There continues to be a need for innovative and inexpensive drugs to treat diseases of the developing world.1,2 It is also important to link academic training and research to critical societal needs. Indiana University−Purdue University Indianapolis (IUPUI) is addressing both these concerns by developing a concept called “Distributed Drug Discovery” (D3).(3) This Perspective describes how D3 can harness combinatorial chemistry, distributed over multiple academic and industrial locations, to educate students while they perform a key role in the early stages of drug lead discovery for developing world and otherwise neglected diseases. Two other articles in this issue of the Journal of Combinatorial Chemistry present case histories implementing the chemistry component of D3. One involves replicated D3 syntheses in the United States, Poland, Russia, and Spain.(4) The second is an application in which students at IUPUI make analogs of a potential anticancer agent.(5) In this Perspective, D3 is discussed in three parts: (I) The Concept of D3, (II) The Role of Combinatorial Chemistry in D3, and (III) Implementation of D3.
Part I. The Concept of D3
It is difficult to find resources to discover drugs to treat diseases in the developing world.1,2 In the developed world, economic incentives have fueled drug discovery, financing the expensive equipment, procedures, and personnel currently required by the pharmaceutical industry. Unfortunately, the burden of disease is disproportionately focused in poor nations, and there is not the same economic incentive for the pharmaceutical industry to discover drugs for diseases of the developing world.(6)
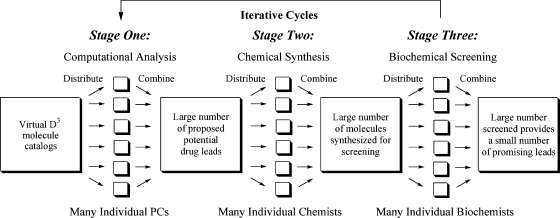
Distributed Drug Discovery (D3) proposes that if simple, inexpensive equipment and procedures are developed for research in each of the core drug-lead discovery stages, computational chemistry, synthetic chemistry, and biochemical screening, this large research challenge can be divided into manageable smaller units and carried out, in parallel, at multiple academic and industrial sites. The academic sites will be located in both the developed and developing world. The industrial locations can be stand-alone nonprofits, private-public partnerships, or nonprofit initiatives within corporations.(7) The coordinated and recombined results of these distributed resources can economically accelerate the identification of leads in the early stages of the drug discovery process. Simultaneously, this effort provides educational and job opportunities in both the developed and developing worlds, while building cultural and economic bridges for the common good. This distribution of problem solving at the three core stages of drug-lead discovery is shown schematically in Figure 1.
Figure 1.
Diagram of distributed problem solving at three stages of Distributed Drug Discovery.
Stage One D3: Distributed Computational Analysis
Independent of the integrated D3 process proposed here, scientists have demonstrated the viability and power of a distributed problem solving approach at the computational stage of drug discovery.(8) The Web site Grid.org(9a) describes how a screen saver surrogate process permits computational screening of a virtual library of over 35 million potential drug molecules, to identify those that might interact with protein targets relevant to the treatment of smallpox. A similar distributed project has been organized by the World Community Grid, with malaria as one of the targeted diseases.(9b) The Schools Malaria Project is an excellent example of how distributed computation and education can be combined to address a developing world disease challenge.(8h) These sites (and the pioneering “SETI” project(9c)) demonstrate the power of dividing a large computational problem into smaller units and distributing them to individual PCs that have the required problem-solving software. In these distributed computational examples, free programs that run on idle PCs are powerfully combined to process large data sets and economically solve resource-intensive computational problems.
Stage Two D3: Distributed Chemical Synthesis
We believe an analogous process can be developed and employed at the synthesis stage of drug-lead discovery. The distributed resources will be individual student-chemists in both undergraduate and graduate academic laboratories, as they receive education and do research in organic synthesis. Temporarily idle resources at private, public, and other nonprofit synthetic laboratories around the world will serve as additional distributed synthetic sites.
By utilizing combinatorial chemistry and the equipment discussed in Part II of this Perspective, D3 will capitalize on a tight integration of Stage One computational analysis with realistic Stage Two synthetic capability. Well-precedented virtual D3 catalogs (discussed in detail in Part II) are key to this integration. Their precedented nature gives assurance that potential drug-lead molecules, computationally selected from these virtual catalogs, will be synthetically accessible by students and researchers utilizing simple, low-cost, globally distributed equipment and procedures.
Stage Three D3: Distributed Biological Screening
All the products from these synthetic efforts can be tested by distributed biological screening, if developed in an analogous fashion and integrated into educational, private, public, or other nonprofit laboratories. Although we are not aware of current examples of a distributed screening process, we believe that the D3 concept, coupled with the documented examples of distributed problem solving in Stage One computation and Stage Two synthesis, will provide incentive for our biological colleagues to develop their own distributed methodology for Stage Three screening. Meanwhile, high-throughput screening resources are available through the NIH Molecular Libraries Initiative.(10)
Integrating All the Stages of D3: The Role of Information Technology
D3 will utilize appropriate information technology, open-access-based when available,11,12 to enable and coordinate the critical elements of all three core disciplines:
-
(1)
Stage One enumeration and computational analysis of virtual D3 catalogs for the identification of potential developing world disease drug-leads. The resulting targeted molecules will be tabulated in a format readily accessible and understandable to chemists.
-
(2)
Facilitation and tracking, at Stage Two, of the distribution and synthetic fate of these selected molecules. Which ones have institutions chosen to make? What are their results? Is their quality acceptable? Are they available for screening? How do biologists obtain them?
-
(3)
Tracking, at Stage Three, of the distribution and screening of the synthesized molecules.
Partnering with the NIH Molecular Library Initiative may provide solutions to many of these shared informatics needs.(10)
The Power and Potential of Distributed Drug Discovery
There has been considerable discussion regarding the benefits and drawbacks of a profit-driven drug discovery process.6,13 On the one hand, the profit motive has provided incentives for the pharmaceutical industry to invest in the human and material resources that have revolutionized the treatment of diabetes, hypertension, cancer, AIDS and other debilitating or deadly diseases. The pharmaceutical industry can be justifiably proud of its role in these life-changing drug discoveries. On the other hand, research requiring a profit results in neglected diseases when patients cannot afford to pay for the drug discovery costs. In addition, because of intellectual property concerns, this profit dependence has restricted the sharing of information and resources that could otherwise facilitate drug discovery.
Distributed Drug Discovery addresses these limitations. Its vision is pragmatic and compelling: by developing and utilizing economical equipment and straightforward procedures, large research problems can be broken down into manageable smaller units to be solved by individual scientists at multiple sites throughout the developing and developed world. Students will have the tools to be educated and do research in important health-related scientific disciplines. While education is taking place, they will simultaneously make and test chemical candidates for drug-lead discovery. This coupling of training and research enables students to clearly understand the value and purpose of their education. By incorporating the initial stages of the drug discovery process into college and university education, costs are minimized while at the same time scientists of the future are trained. With little profit incentive, sharing discovery through the D3 process becomes an asset, not a liability. With the removal of barriers to the global sharing of information and resources, Distributed Drug Discovery provides access to untapped intellectual and practical potential.11,13 Patents will only be pursued in those situations where they facilitate, rather than hinder, information sharing and the delivery of low cost drugs to those in need.(13h)
When spread over global sites, D3 becomes a potent drug-lead discovery process, forging a closer geographical and cultural connection between those in need and those trying to meet those needs. To rephrase a classical saying: “Give people medicine and they will depend on you for their health; teach people to discover medicines and they will develop cures themselves.”(14) It is our hope that D3 will enable the lead discovery phase of this goal, and that other organizations will develop these leads to the final drug stage.2,6,7,11
Part II. The Role of Combinatorial Chemistry in Distributed Drug Discovery
A major focus of this Perspective is to describe the role combinatorial chemistry can perform in enabling distributed synthesis at Stage Two of D3. For successful implementation at this stage there must be well-documented virtual D3 catalogs and cost-effective, reproducible synthetic procedures to make the large number of potential drug-lead molecules selected from them. Clearly combinatorial chemistry is a prime candidate to fulfill this need. The chemical economy and effectiveness of a combinatorial discovery process cannot be disputed. In nature a set of only 24 starting materials (4 nucleotides and 20 amino acids) is sufficient to provide, through combinatorial chemistry, the materials and synthesis instructions to create all the proteins essential for the functioning and protection of life. This includes the huge combinatorial library of unique antibody molecules coded for and expressed by the body’s B-cell repertoire.(15) They are a key component of our endogenous drug discovery factory, the immune system. In a few days’ time, antibodies are selected from this library and scaled up for production, to fight infections and other assaults on the body by foreign substances.
