the most essential functional classes, such as energy metabolism, fatty acid and phospholipid metabolism, protein fate, and protein synthesis.
A portion of the dataset consisted of proteins for which a sufficient number of peptides were recovered to allow reproducible estimates of expression ratios in the absence of isotopic labels [23]. As the number of unique peptides recovered from a particular protein increases, both the qualitative and quantitative reproducibility of the results improve. The distribution of ratios shown in the electronic supplement (see Table S1 and the accompanying frequency histogram, Fig. S1) did not differ significantly from a mean value of 1, and appeared to be approximately lognormal. For the data reported here, ~50% of the qualitative protein identifications were of sufficient quality to yield RSD (relative standard deviation) values of less than 50%, when all MS1 signal intensities observed for peptides mapping to a given protein were averaged. In our analytical scheme MS2 data are used only for qualitative identification and never for estimating expression ratios. Out of the 535 proteins detected qualitatively under both growth conditions, approximately 35 of these showed reproducible evidence of higher expression levels in cKGM, whereas 25 showed evidence of higher expression under the reference condition, KGM. The use of isotopic labeling [14, 24], which would enhance quantitative precision, was avoided in order to maximize qualitative coverage of the proteome. This trade-off is imposed to a large extent by the slow data acquisition rate of our LCQ mass spectrometer relative to the large number of peptides that must be analyzed in a “shotgun” proteomics experiment. Labeling strategies involving stable isotopes typically increase the complexity of the sample when one necessarily must mix extracted proteomes from two defined states together so that the “heavy” and “light “ peptides are analyzed concurrently. More importantly, strategies based on stable isotopic chemical labeling of cysteines, e.g. ICAT [24] are problematic for P. gingivalis for an additional reason, which also impacts qualitative coverage, despite the reduction in peptide mixture complexity inherent in the ICAT method. According to the TIGR annotations, out of 2224 theoretical ORFS, 296 contain no cysteine and 380 contain only one cysteine residue. Hence, if we were to attempt to ICAT label the entire proteome of P. gingivalis, we probably would omit roughly 15% to 30% of the proteome as nondetects. Recent improvements in the speed with which mass spectrometers can acquire peptide collision spectra have largely rendered moot the trade-off between better qualitative coverage (no isotopes used) and better quantitation (use of stable isotopes, e.g. 15N) for organisms with small genomes such as P. gingivalis. Thus, future studies can employ P. gingivalis that has been metabolically labeled, and the analysis can be performed using an LTQ linear ion trap that can acquire three to five times as many useful mass spectra per unit time as was possible under the system used in the work reported here.
Although the true percentage of false positives in the data reported here is unknown at present, several lines of evidence suggest it is quite low. Statistical analysis of the purely qualitative aspect of the dataset suggests we are reasonably well controlled for false positives (Type I error) when the number of high scoring unique peptides for a given ORF is ≥ 3 within a single analysis, even though the number of replicate whole proteome analyses is low compared to what is typical in the transcription microarray field. M. MacCoss has estimated the average reduction in purely random false positive matches using our SEQUEST search conditions to be about a factor of 50 for each peptide used to identify the protein, relative to search strategies that force a fit to tryptic fragments or that report data without cleavage specificity (unpublished observation). A derivation of this estimate is given in the supplementary material. Using this logic, the probability of random false positive protein identifications quickly becomes small as the number of unique peptides identified per ORF increases. Our experience has been that the inherent problem of increased risk of random false positive identifications associated with searching small ORF libraries with large datasets can be minimized by enlarging the database and (or) searching without enzyme specificity, but filtering out non-tryptic sequence assignments at the DTASelect level. We chose not to pursue the first approach in this study due to the impractical computational time required, while using the latter strategy. Data where n = 2 we view with less confidence, but we feel throwing it out would be overly conservative. We do not report protein identifications based on a single unique peptide, regardless of score. The number of replicate non-detects required to adequately control for Type II, or false negative error, is much higher, at least 7 replicates, which is not economically or technically feasible for us at present. In general, Type II error has not been discussed in the proteomics literature, presumably because few labs could afford the time and effort to control it. It is much harder to say with certainty a protein is not present. Users of the data, in our experience, tend not to be much concerned with Type II error. Whether they should be or not depends a great deal on the goals of the study and the potential consequences of such error. Our data collected to date with two model systems, P. gingivalis and Methanococcus maripaludis, suggest that what we are missing is much more of an issue than false positives, i.e. we are essentially uncontrolled at any reasonable confidence level for Type II error when the number of biological replicates is less than 3 or 4. In support of our methods, of the 30 or so most biologically interesting “hits” from the 2D HPLC/MS/MS data discussed in this study, all were subsequently confirmed as true positives by at least one other method.
From the standpoint of gaining insight into P. gingivalis biology, in the majority of cases where differential expression was observed, it was of an “off” or “on” type, with observable signal only in one state or the other but not both. Only in a relatively small number of cases, approximately 60, did we see reproducible quantitative changes for ORFs that were qualitatively detected under both KGM and cKGM conditions. A significant data compression effect is typical of relative abundance measurements acquired under our conditions. Similarly to transcription microarrays, relatively subtle changes in expression ratios may represent much larger changes in the “true” level of expression. Although this relationship is believed to scale nonlinearly, the precise relationship between measured protein relative abundance and true expression levels is unknown. The caveats with respect to the interpretation of these numbers are defined largely by the limitations of current technology and methods. Protein expression has been cited in the literature to have a dynamic range of as high as six or more orders of magnitude [10, 14]. However, the methodology for measurement is currently limited to perhaps two or three orders at best. A qualitative “nondetect” means that at an instrumentally defined threshold value (~40,000 counts of signal intensity in the MS1 spectra) we failed to observe two or more peptides unique for the ORF in question. That threshold is determined by the signal/noise ratio for a parent ion (MS1) that is required in the average case to yield a potentially interpretable collision spectrum (MS2), from which the sequence information is derived, either by means of a database search or de novo. Thus, it is quite possible that certain ORFs are expressed at low levels that we failed to detect under our experimental conditions. Other limiting aspects of the analytical scheme are described below as they apply to classes of proteins that share certain physicochemical properties. In addition, it is formally possible that some changes observed could result from the absence of components from the growth media as a result of GEC metabolism.
3.3 Characteristics of detected proteins
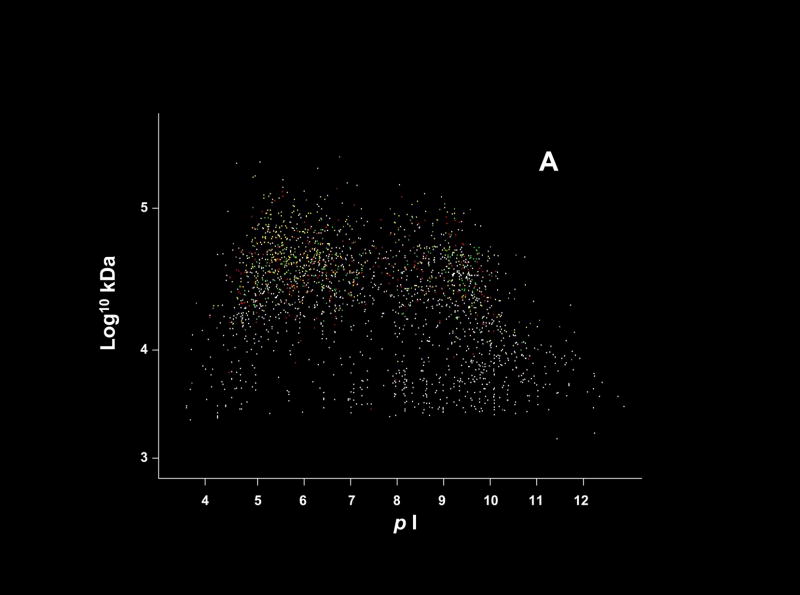
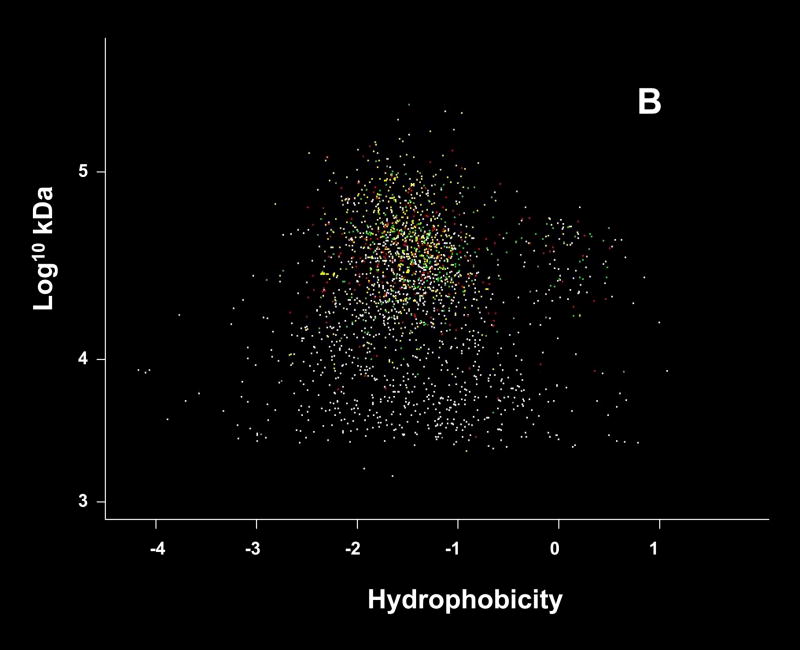
As shown in the plots of isoelectric point or hydrophobicity against molecular weight (Fig. 2), there was no obvious pattern of discrimination in the results based on these two criteria. With respect to hydrophobicity, the lack of discrimination suggested by these data may appear counter-intuitive as the solution chemistries used in our methods were not expected to yield efficient recoveries for membrane proteins [15]. However, upon closer inspection of the ~20 to 30% of the P. gingivalis proteins that are predicted to contain hydrophobic membrane domains, the majority contain regions that are relatively more hydrophilic and thus potentially available for proteolysis and subsequent analysis under our experimental conditions. Our preliminary results (Xia, Q., Zhang, Y., and Hackett, M., unpublished) modeling transmembrane domains in P. gingivalis