Chemists have attempted to learn from nature’s powerful combinatorial examples and implement an analogous process in an organic chemistry laboratory setting. Solutions to some key challenges shared by the combinatorial chemistry discipline and D3 are discussed in the following sections.(16)
Rationally Selecting Candidate Molecules: The Role of Virtual D3 Catalogs
Computational analysis is now commonly used to select or design potential drug leads prior to synthesis. However, often this process does not consider if the selected molecules can be made by any simple, known procedure. On the other hand, combinatorial chemistry can enable the synthesis of many molecules, but sometimes without regard for theoretical activity. Researchers who are expert in both of these areas can help narrow this communication gap. In general, however, computational specialists cannot be expected to be experts at synthesis, nor can the synthetic chemists be experts in computational analysis.
An effective way of bridging this divide is to provide computational chemists with comprehensive information that captures and translates for them the knowledge and capability of the synthetic chemist. Currently, one of the most direct ways of accomplishing this goal is to encode, for the computational chemists, synthetic expertise in the form of virtual libraries (or, as we refer to them in D3, virtual “catalogs”):(17) collections of theoretical molecules accessible by known chemistry. Virtual libraries can readily be constructed with available enumeration packages based on known synthetic transformations operating on available reagents.(18) They are especially valuable when the classes of molecules constructed are “biased”, that is, they resemble structures with known or expected biological activity. When these virtual libraries have been enumerated using reagents with predictable reactivity and synthetic routes that are well documented, the computational chemist, with no knowledge of synthesis, can use virtual screening tools to confidently select target molecules that can realistically be prepared.(19)
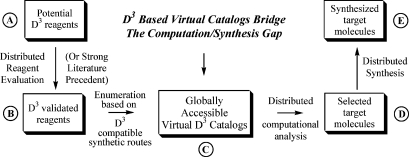
Unfortunately, the utility of virtual libraries often suffers from major limitations: (1) They may not be representative of classes of molecules with promising biological activity. (2) Because of intellectual property concerns and technical issues, such libraries are not universally available for comprehensive computational analyses. (3) Depending on documentation and precedent, the successful synthesis of any selected “virtual” molecule can be wildly unpredictable. This is where D3 comes into play. As depicted in Figure 2, the following steps integrate computational analysis and realistic D3 synthesis:
-
(1)
Combinatorial chemistry based on straightforward methodologies and inexpensive equipment is developed to enable the synthesis of promising structural types. All the reagents compatible with this chemistry are identified from commercial or other sources (Figure 2A).
-
(2)
With D3 equipment and procedures, many of these potential reagents are tested at the appropriate step in the combinatorial synthesis (Figure 2A and B). This rehearsal(20) can be an integral part of the educational process, with replicated evaluation at globally distributed educational laboratories.(4)
-
(3)
Virtual D3 catalogs are enumerated(18) from these and other validated reagents (Figure 2B and C). We have chosen to term these collections of molecules virtual “catalogs” rather than “libraries” to distinguish them from enumerated libraries based on more hypothetical or complicated chemistry or on unvalidated reagents and procedures. The term “catalog” emphasizes the greater likelihood of synthetic accessibility for molecules contained within a virtual D3 library. These catalogs are made freely available globally for analysis by the entire scientific community.
-
(4)
Distributed computational analysis is performed on these large virtual catalogs (possibly >106 compounds), against models appropriate for developing world or neglected disease targets.(8) This yields a smaller list of potential drug lead candidates (Figure 2C and D) which are accessible by D3 syntheses.
-
(5)
This list of proposed drug leads (D) is subdivided into smaller sets, which can be chosen by different laboratories for replicated distributed synthesis (Figure 2D and E). For example, a list of 1000 compounds step D could be divided into 50 sets of 20 compounds each. The earlier rehearsal process (A to B) will have already confirmed the viability of the required chemistry. The inexpensive equipment (see Part III) can be manufactured locally and the distributed synthesis (i.e., D to E) conducted, at 50 sites, as part of educational programs that train students in organic synthesis while simultaneously making potential drug leads.
Figure 2.
Role of D3-based virtual catalogs in integrating computational analysis with synthesis.
The computational component of D3 will take full advantage of open-source, web-based, internationally distributed methodologies(8) to analyze these large, global, virtual D3 catalogs, using computational models for developing world disease targets.(8e) Specific examples of distributed computation applied to neglected diseases are the “WISDOM” project,(21) and Schools Malaria Project(8h) which search libraries of molecules for potential antimalaria drugs.
On the synthetic side, these virtual D3 catalogs will be constructed with the significant constraint that they be realistic virtual libraries, based on reagents whose synthetic suitability has various levels of precedent. They will be either completely rehearsed (i.e., that actual reagent has been successfully employed using a D3 protocol) or well-precedented, based on a close resemblance to completely rehearsed reagents or published work using related chemistry. By this more comprehensive integration of computational tools with realistic and cost-effective synthetic capability, D3 should greatly advance both the selection and synthesis of appropriate molecular candidates. This is possible because such target molecules will be simultaneously computationally desirable, synthetically accessible, and resource enabled. The expertise of the computational chemists will determine the structures to be made. Molecules will be synthesized and tested in a distributed fashion and the data captured and analyzed to enable iterative computational analysis and subsequent development work.
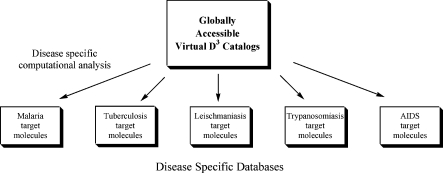
D3 will establish a coordinated information network of computational, synthetic, and biochemical screening resources. To link computational expertise and synthesis goals, we envision this network as a globally accessible resource for both virtual D3 catalogs and the desired disease specific target structures derived from their computational analysis (Figure 3). It will facilitate distributed selection, synthesis, and screening of molecules and gathering and analyzing the resulting information. This network will require careful design for easy use by each scientific discipline within the global community. To encourage participation of all the respective scientific expertise, it should incorporate an advocacy component for the potential of D3. It will be an excellent candidate for sponsorship by nonprofits that want to facilitate the discovery of drugs for developing world and otherwise neglected diseases.2,7
Figure 3.
Globally accessible databases of D3 target molecules.
Synthesizing Targeted Molecules by Reproducible Synthetic Routes that Permit Ready and Comprehensive Analog Synthesis and Follow-up Work
Combinatorial D3 chemists should have confidence that they can synthesize any individual member of a virtual library. In addition, in lead discovery, they need to be able to quickly remake, modify, and scale-up active molecules identified through the screening process, both for secondary screening and to improve on their properties through analog synthesis. For in vitro enzyme, receptor, or cell-based assays, this can involve the synthesis (and resynthesis) of anywhere from a few milligrams to several grams of compound. Accomplishing this has sometimes been a difficult or impossible task, and negative examples are cited in arguments for avoiding combinatorial chemistry altogether.(16) Especially problematic has been batch to batch variability in the resynthesis of compounds made “in house” or problems resynthesizing, from poorly documented or unavailable protocols, compounds obtained from external sources. To avoid the resulting challenges of rapid, reproducible resynthesis, or small quantity scale-up, industry is increasingly preparing or purchasing larger quantities of purified compounds.
D3 addresses these issues by placing a strong emphasis on reproducibility and wide synthetic capability to a degree that we believe is unprecedented in the synthetic world. At its core is the ability to reproducibly synthesize, in a worldwide distributed network, large numbers of molecules. This reproducibility challenge is met because, by virtue of its low expense and distributed nature, all syntheses are replicated by at least one other student-chemist, often at more than one site and globally distant locations. This replication process is done in both the assessment of potential D3 reagents (reagent rehearsal) and targeted synthesis.
Replicated reagent rehearsals simultaneously validate the reproducibility of the synthetic procedures and the acceptability of reagents, while increasing the likelihood that any individual member of a virtual, combinatorial D3 catalog can be made and remade. Although reagent rehearsal is currently being done in combinatorial chemistry work,(22) it can be taken to a new level of certainty through distributed replication in student hands in different locations throughout the world. This distributed process has now taken place in the United States, Poland, Russia, and Spain, and is documented in an accompanying article.(4) It provides powerful validation of the distributed approach.
When target molecules are identified from virtual D3 catalogs, their syntheses will similarly be replicated. This replication provides the precedent required to give confidence in the ability, should it be needed, to resynthesize these hits and to use the same synthetic procedures and reagent sets to produce analogs in follow-up work. Since multiple researchers were involved in replicated syntheses of a promising lead, even the scale-up work can be distributed, for example by multiplying the replication of a given molecule in subsequent distributed laboratories. Because the D3 process can quickly and reproducibly resynthesize molecules, only those small quantities required for initial in vitro biological screening need be made. This is in contrast to the more expensive approach used in industry, which often synthesizes and purifies larger amounts of molecules even before they show any interesting biological activity.
Attaining Appropriate Analytical Quality
An early issue in combinatorial chemistry was the questionable analytical quality of the molecules prepared. Some collections contained either intentional mixtures or single molecules of varying purities and characterization. Through inappropriate follow-up work, these samples sometimes led to “false positives”(23) and wasted effort. As a result, many research programs now require the purification, to >90−95% purity, of all molecules prior to any screening.(24)
There is no disputing the value of isolating substantially pure and well-characterized molecules for testing in follow-up structure/activity work. However, a universal, single-compound purity criteria may not always be necessary at the initial screening level. For example, with some screens it may be reasonable, even desirable, to test mixtures (enantiomers, diastereomers, or even more complicated mixtures(25)) from a single synthesis because there are sophisticated and effective tools (mainly pioneered by natural products chemists(26)) to identify the active component(s) of a mixture.(27) Bioassay-guided fractionation is especially effective in this regard.(28) When needed, it can be performed either at an underutilized centralized site, or as part of an educational program in analytical laboratories that teach students these skills. The important point at this stage is to have a careful coordination of analytical and screening expertise for quick and positive identification of the active component of a screen hit, regardless of the initial purity or complexity of the tested sample. It is through this coordinated effort that purity criteria should be established and hits identified. In D3, all products made will be analyzed for purity, usually by a combination of liquid chromatography and mass spectral identification techniques.(29) When further purification is required prior to testing, this will be carried out either in an academic laboratory (for example by students as part of the educational process(5)) or in a more centralized facility.
Managing Expense
It is inherent in the combinatorial problem solving process that the likelihood of finding a solution is increased by performing multiple experiments. This inevitably means making more molecules than required for traditional problem solving. If D3 is to be successfully carried out using combinatorial chemistry, the financial issues accompanying this increased experimentation must be addressed.
Critical cost categories in organic syntheses are personnel, equipment, laboratory space, reagents, and waste disposal. However, where the distributed computational screen saver analogy applies, the implementation of D3 synthesis requires little or no additional expense. For example, undergraduate students often seek the opportunity to participate in independent research, facilitating the development of these distributed laboratories. When these laboratories are implemented in the educational process (at IUPUI as part of the second semester undergraduate organic laboratory), the costs of equipment, reagents, waste disposal, and teaching personnel are partially or completely covered by tuition and other funding sources. As described in Part III, by having each student perform six syntheses on a small scale (typically 50 μmol), and minimizing the need for intermediate purification steps, the cost can be kept close to the expense of a laboratory in which each student conducts only one synthetic sequence but on a scale 10−100 times larger (see Part III for a more complete discussion of equipment and reagent needs).
Financial support for development of D3 currently comes from a variety of sources. The National Institutes of Health (NIH) has funded the ongoing basic solution- and solid-phase research subsequently adapted to D3 chemistry. As mentioned above, a large part of the student laboratory expense is covered by tuition and private/public-supported funds. In our laboratories, undergraduate research was funded by IUPUI student tuition, scholarships, and grants, and by The Camille and Henry Dreyfus Foundation. A private fund for Distributed Drug Discovery(30) finances the purchase of equipment and supplies when educational money is limited. Eli Lilly and Company has actively supported the Distributed Drug Discovery effort. Future funding could come from non- and for-profit organizations that have already demonstrated interest in supporting the discovery of new drugs for developing world diseases, and in increasing the level and accessibility of science education in both the developed and developing world.2,7,11e
Part III. Implementation of D3 Chemistry in the United States, Poland, Russia, and Spain
Development of D3 Combinatorial Chemistry Procedures that Enable the Synthesis of any Member of Large Virtual D3 Catalogs of Potential Drug Leads
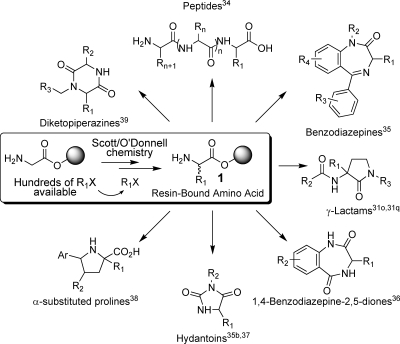
Over the past twelve years a major objective in our research program has been the development of solid-phase synthetic methodology to varied classes of potentially biologically active molecules.(31) It was natural to consider some of this chemistry for the D3 project. We focused first on the methodology we developed to synthesize a wide range of resin-bound unnatural amino acids 1.31a,31c This generic structure (including resin-bound naturally occurring amino acids) is one of the most common types of intermediates used in combinatorial chemistry. It is often embedded in subsequent molecular scaffolds.32,33 Scheme 1 gives just a few of the many documented examples of libraries based on 1.31o,31q,34−39
Scheme 1. Role of Resin-Bound Amino Acids 1 as a Key Combinatorial Intermediate.
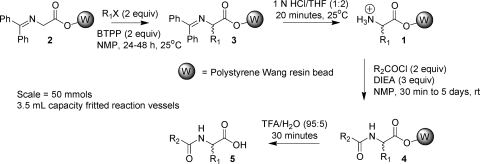
Our route to resin-bound analogs of 1(31) and their acylated derivatives 5 (Scheme 2) offered an opportunity to document the ability of students to make and use 1 in multiple D3 projects.
Scheme 2. Preparation of an Acylated Unnatural Amino Acid (5) Library4,5.