suggest that the peptides we recover are almost exclusively from these more hydrophilic regions. More complete recovery studies with the methanogenic archaeon Methanococcus maripaludis (Zhang, Y., Xia, Q., Hackett, M., unpublished) also support this interpretation. Thus, protein recovery is sufficient to get a qualitative identification, but coverage on a peptides recovered per protein basis remains poor. This observation has consequences for the quantitative analysis of hydrophobic proteins and the generation of expression ratios. Recoveries of hydrophobic peptides must be improved significantly in order to generate more quantitatively reproducible data.
Figure 2.
The P. gingivalis proteome plotted by predicted values for isoelectric point (pI) and molecular weight (A) or hydrophobicity and molecular weight (B). These computations were based on primary structure only as determined from the ORF predictions, and do not take into account modifications and folding. The color coding scheme is the same as that used for Fig. 1. The hydrophobicity calculations were based on the scale published by Engelman, Steitz and Goldman [37]. In (B) the more hydrophobic proteins have positive values that graph to the right side of the image.
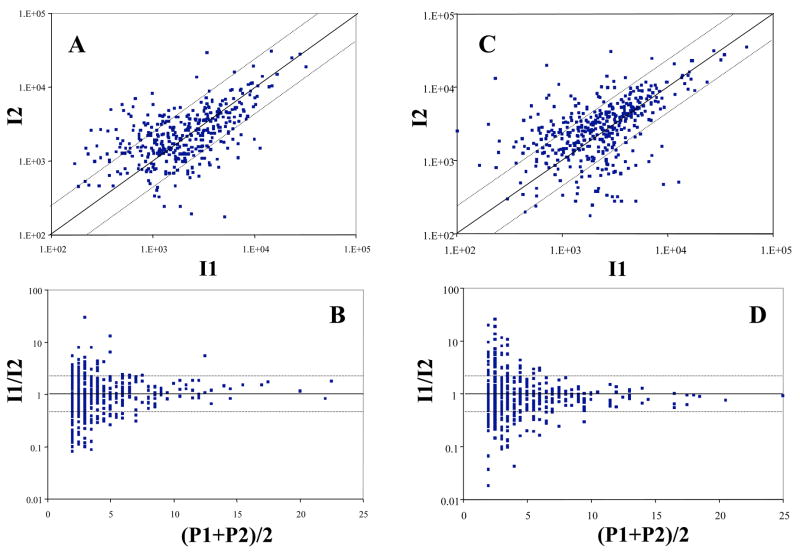
As with 2D gels, there is some evidence for discrimination against short proteins (see Fig. 2), especially those with predicted molecular weights below ~ 7,000 Da. This is to be expected in an analytical scheme that is ultimately limited by mass spectrometer duty cycle. Of all the MS1 mass spectra acquired with sufficiently high signal/noise to potentially yield an interpretable MS2 spectrum, only about 3% were actually being recorded as MS2 spectra, due to the large number of peptide ions flooding the mass spectrometer. Thus, detection probability, as well as quantitative reproducibility, improves dramatically as the number of proteolytic fragments observed for a given protein increases. The relationships shown graphically with data from P. gingivalis in Fig. 3 are the key to generating an estimate of protein expression ratios in the absence of isotopic labels. Although only a small percentage of the total gene products reported here had sufficient coverage to make practical use of this relationship, incremental advances in the technology should soon allow much greater coverage. These developments, especially with respect to faster data acquisition times and improved mass spectrometer duty cycle, may reduce the need for stable isotopes and allow such shotgun proteomics experiments to be both more complete qualitatively and quantitative at the same time. More sophisticated error models are badly needed in this field [25, 26], but they are not a complete solution to the problem of data sets that are still fundamentally “noisy.” However, such improved statistical treatments can serve to decrease the overlap region between the “false positive” and “true positive” score distributions in a given set of data, leading to an incremental increase in the amount of usable information that can be extracted, and the probability that it is correct.