In July of 2003, with strong support and encouragement from the organizers of an NSF Workshop at Miami University,(40) early adaptations of this chemistry and equipment were tested by a set of educators from small and large institutions. With the success of this workshop, and further encouragement and input from the participating educators (one of whom helped coin the term “D3”), we decided to make a concerted effort to enable the D3 concept. We enlisted the help of undergraduate, graduate level and postdoctoral researchers at IUPUI, in collaborative and independent research, to adapt our published procedures to the scale, equipment, and simplicity that would be required for D3 incorporation into a standard second semester undergraduate organic chemistry laboratory. It was assumed that students in such a laboratory would have no prior exposure to combinatorial chemistry or solid-phase organic synthesis. A number of practical issues were considered, among them reproducibility, time constraints, and solvent/reagent expenses. From this developmental work, a formal laboratory was designed and implemented at IUPUI in Fall 2004. Variations of this laboratory have now been carried out over twelve semesters at IUPUI, as well as at locations in Poland, Russia, and Spain. This work is described in more detail below, and in the following article in this Journal.(4)
Design and Manufacture of Simple, Inexpensive Equipment to Carry Out These Syntheses
Carrying out D3 chemistry requires simple and low-cost equipment. While there are numerous devices for conducting solid-phase reactions, most do not meet the D3 constraint of simplicity, low cost, reusability, and especially, appropriate student scale. We initially explored, at the Miami University NSF Workshop, the use of equipment capable of conducting 24 combinatorial reactions at a time. However, it was clear that it would be an overwhelming challenge to use in most undergraduate laboratories. To proceed further, the equipment, which was originally developed in industry,(41) was redesigned to carry out six solid-phase reactions at a time, on a 25 to 200 μmol scale, in a 2 × 3 combinatorial grid. The 3.5 mL reaction vessels, with screw caps on both ends and a fused frit at one end, are made out of glass. This provides inertness, durability, and long-term vessel reusability, making these reaction vessels cost competitive with disposable plastic cartridges. A picture of the kit, known as a “Bill-Board 6-pack”, is shown in Figure 4. The Bill-Board 6-pack kit is manufactured locally at modest cost.(42)
Figure 4.
Bill-Board equipment for Distributed Drug Discovery.
It should be emphasized that inherent in the D3 concept is an open-access approach to meeting all needs. This project will fail if expensive synthesis equipment is required. In the spirit of open access D3, any alternative low-cost equipment that would be more readily manufactured locally or enable an alternative D3 chemistry (solution or solid-phase based) will be welcomed, used, and shared.
Successful Demonstration of Globally Replicated D3 Synthesis
With the required chemical procedures and equipment in place, it remained to be shown that the distributed combinatorial synthesis that D3 requires is practically feasible in everyday working educational laboratories throughout the world. This goal has been accomplished. Participating laboratories engage in either of two activities: “reagent rehearsal” or “targeted library synthesis.” In the rehearsal laboratories, diversity reagents are evaluated in a replicated fashion for potential use at each step of a D3 combinatorial synthesis and subsequent use in the enumeration of virtual libraries. The targeted library synthesis laboratories then synthesize a subset of the virtual D3 catalogs.
For our first exemplification of the “reagent rehearsal laboratory”, we chose to evaluate alkylating agents R1X and acylating agents R2COCl (or R2CO2H), in the synthesis of acylated unnatural amino acid libraries (5, Scheme 2). This enables the creation of a global database of reagents for enumeration of a large, rehearsed (or otherwise precedented) virtual catalog of acylated unnatural amino acids 5 or other combinatorial libraries based on intermediate 1 (Scheme 1). This laboratory was piloted at IUPUI in fall 2004. Second semester beginning organic chemistry laboratory students conducted a rehearsal of alkylating agents R1X. The twenty participating students successfully carried out a total of 120 separate solid-phase reaction sequences. This laboratory was expanded in spring 2005 to four sections with a total of sixty-five students conducting 198 separate reactions. Later that spring, the laboratory was conducted at the University of Barcelona (Spain), and finally, in the summer of 2005, at Moscow State University (Russia) and the Lublin School of Pharmacy (Poland). With few exceptions, all rehearsal syntheses were replicated satisfactorily, locally, and globally, and a control molecule synthesized at multiple sites was obtained with consistent purity. Currently over 40 alkylating agents and 60 acylating agents have been evaluated. Part of this work is reported in the following article, “Distributed Drug Discovery, Part 2: Global Rehearsal of Alkylating Agents for the Synthesis of Resin-Bound Unnatural Amino Acids and Virtual D3 Catalog Construction.”(4) New molecules produced in this project have been submitted for biological evaluation to the Small Molecules Library Repository created as part of the NIH Molecular Libraries initiative.(10)
The challenge of a “targeted library synthesis” laboratory has also been met. It is crucial to the Distributed Drug Discovery concept that we show that students can be educated, at the same time they make, in a distributed fashion, new molecules that have been rationally chosen (targeted), from virtual D3 catalogs. While waiting for neglected disease drug-lead candidates to emerge from the computational analysis of virtual D3 catalogs, we wanted to immediately demonstrate the ability of students to make targeted, high quality, potentially biologically active molecules in the course of their normal educational laboratory training.
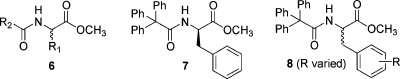
In this regard, a biological target and class of molecules (albeit for cancer and not uniquely a developing world disease) was chosen, from our virtual catalog, for proof of concept. An article from the Hergenrother group reported that the (R)-phenylalanine derivative 7 induced apoptosis in a melanoma (skin cancer) cell line (Figure 5).(43)
Figure 5.
Generic structure 6, anti-melanoma lead 7, and analogs 8 accessible through D3.
Students had already rehearsed alkylating agents that produced racemic acylated phenylalanine analogs (Scheme 2, 5a, R1 = CH2Ar).(4) Thus, it was readily apparent that a simple modification of our current synthetic route could provide for the rapid production, in a distributed fashion, of the most active antimelanoma compound (7), along with new analogs 8. As in the preparation for our first rehearsal laboratories, we enlisted the help of undergraduates, graduate students, and postdoctoral researchers at IUPUI, in collaborative and independent research, to adapt our published procedures to the scale, equipment, and simplicity that would be required for incorporation of a “targeted library synthesis” laboratory into a routine undergraduate organic chemistry laboratory. This was accomplished, and the laboratory entitled “Solid-Phase Combinatorial Synthesis of Analogs of an Anti-Melanoma Compound” was implemented at IUPUI. The 20 undergraduate organic laboratory students synthesized, in duplicate, 38 new analogs. These were then purified by a single undergraduate research student. The article describing this endeavor, entitled “Distributed Drug Discovery, Part 3: Using D3 Methodology to Synthesize Analogs of an Anti-Melanoma Compound,” is the second article following this Perspective.(5)
D3 Enumeration and Virtual Catalogs
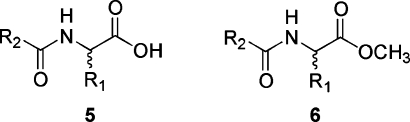
A key component of Distributed Drug Discovery is the open access availability of synthetically precedented virtual D3 catalogs for global computational modeling. This modeling process can identify subsets of molecules, capable of ready D3 synthesis, as potential developing world drug-leads. With documented D3 synthetic procedures now in place, we have constructed two of these virtual catalogs and are making them freely available to the worldwide community. The two catalogs are based on generic structures 5 and 6 (Figure 6).
Figure 6.
Generic Structures for the First Virtual D3 Catalogs.
The first, a 24 416 compound catalog based on acylated unnatural amino acids 5, is precedented by the work reported in “Distributed Drug Discovery, Part 2: Global Rehearsal of Alkylating Agents for the Synthesis of Resin-Bound Unnatural Amino Acids and Virtual D3 Catalog Construction.” This article immediately follows the Perspective.(4) The second, a 24 192 member catalog based on acylated unnatural amino acid methyl esters 6, is precedented by the work reported in the subsequent article, “Distributed Drug Discovery, Part 3: Using D3 Methodology to Synthesize Analogs of an Anti-Melanoma Compound.”(5) For enumeration of each catalog, 100 alkylating agents or Michael acceptors were used in the alkylation step (Scheme 2, 2−3), and 100 carboxylic acids (Scheme 2, 1−4 using carboxylic acids instead of acid chlorides) were used in the acylation step. The 24 416 theoretical acylated amino acids 5 would arise from cleavage of the resin-bound products 4 with TFA, while the 24 192 acylated amino acid methyl esters 6 would come from transesterification cleavage with methanol. These combined results afford a virtual D3 catalog of 48 608 molecules. Since the stereochemistry is not controlled in the alkylation step, both stereoisomers are obtained at the α-carbon. Additional stereoisomers, obtained when racemic or prochiral reactants are used, are each represented uniquely in the virtual D3 catalog. This stereochemical richness accounts for the greater number of compounds in the catalogs than would result from a simple 100 × 100 enumeration in each case.
All the reagents used in the enumeration of these catalogs are commercially available and their compatibility with the D3 synthetic procedures is either documented in our two subsequent articles or well-precedented in the literature. The enumerations were conducted using commercial software,(18b) and the complete virtual D3 catalog is freely available online through the Collaborative Drug Discovery (CDD) interface.12,44
Future Directions and Needs
We have identified six major goals to further the development of D3:
-
(1)
Cataloging, with global open-access, biochemical targets for developing world diseases, along with the identification of the molecule scaffolds (and functionalization) appropriate for binding to these targets.
-
(2)
Continued adaptation of solid- and solution-phase combinatorial methodologies, from the basic research level to D3 compatible equipment and procedures, to enable the simple, inexpensive distributed synthesis of large numbers of these structurally diverse potential drug-lead molecules.
-
(3)
Establishment of a network to facilitate the creation of additional virtual D3 catalogs based on this D3 chemistry, and to provide coordinated information sharing and analysis, with global links, to computational chemists, distributed synthetic chemists and biochemical screening resources.
-
(4)
Development and/or identification of computational models relevant to developing world and other neglected disease targets, and their utilization in the analysis of virtual D3 catalogs.
-
(5)
Development and implementation of D3 compatible biochemical screens appropriate for these disease targets.
-
(6)
Identification and establishment of the purification, registration, storage and sample submission resources required to expedite the testing and tracking of compounds made at global sites.
Summary
This Perspective describes, both conceptually and in practical embodiment, a project developed at IUPUI that we call Distributed Drug Discovery (D3). This cross-disciplinary program is grounded in the conviction that the major challenge of developing drug leads for neglected diseases can be addressed, at low cost and significant educational benefit, through a distributed combinatorial discovery process. This will be accomplished by dividing the computational, synthesis, and screening stages of drug discovery into smaller units and developing simple procedures and inexpensive equipment to permit students, worldwide, to be the problem solvers during their normal educational studies. This connects them, in their training, to an ultimate application of the skills and expertise they are learning. At the same time it enables the problem of drug-lead discovery to be economically addressed. Once drug leads are identified, other nonprofit initiatives can shepherd them through the numerous steps remaining to convert these leads into approved drugs, and make them available, at low cost, to those in need.
Distributed computation is now well documented in drug discovery. This Perspective focused on how combinatorial chemistry can be utilized to enable the virtual catalog and synthesis components of D3. When distributed screening methodologies are developed, the overall integrated process will be in place. The chemistry component of D3 envisions two students (perhaps one in an undergraduate organic laboratory in a developing world country and the other in a laboratory in the developed world) synthesizing, in duplicate, the lead molecule that is subsequently turned into a drug to treat malaria, AIDS, tuberculosis, leischmaniasis, trypanosomiasis, or some other disease widely affecting the developing world. It is our hope that D3 will be a unifying concept encouraging our colleagues throughout the world to join us in developing and harnessing distributed global resources for education, human development, and the discovery of drug leads for developing world and other neglected diseases.
Acknowledgments
We wish to thank Donald B. Boyd, Guillermo Morales, and Daniel H. Robertson for helpful discussions. We gratefully acknowledge Frank Jasper for the photograph of the Bill-Board equipment. We acknowledge the National Institutes of Health (R01 GM028193), The Camille and Henry Dreyfus Foundation, and the Lilly Research Laboratories for their financial support.
Funding Statement
National Institutes of Health, United States
References
- Disease Analysis/Prevalence:; a The World Health Report 2004: Changing History.http://www.who.int/whr/2004/en/ (accessed November 2, 2008).; b The World Health Report 2005: Make Every Mother and Child Count. http://www.who.int/whr/2005/en/ (accessed November 2, 2008).; c The World Health Report 2006: Working Together for Health. http://www.who.int/whr/2006/en/ (accessed November 2, 2008).; d Mathers C. D.; Loncar D. PLoS Med. 2006, 3, 2011–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]; e The World Health Report 2007: A Safer Future: Global Public Health Security in the 21st Century. http://www.who.int/whr/2007/en/ (accessed November 2, 2008).; f World Malaria Report 2008. http://www.who.int/malaria/wmr2008/ (accessed November 2, 2008).; g The World Health Report 2008: Primary Health Care—Now More Than Ever. http://www.who.int/whr/2008/en/index.html (accessed November 2, 2008).
- Focusing Attention on Developing World Diseases:; a Varmus H.; Klausner R.; Zerhouni E.; Acharya T.; Daar A. S.; Singer P. A. Science 2003, 302, 398–399. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yach D.; Hawkes C.; Gould C. L.; Hofman K. J. J. Am. Med. Assoc. 2004, 291, 2616–2622. [DOI] [PubMed] [Google Scholar]; c Birn A.-E. Lancet 2005, 366, 514–519. [DOI] [PubMed] [Google Scholar]; d Morel C. M.; Acharya T.; Broun D.; Dangi A.; Elias C.; Ganguly N. K.; Gardner C. A.; Gupta R. K.; Haycock J.; Heher A. D.; Hotez P. J.; Kettler H. E.; Keusch G. T.; Krattiger A. F.; Kreutz F. T.; Lall S.; Lee K.; Mahoney R.; Martinez-Palomo A.; Mashelkar R. A.; Matlin S. A.; Mzimba M.; Oehler J.; Ridley R. G.; Senanayake P.; Singer P.; Yun M. Science 2005, 309, 401–404. [DOI] [PubMed] [Google Scholar]; e Nwaka S.; Hudson A. Nat. Rev. Drug Discovery 2006, 5, 941–955. [DOI] [PubMed] [Google Scholar]; f Omenn G. S. Science 2006, 314, 1696–1704. [Google Scholar]; g O’Connell D. Nature 2007, 449, 157. [Google Scholar]; h Butler D. Nature 2007, 449, 158–159. [DOI] [PubMed] [Google Scholar]; i Callan B.; Gillespie I. Nature 2007, 449, 164–165. [DOI] [PubMed] [Google Scholar]; j Hopkins A. L.; Witty M. J.; Nwaka S. Nature 2007, 449, 166–169. [DOI] [PubMed] [Google Scholar]; k Yamada T. Nat. Rev. Drug Discovery 2007, 6, 950. [DOI] [PubMed] [Google Scholar]; l McNeil D. G.,. Jr. Jump-Start on Slow Trek to Treatment for a Disease. The New York Times, January 8, 2008, pp D5−D7 [Google Scholar]; m Open-Access Public Library of Science (PloS) Journal PloS Neglected Tropical Diseases. http://www.plosntds.org/home.action (accessed November 2, 2008
- The concept of “Distributed Drug Discovery” was first presented by WLS at a seminar at the Department of Biochemistry, Biophysics and Biotechnology at the Jagiellonian University in Kraków, Poland on April 7, 2003.