Figure 3.
(A) and (C ) are average signal intensity correlation plots for the KGM and cKGM samples, respectively, that indicate the level of signal reproducibility achieved to date in our work with the P. gingivalis proteome. Each single spot represents the average signal intensity for all the peptides observed for that protein. Unlike transcription arrays, at present only a small number of replicates can realistically be generated for an entire proteome analysis, which may take months to generate with current technology. Each spot represents an ORF with at least 2 peptides qualitatively identified from each run. I1 and I2 are the calculated intensity values for replicate 1 and replicate 2 of the same sample in units of 10,000 counts from the LCQ data system. 385 ORFs are shown in (A). 254 of them show intensity differences of less than 50% (I1/I2 [0.5, 2.0]), as indicated by the area within the confidence band. 53 of them show higher intensities in replicate 1 (I1/I2 (2.0, )). 78 of them show higher intensities in replicate 2 (I1/I2 (0,0.5)). 426 ORFs are shown in (C) . 287 of them show intensity differences of less than 50% (I1/I2 [0.5, 2.0]). 57 of them show higher intensities in replicate 1 (I1/I2 (2.0, )). 87 of them show higher intensities in replicate 2 (I1/I2 (0,0.5)). Panels (B) and (D)are intensity ratio versus peptide number plots for KGM and cKGM sample respectively. Each spot represents an ORF with at least 2 unique peptides identified qualitatively from each run. For a given ORF, I1/I2 is the ratio of average intensity values, calculated by summing all signals associated with a particular ORF and dividing by the number of peptides used in the summation. P1 and P2 are the numbers of peptides per ORF database entry identified from replicate 1 and replicate 2 of the same KGM or cKGM sample. 385 ORFs are shown in (B). 426 ORFs are shown in (D). Note that the reproducibility of the average intensity calculation improves dramatically as the number of observed peptides per ORF increases.
Coverage becomes an especially significant issue for small proteins with only a few cleavage sites, which are less likely to be detected. It is also unknown how many of the small ORFs predicted for P. gingivalis are real genes, hence the difficulty in estimating how much of the apparent shortage of low molecular weight gene products is due to analytical discrimination, as opposed to the possibility that the predicted ORFs may not correspond to real genes. ORFs with limited unique sequence also present detection problems, because a unique peptide is required to unambiguously identify the gene product. In all, 37 proteins, (supplementary material, Table S2) mainly insertion sequence elements and haemagglutinins, could not be definitively identified with a unique ORF and were thus excluded from Table S1. However, as some large percentage of these proteins are likely being expressed, these ORFs were not excluded from Figs. 1 and 2. The long-term solution to the non-unique peptide issue is to increase coverage of the proteome, and therefore the probability of detecting peptides unique to single ORFs. However, distinguishing between proteins that may differ by only a few amino acids in a long sequence is likely to remain problematic, and will continue to require spatial resolution at the intact protein level prior to digestion. Alternatively, closely related genes can be grouped and treated as a single ORF for purposes of reporting protein expression data based on peptide fragments [14].
3.4 2D gel electrophoresis analysis of differential expression of proteins in KGM and cKGM
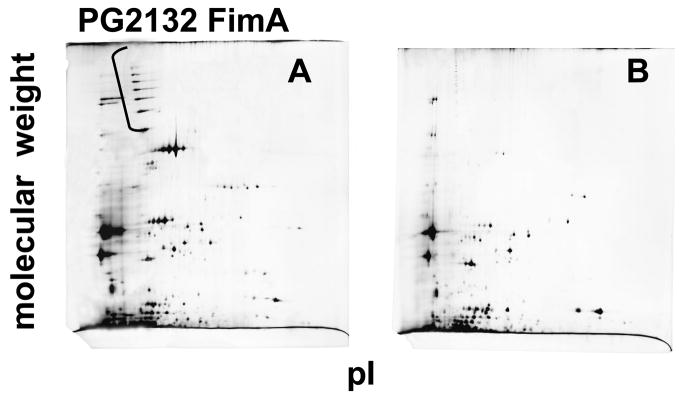
One of the differentially expressed proteins detected by MudPIT was the major fimbrial protein FimA. Consistent with this, an alternative technique for protein analysis, silver-stained 2D gels, were dominated by the presence of multiple variants of FimA in the KGM samples (Fig. 4A). Also in agreement with the MudPIT data, this protein was absent or detected at very low abundance in cKGM samples (Fig. 4B). However, the proteins represented in these gels, as identified by mass spectrometry, were derived from only about 1.4 % of the predicted ORFs. The total number of spots was larger, due to multiple post-translationally processed forms of proteins derived from a relatively small number of genes. The use of 2D gel electrophoresis alone, therefore, would provide a very limited number of the differentially expressed proteins in this model system. This is consistent with observations made in a number of laboratories for a variety of organisms, perhaps most notably for the yeast, Saccharomyces cerevisiae [10, 13]. Corroborration of the down regulation of FimA was also obtained at the transcriptional level. RT-PCR (reverse transcriptase-polymerase chain reaction) of mRNA extracted from P. gingivalis cells under the conditions used for proteomic analysis showed a reduced expression of fimA in mRNA from cells exposed to cKGM (Fig. 4C). However the relationship between global protein and mRNA levels is undoubtedly a complex one [27], and there is unlikely to be a concordance for all the regulated proteins, particularly those having posttranscriptional regulatory mechanisms.