- See first article following this Perspective:; Scott W. L.; Alsina J.; Audu C. O.; Babaev E.; Cook L.; Dage J. L.; Goodwin L. A.; Martynow J. G.; Matosiuk D.; Royo M.; Smith J. G.; Strong A. T.; Wickizer K.; Woerly E. M.; Zhou Z.; O’Donnell M. J. J. Comb. Chem. 2009, 11, 14–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See second article following this Perspective:Scott W. L.; Audu C. O.; Dage J. L.; Goodwin L. A.; Martynow J. G.; Platt L. K.; Smith J. G.; Strong A. T.; Wickizer K.; Woerly E. M.; O’Donnell M. J. J. Comb. Chem. 2009, 11, 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little Economic Incentive:; a Reich M. R. Science 2000, 287, 1979–1981. [DOI] [PubMed] [Google Scholar]; b Yamey G. Brit. Med. J. 2002, 325, 176–177.12142292 [Google Scholar]; c Pécoul B. PLOS Med. 2004, 1, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Sharp D. Lancet 2004, 364, 1472–1474. [DOI] [PubMed] [Google Scholar]; e Hale V. G.; Woo K.; Lipton H. L. Health Affairs 2005, 24, 1057–1063. [DOI] [PubMed] [Google Scholar]; f White N. J. Trop. Med. Int. Health 2006, 11, 383–384. [DOI] [PubMed] [Google Scholar]
- Non-Profits and Public Private Partnerships:; a Institute for OneWorld Health. http://oneworldhealth.org/ (accessed November 2,2008).; b Walgate R. Bull. World Health Org. 2002, 80, 842–843. [PMC free article] [PubMed] [Google Scholar]; c Moran M. PLOS Med. 2005, 2, 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Groopman J. Buying a Cure. The New Yorker, Jan 28, 2008, pp38−43 [PubMed] [Google Scholar]
- Distributed Computation in Drug Discovery:; a Chien A.; Foster I.; Goddette D. Drug Discovery Today 2002, 7, S176–S180. [DOI] [PubMed] [Google Scholar]; b Richards W. G. Nat. Rev. 2002, 1, 551–555. [DOI] [PubMed] [Google Scholar]; c Buyya R.; Branson K.; Giddy J.; Abramson D. Concurrency Computat.: Pract. Exp. 2003, 15, 1–25. [Google Scholar]; d Richards W. G.; Grant G. H.; Harrison K. N. J. Mol. Graphics Model. 2004, 22, 473–478. [DOI] [PubMed] [Google Scholar]; e Birkholtz L.-M.; Bastien O.; Wells G.; Grando D.; Joubert F.; Kasam V.; Zimmermann M.; Ortet P.; Jacq N.; Saïdani N.; Roy S.; Hofmann-Apitius M.; Breton V.; Louw A. I.; Maréchal E. Malar. J. 2006, 5, 110–134. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Jiang Z.; Lu F.; Zhang J. J. Drug Targeting 2006, 14, 623–631. [DOI] [PubMed] [Google Scholar]; g Sild S.; Maran U.; Lomaka A.; Karelson M. J. Chem. Inf. Model. 2006, 46, 953–959. [DOI] [PubMed] [Google Scholar]; h Gledhill R.; Kent S.; Hudson B.; Richards W. G.; Essex J. W.; Frey J. G. J. Chem. Inf. Model. 2006, 46, 960–970. [DOI] [PubMed] [Google Scholar]; i Spreading the Load. The Economist (Technology Quarterly), Dec 8, 2007, pp19−21 [Google Scholar]
- a Grid.org web site. http://www.grid.org/ (accessed November 2, 2008).; b World Community Grid web site. http://www.worldcommunitygrid.org/ (accessed November 2, 2008).; c SETI project web site. http://setiathome.berkeley.edu/ (accessed November 2, 2008).
- NIH Molecular Libraries Initiative. http://nihroadmap.nih.gov/molecularlibraries/ (accessed November 2, 2008).; a Austin C. P.; Brady L. S.; Insel T. R.; Collins F. S. Science 2004, 306, 1138–1139. [DOI] [PubMed] [Google Scholar]; b Livingston D.; Lease T. Behind the Scenes at the NIH Molecular Libraries Small Molecule Repository. Society for Biomolecular Sciences News, December, 2005, pp 1−4 [Google Scholar]; c Kaiser J. Science 2008, 321, 764–766. [DOI] [PubMed] [Google Scholar]
- Open Source Research:; a Taylor G.Open Source Biomedical Research for the 21st Century. The Synaptic Leap:http://thesynapticleap.org/ (accessed November 2, 2008).; b Kepler T. B.; Marti-Renom M. A.; Maurer S. M.; Rai A. K.; Taylor G.; Todd M. H. Aust. J. Chem. 2006, 59, 291–294. [Google Scholar]; c Chokshi D. A.; Parker M.; Kwiatkowski D. P. Bull. World Health Org. 2006, 84, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Wild D. J.; Wiggins G. D. Drug Discovery Today 2006, 11, 436–439. [DOI] [PubMed] [Google Scholar]; e Munos B. Nat. Rev. Drug Discovery 2006, 5, 723–729. [DOI] [PubMed] [Google Scholar]; f Edwards A. Drug Discovery Today 2008, 13, 731–733. [DOI] [PubMed] [Google Scholar]
- Collaborative Drug Discovery, Inc. (CDD). http://www.collaborativedrug.com/ (accessed November 2, 2008).
- Intellectual Property and Patent Issues:; a Benkler Y. Science 2004, 305, 1110–1111. [DOI] [PubMed] [Google Scholar]; b Triggle D. J. Drug Dev. Res. 2005, 63, 139–149. [Google Scholar]; c Benatar S. R. PLoS Med. 2005, 2, 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Everts S. Chem. Eng. News 2006, 84 (30), 34–35. [Google Scholar]; e Mintzberg H. Can. Med. Assoc. J. 2006, 175, 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Lipinski C. A. Curr. Opin. Chem. Biol. 2006, 10, 380–383. [DOI] [PubMed] [Google Scholar]; g Winters D. J. Bull. World Health Org. 2006, 84, 414–416. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Herrling P. Nature 2007, 449, 174–175. [DOI] [PubMed] [Google Scholar]; i Breitstein, J. R&D: Learning to Share. PharmExec.Com. http://pharmexec.findpharma.com/pharmexec/Articles/RampD-Learning-to-Share/ArticleStandard/Article/detail/483069?contextCategoryId=39724&ref=25/ (accessed November 2, 2008).
- “Give a man a fish and you feed him for a day. Teach a man to fish and you feed him for a lifetime”, Maimonides, Spanish philosopher (1135−1204).