Figure 4.
Validation of differential protein expression by 2D polyacrylamide gel electrophoresis (A), (B) and RT-PCR (C). Representative 2D gel images of silver-stained proteins from P. gingivalis 33277 exposed to KGM (A) showing a prominent “stair step” pattern in the upper left corner (high molecular weight, more acidic) that consists of polymerized FimA of various lengths. FimA almost completely disappears from P. gingivalis exposed to cKGM (B), as was observed for a number of other proteins (see text). Gels loaded with an increased level of protein will reveal many more spots, as is common in the literature, however such overloaded gels are not optimal for protein identification using our methods, as resolution of the spots deteriorates. (C) Differential expression of fimA in P. gingivalis exposed to cKGM or KGM corroborated at the transcriptional level by RT-PCR. 16S rRNA is a control for total mRNA levels.
3.5 Functionality of differentially expressed proteins
Identified proteins that were qualitatively differentially expressed between cKGM and KGM samples, i.e. detectable in one state but not the other (see Fig. 1 and the supplementary material, Table S1), spanned all the functional classes as defined by TIGR (Table 1). These proteins included 51 cell envelope proteins, 13 regulatory proteins, and 6 proteins involved in signal transduction.
Table 1.
P. gingivalis Protein Functional Classes1
| Functional Class | DB2 | KGM-only | cKGM-only | Common3 | Total4 |
|---|---|---|---|---|---|
| energy metabolism | 126 | 11 | 6 | 82 | 99 |
| fatty acid and phospholipid metabolism | 16 | 1 | 1 | 9 | 11 |
| purines, pyrimidines, nucleosides and nucleotides | 44 | 3 | 4 | 21 | 28 |
| amino acid biosynthesis | 18 | 3 | 0 | 7 | 10 |
| protein synthesis | 114 | 11 | 15 | 56 | 82 |
| transcription | 32 | 3 | 5 | 9 | 17 |
| protein fate | 75 | 14 | 11 | 37 | 62 |
| DNA metabolism | 75 | 9 | 9 | 24 | 42 |
| unknown function | 198 | 27 | 18 | 60 | 105 |
| cellular processes | 50 | 8 | 7 | 16 | 31 |
| central intermediary metabolism | 24 | 3 | 5 | 7 | 15 |
| biosynthesis of cofactors, prosthetic groups, and carriers | 74 | 10 | 12 | 18 | 40 |
| hypothetical proteins-conserved | 197 | 22 | 27 | 38 | 87 |
| transport and binding proteins | 110 | 22 | 20 | 28 | 70 |
| cell envelope | 119 | 26 | 25 | 30 | 81 |
| hypothetical proteins | 808 | 75 | 48 | 66 | 189 |
| regulatory functions | 44 | 7 | 6 | 5 | 18 |
| other categories | 133 | 5 | 3 | 3 | 11 |
| signal transduction | 12 | 1 | 5 | 1 | 7 |
| disrupted reading frame | 41 | 0 | 1 | 0 | 1 |
| Total | 23106 | 261 | 228 | 517 | 10065 |
From www.tigr.org. Proteins without observed unique peptides were not listed, i.e. those proteins with observed peptides that could be mapped to two or more ORFs were eliminated. These 37 additional putative proteins are listed in Table S2 (supplementary material).
TIGR P. gingivalis ORF database (2224 ORFs)
The number of ORFs present in both KGM and cKGM samples.
The total number of ORFs identified in both KGM and cKGM samples.
29 of the ORFs observed to express protein belong to two different functional class groups according to the TIGR classification system, hence the total of 1006.
This number is higher than the total number of theoretical ORFs (2224) due to the listing of ORFs in multiple functional classes.
Several hundred ORFs were identified in functional classes with unclear function, such as hypothetical proteins and proteins of unknown function. Indeed, one accomplishment of our work is that 276 proteins classified as hypothetical were identified. This represents the first tangible evidence that these proteins are actually expressed by P. gingivalis and paves the way for future functional studies. Furthermore, this information should help resolve conflicts between the two existing annotations of the P. gingivalis genome at www.tigr.org (2224 putative proteins) and www.stdgen.lanl.gov/oragen/ (1949 putative proteins). Hypothetical proteins constitute a significant portion of the annotated databases for many bacteria; therefore, this protein detection methodology has broad application as a bacterial genome annotation tool.
3.6 Insights into pathogenicity
Under laboratory culture conditions, P. gingivalis expresses a number of well studied putative virulence factors. These include major fimbriae (comprising FimA protein), minor fimbriae (Mfa1), and proteinases such as RgpA. RgpB and Kgp [1]. Of these, FimA was not detected in cKGM, Mfa1 was quantitatively downregulated, while the proteinases were detected but not regulated. FimA induces invasion of epithelial cells through engagement of an integrin receptor and subsequent signaling mediated by integrin associated focal adhesions [28]. This is considered the major pathway of entry of P. gingivalis, although a residual level of invasion does occur in the absence of fimbriae [8]. Down-regulation of FimA suggests that the presence of this invasin is no longer required after the uptake process has been triggered. Furthermore, as FimA is immunostimulatory [29], reduced expression may aid in immune avoidance.
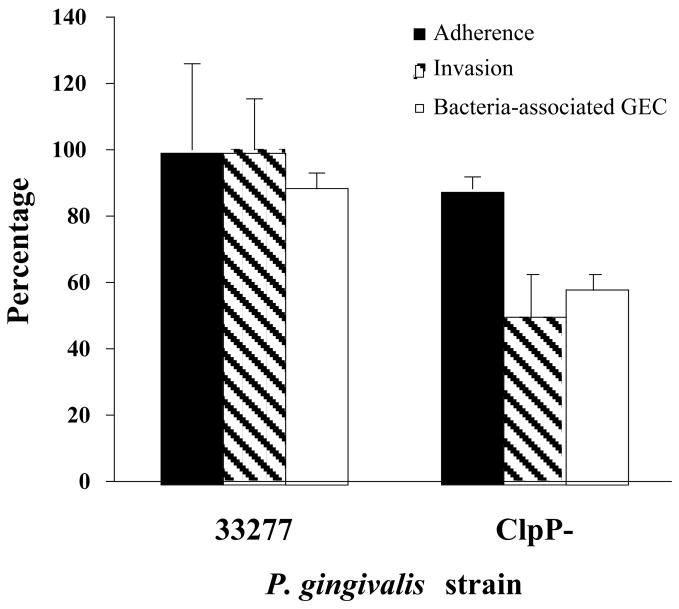
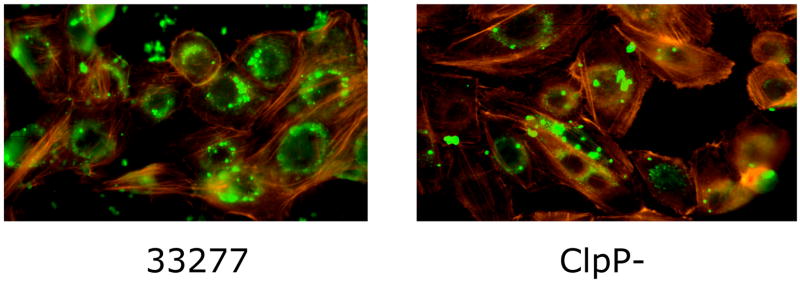
The pathogenic mechanisms of many gram-negative organisms, with regard to their interactions with epithelial cells, revolve around a specialized type III secretion system that injects effector proteins directly into the host cell cytosol. Proteins secreted and translocated by the type III machinery have the capacity to modulate a variety of host cell functions, including the signal transduction pathways and cytoskeletal rearrangements required for bacterial entry [30]. One of the features of P. gingivalis invasion of GEC is that it is accomplished without any obvious homologs of the type III secretion apparatus or effector proteins. Gram-positive pathogens such as Listeria monocytogenes have also evolved an intracellular lifestyle without the involvement of type III secretion systems. Interestingly, the results of this study indicate that P. gingivalis may have devised a means to enter and survive within epithelial cells based on orthologous components of the Listeria system. A number of differentially regulated proteins in P. gingivalis were found to possess homology with invasion-related proteins of L. monocytogenes (Table 2). P. gingivalis PG0350 shares homology with the leucine rich L. monocytogenes internalin (Inl) surface proteins, albeit mostly over the leucine rich region so similarity of function remains to be established. The Listeria InlA and InlB proteins engage receptors on epithelial cells and trigger bacterial invasion [31]. Expression of PG0350 was upregulated in cKGM indicating a possible role for this protein in the fimbriae-independent invasion [8], although this does not exclude a role for other potential invasins of P. gingivalis in this process. In Listeria, expression of InlA and InlB is controlled by the transcriptional regulator PrfA, a member of the Crp/Fnr family [32], and synthesis of PrfA can be induced by epithelial cell extracts [33]. P. gingivalis PG0396 is a putative Crp/Fnr transcription regulator and was also upregulated in cKGM. The possible role of PG0350 in invasion and its regulation by PG0396 is currently under investigation. A pg0350 mutant showed similar invasion characteristics to the parent (not shown); however, our model would predict that a phenotype for a PG0350 mutant would only be discernible in a FimA minus background. Another parallel between P. gingivalis and L. monocytogenes was provided by the ClpC and ClpP chaperones. In Listeria, the ClpC ATPase is