- Antibodies and the Immune System:; a Schultz P. G.; Lerner R. A. Science 1995, 269, 1835–1842. [DOI] [PubMed] [Google Scholar]; b Marchalonis J. J.; Kaveri S.; Lacroix-Desmazes S.; Kazatchkine M. D. FASEB J. 2002, 16, 842–848. [DOI] [PubMed] [Google Scholar]
- Combinatorial Chemistry Issues:; a Lahana R. Drug Discovery Today 1999, 4, 447–448. [DOI] [PubMed] [Google Scholar]; b Ramesha C. S. Drug Discovery Today 2000, 5, 43–44. [DOI] [PubMed] [Google Scholar]; c Borman S. Chem. Eng. News 2004, 82 (40), 32–40. [Google Scholar]; d Klebe G. Drug Discovery Today 2006, 11, 580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Kennedy J. P.; Williams L.; Bridges T. M.; Daniels R. N.; Weaver D.; Lindsley C. W. J. Comb. Chem. 2008, 10, 345–354. [DOI] [PubMed] [Google Scholar]
- Virtual Libraries:; a Leach A. R.; Hann M. M. Drug Discovery Today 2000, 5, 326–336. [DOI] [PubMed] [Google Scholar]; b Barnard J. M.; Downs G. M.; Scholley-Pfab A. v.; Brown R. D. J. Mol. Graph. Model. 2000, 18, 452–463. [PubMed] [Google Scholar]; c Lobanov V. S.; Agrafiotis D. K. J. Mol. Graph. Model. 2001, 19, 571–578. [DOI] [PubMed] [Google Scholar]; d Lobanov V. S.; Agrafiotis D. K. Comb. Chem. High Throughput Screening 2002, 5, 167–178. [DOI] [PubMed] [Google Scholar]; e Green D. V. S.; Pickett S. D. Mini-Rev. Med. Chem. 2004, 4, 1067–1076. [DOI] [PubMed] [Google Scholar]
- Enumeration Packages:; a ChemAxon (free academic license available) from ChemAxon Kft., Máramaros köz 3/a, Budapest, 1037 Hungary. Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions (Marvin 5.1.02, ChemAxon. http://www.chemaxon.com (accessed November 2, 2008)).; b CombiChem/Excel from CambridgeSoft. http://www.cambridgesoft.com/ (accessed November 2, 2008).
- Virtual Drug Discovery:; a Nwaka S.; Ridley R. G. Nat. Rev. Drug Discovery 2003, 2, 919–928. [DOI] [PubMed] [Google Scholar]; b Lipinski C.; Hopkins A. Nature 2004, 432, 855–861. [DOI] [PubMed] [Google Scholar]; c Chin D. N.; Chuaqui C. E.; Singh J. Mini-Rev. Med. Chem. 2004, 4, 1053–1065. [DOI] [PubMed] [Google Scholar]
- The term “rehearsal” is often used to describe the practice of conducting a trial reaction of every reagent candidate at a particular diversity step in a complete combinatorial scheme. This process eliminates clearly problematic reagents and increases the confidence that any member of a virtual library based on these rehearsed reagents can be successfully synthesized.
- WISDOM Project for Malaria:; a WISDOM: Initiative for Grid-Enabled Drug Discovery Against Neglected and Emergent Diseases. http://wisdom.eu-egee.fr/ (accessed November 2, 2008).; b From Sheffield to Singapore, International Computing Grid Battles Malaria. Science Daily, February 2, 2007; http://www.sciencedaily.com/releases/2007/01/070131090602.htm (accessed November 2, 2008).; c More than 4.3 Million Malaria Medicines Tested Thanks to Calculation Grids. Medical News Today, February 13, 2007; http://www.medicalnewstoday.com/medicalnews.php?newsid=62700 (accessed November 2, 2008).
- Rehearsal example:El-Araby M.; Guo H.; Pottorf R. S.; Player M. R. J. Comb. Chem 2004, 6, 789–795. [DOI] [PubMed] [Google Scholar]
- Unfortunately, the term “false positive” is often used when the principal, desired component of a sample is not confirmed as the active compound. In fact, the screen may have accurately shown activity in the test solution, but from a minor, even “undesired,” compound. In such cases, bioassay-guided fractionation follow-up can be used to assist in identifying a “true positive” present in the test solution. Notably, when a minor compound is the active component, the level of activity will be considerably higher following purification and retesting. Alternatively, bioassay-guided fractionation can be used to identify a “false-positive” sample that should not be pursued.
- High Throughput Purification:; a Yan B.; Fang L.; Irving M.; Zhang S.; Boldi A. M.; Wollard F.; Johnson C. R.; Kshirsagar T.; Figliozzi G. M.; Krueger C. A.; Collins N. J. Comb. Chem 2003, 5, 547–559. [DOI] [PubMed] [Google Scholar]; b Popa-Burke I. G.; Issakova O.; Arroway J. D.; Bernasconi P.; Chen M.; Coudurier L.; Galasinski S.; Jadhav A. P.; Janzen W. P.; Lagasca D.; Liu D.; Lewis R. S.; Mohney R. P.; Sepetov N.; Sparkman D. A.; Hodge C. N. Anal. Chem. 2004, 76, 7278–7287. [DOI] [PubMed] [Google Scholar]; c Isbell J. J.; Zhou Y.; Guintu C.; Rynd M.; Jiang S.; Petrov D.; Micklash K.; Mainquist J.; Ek J.; Chang J.; Weselak M.; Backes B. J.; Brailsford A.; Shave D. J. Comb. Chem. 2005, 7, 210–217. [DOI] [PubMed] [Google Scholar]; d Isbell J. J. Comb. Chem. 2008, 10, 150–157. [DOI] [PubMed] [Google Scholar]
- Combinatorial Chemistry with Mixtures:Houghten R. A.; Pinilla C.; Giulianotti M. A.; Appel J. R.; Dooley C. T.; Nefzi A.; Ostresh J. M.; Yu Y.; Maggiora G. M.; Medina-Franco J. L.; Brunner D.; Schneider J. J. Comb. Chem 2008, 10, 3–19. [DOI] [PubMed] [Google Scholar]
- Natural Products and Drug Discovery:; a Butler M. S. J. Nat. Prod 2004, 67, 2141–2153. [DOI] [PubMed] [Google Scholar]; b Newman D. J.; Cragg G. M. J. Nat. Prod. 2007, 70, 461–477. [DOI] [PubMed] [Google Scholar]
- This potential is demonstrated by the numerous examples of drugs originating from natural products, in which the active molecule or lead was present at a level of less than 1% of the initial tested sample. Unfortunately, these natural product derived tools, such as bioassay-guided fractionation,(28) have not always been fully utilized in combinatorial chemistry, resulting in “false positives” (sometimes true positives, just not the intended molecule!) and statements such as “garbage in, garbage out”. It is noted, with some irony, that one of the most important classes of antibiotics, cephalosporins, was discovered as a minor component in a sewage outfall off the coast of Sardinia, see:; a Brotzu G. Lavori Instituto Igiene Cagliari 1948, 1. [Google Scholar]; b Mandell G. L.; Sande M. A.. Antimicrobial Agents. In The Pharmacological Basis of Therapeutics, 8th ed; Gilman A. G., Rall T. W., Nies A. S., Taylor P., Eds.; Pergamon Press: New York, 1990; p 1085. [Google Scholar]
- Bio-Assay Guided Fractionation:; a Phillipson D. W.; Milgram K. E.; Yanovsky A. I.; Rusnak L. S.; Haggerty D. A.; Farrell W. P.; Greig M. J.; Xiong X.; Proefke M. L. J. Comb. Chem 2002, 4, 591–599. [DOI] [PubMed] [Google Scholar]; b Peake D. A.; Duckworth D. C.; Perun T. J.; Scott W. L.; Kulanthaivel P.; Strege M. A. Comb. Chem. High Throughput Screening 2005, 8, 477–487. [DOI] [PubMed] [Google Scholar]; c Inglese J.; Auld D. S.; Jadhav A.; Johnson R. L.; Simeonov A.; Yasgar A.; Zheng W.; Austin C. P. Proc. Nat. Acad. Sci. U. S. A. 2006, 103, 11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The two following articles in this issue of Journal of Combinatorial Chemistry(4,5) illustrate how a collaboration with industry makes this possible. In these cases Eli Lilly and Company donated their analytical resources by incorporating, when time was available, the LC/MS analysis into the daily work schedule.
- The Distributed Drug Discovery fund is administered by the Central Indiana Community Foundation, 615 North Alabama Street, Suite 119, Indianapolis, Indiana 46204.