required for adhesion and invasion and the protein modulates the expression of other virulence factors including InlA and InlB [34]. ClpC also promotes the early escape of Listeria from the phagosomal compartment of macrophages [35]. The serine protease ClpP is involved in the rapid adaptive response of Listeria within macrophages [36]. P. gingivalis possess ClpC, ClpP and ClpX homologs, all of which are up-regulated in cKGM. These stress induced proteins may thus play an important role in P. gingivalis invasion and intracellular survival. Consistent with this, proteins homologous to heat shock proteins 15 (PG0165) and 90 (HtpG, PG0045) were also up-regulated in cKGM. As a representative of the Clp system, a ClpP deficient mutant was constructed in P. gingivalis and tested for association with gingival epithelial cells. In an antibiotic protection assay, invasion/intracellular survival of the clpP mutant was decreased by approximately 50% in comparison to the parent (Fig. 5A). Immunofluorescence microscopy revealed that fewer mutant cells located intracellularly compared to the parent (Fig. 5B) and that fewer epithelial cells contained mutant P. gingivalis above the threshold detection level (Fig. 5A). This defect was not the result of reduced adhesion to the epithelial cells as mutant and parent showed similar adhesive capacities (Fig. 5A). Thus, the clpP mutant has a diminished invasive, but not adhesive, capacity for GEC. As with any insertional mutation there is a possibility of a polar effect on the downstream gene. In the case of clpP, however, the downstream gene is clpX and thus it is reasonable to conclude that a functional Clp system is required for optimal invasion by P. gingivalis.
Table 2.
Proteins differentially expressed in P. gingivalis that have homology to Listeria invasion-related proteins
| ORF no.1 | Protein1 | L. monocytogenes homolog | e value |
|---|---|---|---|
| PG0010 | CLP protease ATP-binding subunit ClpC | ClpC | 0 |
| PG0350 | Internalin-related protein | Protein similar to internalin | 1e-38 |
| PG0417 | ATP-dependent Clp protease, ATP-binding subunit ClpX | ClpX | e-116 |
| PG0418 | ATP-dependent Clp protease proteolytic subunit | ClpP | 9e-45 |
Numbers and annotation from www.tigr.org.
Figure 5.
Invasion characteristics of the ClpP mutant. (A) Relative percentage (compared to 33277) of the ClpP mutant adhered to GEC (black bars) and recovered intracellularly from GEC (gray bars). White bars show percent of GEC containing intracellular bacteria by immunofluorescence microscopy. (B) Immunofluorescence microscopy of GEC infected with 33277 or the ClpP mutant. P. gingivalis cells are green, and actin microfilaments appear red.
Among the cKGM down-regulated proteins were a homolog of FeoB (PG1043), a ferrous iron transporter, and HemR/HmuR (PG1552) a TonB-dependent hemoglobin receptor and a component of one of the major pathways for hemin acquisition by the organism. Hence, in the nutritionally altered cKGM environment P. gingivalis may have less need of iron uptake pathways, or may fine tune uptake mechanisms according to the sources of iron available.
4 Concluding remarks
While considered an important pathogen in periodontal diseases, P. gingivalis can also exist in the oral cavity in the absence of overt disease, indicating a balance between pathogenic potential and host defense. Moreover, while the organism possesses a number of adhesins, invasins, modulins and tissue destructive enzymes, no single virulence factor alone fully accounts for pathogenicity. Thus, the pathophysiology of the organism may depend on a number of factors acting in concert and regulated by environmental cues, a concept that is supported by the broadly based changes in protein expression exhibited in response to epithelial cell components.
Supplementary Material
Acknowledgments
Supported by the NIH through DE14372, DE11111, DE14168 and DE14955. Additional support was provided by Michrom Bioresources. We thank Michael MacCoss, Martin Sadilek, Mary Lidstrom, Norm Dovichi, John Leigh, Fred Taub, Kobi Alfandari, Luping Fan and Qiangwei Xia for their assistance. We thank TIGR for prepublication use of databases and annotations for the P. gingivalis genome. Additional annotations were provided by Los Alamos National Laboratory (LANL).
List of Abbreviations
- cKGM
conditioned keratinocyte growth medium
- GEC
gingival epithelial cells
- KGM
keratinocyte growth medium
- MS1
first dimension of mass spectrometry
- MS2
second dimension of mass spectrometry
- MudPIT
multidimensional protein separation technology
References
- 1.Lamont RJ, Jenkinson HF. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scannapieco FA, Genco RJ. J Periodont Res. 1999;34:340–345. doi: 10.1111/j.1600-0765.1999.tb02263.x. [DOI] [PubMed] [Google Scholar]
- 3.Deshpande RG, Khan MB, Genco CA. Infect Immun. 1998;66:5337–5343. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorn BR, Dunn WA, Progulske-Fox A. Infect Immun. 1999;67:5792–5798. doi: 10.1128/iai.67.11.5792-5798.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Cell Microbiol. 1999;1:215–224. doi: 10.1046/j.1462-5822.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- 6.Lamont RJ, Chan A, Belton CM, Izutsu KT, et al. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, et al. FEMS Microbiol Lett. 2001;200:145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 8.Yilmaz O, Young PA, Lamont RJ, Kenny GE. Microbiol. 2003;149:2417–2426. doi: 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 9.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, et al. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 11.Griffin TJ, Aebersold R. J Biol Chem. 2000;276:45497–45500. doi: 10.1074/jbc.R100014200. [DOI] [PubMed] [Google Scholar]
- 12.Washburn MP, Yates JR., III Curr Opin Microbiol. 2000;3:292–297. doi: 10.1016/s1369-5274(00)00092-8. [DOI] [PubMed] [Google Scholar]
- 13.Washburn MP, Wolters D, Yates JR., III Nature Biotech. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 14.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., III Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Zhang Z, Chen W, Park Y, et al. The Analyst. 2002;127:1450–1456. doi: 10.1039/b206157k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watari M, Andrews HL, Isberg RR. Mol Microbiol. 2001;39:313–329. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 17.Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- 18.Chung WO, Park Y, Lamont RJ, McNab R, et al. J Bacteriol. 2001;183:3903–3909. doi: 10.1128/JB.183.13.3903-3909.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng JK, McCormack AL, Yates JR., III J Amer Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, Laidig KE, Park Y, Park K, et al. The Analyst. 2001:52–57. doi: 10.1039/b008012h. [DOI] [PubMed] [Google Scholar]
- 21.Tabb DL, McDonald WH, Yates JR., III J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guina T, Purvine SO, Yi E, Eng J, et al. Proc Natl Acad Sci USA. 2003;100:2771–2776. doi: 10.1073/pnas.0435846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bondarenko PV, Chelius D, Shaler TA. Anal Chem. 2002;74:4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 24.Gygi SP, Rist B, Gerber SA, Turecek F, et al. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 25.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 26.Sadygov RG, Yates JR., III Anal Chem. 2003;75:3792–3798. doi: 10.1021/ac034157w. [DOI] [PubMed] [Google Scholar]
- 27.Ideker T, Vesteinn T, Ranish JA, Christmas R, et al. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 28.Yilmaz O, Watanabe K, Lamont RJ. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa T, Ogo H, Uchida H, Hamada S. J Med Microbiol. 1994;40:397–402. doi: 10.1099/00222615-40-6-397. [DOI] [PubMed] [Google Scholar]
- 30.Hueck CJ. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cossart P, Pizarro-Cerda J, Lecuit M. Trends Cell Biol. 2003;13:23–31. doi: 10.1016/s0962-8924(02)00006-5. [DOI] [PubMed] [Google Scholar]
- 32.Dramsi S, Lebrun M, Cossart P. Curr Top Microbiol Immunol. 1996;209:61–77. doi: 10.1007/978-3-642-85216-9_4. [DOI] [PubMed] [Google Scholar]
- 33.Renzoni A, Cossart P, Dramsi S. Mol Microbiol. 1999;34:552–561. doi: 10.1046/j.1365-2958.1999.01621.x. [DOI] [PubMed] [Google Scholar]
- 34.Nair S, Milohanic E, Berche P. Infect Immun. 2000;68:7061–7068. doi: 10.1128/iai.68.12.7061-7068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouquette C, de Chastellier C, Nair S, Berche P. Mol Microbiol. 1998;27:1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 36.Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. Mol Microbiol. 2000;35:1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x. [DOI] [PubMed] [Google Scholar]
- 37.Engelman DM, Steitz TA, Goldman A. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 38.McCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR., III Anal Chem. 2003;75:6912–6921. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen S. Guide to Analysis of DNA Microarray Data. 2. Wiley; Hoboken: 2004. pp. 33–49. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.