- References from the authors’ laboratory concerning the solid-phase synthesis of unnatural amino acids, peptides, and peptidomimetics:; a O’Donnell M. J.; Zhou C.; Scott W. L. J. Am. Chem. Soc. 1996, 118, 6070–6071. [Google Scholar]; b Scott W. L.; Zhou C.; Fang Z.; O’Donnell M. J. Tetrahedron Lett. 1997, 38, 3695–3698. [Google Scholar]; c O’Donnell M. J.; Lugar C. W.; Pottorf R. S.; Zhou C.; Scott W. L.; Cwi C. L. Tetrahedron Lett. 1997, 38, 7163–7166. [Google Scholar]; d Griffith D. L.; O’Donnell M. J.; Pottorf R. S.; Scott W. L.; Porco J. A. Jr. Tetrahedron Lett. 1997, 38, 8821–8824. [Google Scholar]; e Domínguez E.; O’Donnell M. J.; Scott W. L. Tetrahedron Lett. 1998, 39, 2167–2170. [Google Scholar]; f O’Donnell M. J.; Delgado F.; Drew M. D.; Pottorf R. S.; Zhou C.; Scott W. L. Tetrahedron Lett. 1999, 40, 5831–5835. [Google Scholar]; g O’Donnell M. J.; Delgado F.; Pottorf R. S. Tetrahedron 1999, 55, 6347–6362. [Google Scholar]; h O’Donnell M. J.; Drew M. D.; Pottorf R. S.; Scott W. L. J. Comb. Chem. 2000, 2, 172–181. [DOI] [PubMed] [Google Scholar]; i O’Donnell M. J.; Scott W. L.. Unnatural Amino Acid and Peptide Synthesis (UPS). In Peptides 2000; Proceedings of the 26th European Peptide Symposium; Martinez J., Fehrentz J.-A., Eds.; EDK: Paris, 2001; pp 31−36 [Google Scholar]; j O’Donnell M. J.; Delgado F.; Domínguez E.; de Blas J.; Scott W. L. Tetrahedron: Asymmetry 2001, 12, 821–828. [Google Scholar]; k Scott W. L.; Delgado F.; Lobb K.; Pottorf R. S.; O’Donnell M. J. Tetrahedron Lett. 2001, 42, 2073–2076. [Google Scholar]; l Scott W. L.; O’Donnell M. J.; Delgado F.; Alsina J. J. Org. Chem. 2002, 67, 2960–2969. [DOI] [PubMed] [Google Scholar]; m Scott W. L.; Alsina J.; O’Donnell M. J. J. Comb. Chem. 2003, 5, 684–692. [Google Scholar]; n O’Donnell M. J.; Alsina J.; Scott W. L. Tetrahedron Lett. 2003, 44, 8403–8406. [Google Scholar]; o Scott W. L.; Alsina J.; Kennedy J. H.; O’Donnell M. J. Org. Lett. 2004, 6, 1629–1632. [DOI] [PubMed] [Google Scholar]; p Alsina J.; Scott W. L.; O’Donnell M. J. Tetrahedron Lett. 2005, 46, 3131–3135. [Google Scholar]; q Scott W. L.; Martynow J. G.; Huffman J. C.; O’Donnell M. J. J. Am. Chem. Soc. 2007, 129, 7077–7088. [DOI] [PubMed] [Google Scholar]
- The α-amino acid substructure occurs in 28% of the structures cited in a recent “Comprehensive Survey of Library Synthesis.” Search strategy: nitrogen-carbon-carbonyl substructure contained in 103 of the 365 structures in Tables 1−10 in ref (33i).
- For an excellent series of structurally based comprehensive reviews on combinatorial library syntheses, see:; a Dolle R. E. Mol. Diversity 1998, 4, 233–256. [DOI] [PubMed] [Google Scholar]; b Dolle R. E.; Nelson K. H. Jr. J. Comb. Chem. 1999, 1, 235–282. [DOI] [PubMed] [Google Scholar]; c Dolle R. E. J. Comb. Chem. 2000, 2, 383–433. [DOI] [PubMed] [Google Scholar]; d Dolle R. E. J. Comb. Chem. 2001, 3, 477–517. [DOI] [PubMed] [Google Scholar]; e Dolle R. E. J. Comb. Chem. 2002, 4, 369–418. [DOI] [PubMed] [Google Scholar]; f Dolle R. E. J. Comb. Chem. 2003, 5, 693–753. [DOI] [PubMed] [Google Scholar]; g Dolle R. E. J. Comb. Chem. 2004, 6, 623–679. [DOI] [PubMed] [Google Scholar]; h Dolle R. E. J. Comb. Chem. 2005, 7, 739–798. [DOI] [PubMed] [Google Scholar]; i Dolle R. E.; Le Bourdonnec B.; Morales G. A.; Moriarty K. J.; Salvino J. M. J. Comb. Chem. 2006, 8, 597–635. [DOI] [PubMed] [Google Scholar]; j Dolle R. E.; Le Bourdonnec B.; Goodman A. J.; Morales G. A.; Salvino J. M.; Zhang W. J. Comb. Chem. 2007, 9, 855–902. [DOI] [PubMed] [Google Scholar]; k Dolle R. E.; Le Bourdonnec B.; Goodman A. J.; Morales G. A.; Thomas C. J.; Zhang W. J. Comb. Chem. 2008, 10, 753–802. [DOI] [PubMed] [Google Scholar]
- Peptides:; a Merrifield R. B. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar]; b Merrifield B. Science 1986, 232, 341–347. [DOI] [PubMed] [Google Scholar]
- Benzodiazepines:; a Bunin B. A.; Ellman J. A. J. Am. Chem. Soc. 1992, 114, 10997–10998. [Google Scholar]; b DeWitt S. H.; Kiely J. S.; Stankovic C. J.; Schroeder M. C.; Reynolds Cody D. M.; Pavia M. R. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 6909–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kamal A.; Reddy K. L.; Devaiah V.; Shankaraiah N.; Reddy D. R. Mini-Rev. Med. Chem. 2006, 6, 53–69. [DOI] [PubMed] [Google Scholar]
- 1,4-Benzodiazepine-2,5-diones:; a Mayer J. P.; Zhang J.; Bjergarde K.; Lenz D. M.; Gaudino J. J. Tetrahedron Lett. 1996, 37, 8081–8084. [Google Scholar]; b Boojamra C. G.; Burow K. M.; Thompson L. A.; Ellman J. A. J. Org. Chem. 1997, 62, 1240–1256. [Google Scholar]
- Hydantoins:Meusel M.; Gütschow M. Org. Prep. Proced. Int. 2004, 36, 391–443. [Google Scholar]
- α-Substituted Prolines:Murphy M. M.; Schullek J. R.; Gordon E. M.; Gallop M. A. J. Am. Chem. Soc. 1995, 117, 7029–7030. [Google Scholar]
- Diketopiperazines:; a Gordon D. W.; Steele J. Bioorg. Med. Chem. Lett. 1995, 5, 47–50. [Google Scholar]; b Scott B. O.; Siegmund A. C.; Marlowe C. K.; Pei Y.; Spear K. L. Mol. Diversity 1995, 1, 125–134. [DOI] [PubMed] [Google Scholar]; c Dinsmore C. J.; Beshore D. C. Tetrahedron 2002, 58, 3297–3312. [Google Scholar]; d Fischer P. M. J. Peptide Sci. 2003, 9, 9–35. [DOI] [PubMed] [Google Scholar]; e Martins M. B.; Carvalho I. Tetrahedron 2007, 63, 9923–9932. [Google Scholar]
- NSF Workshop “Solid Phase Synthesis and an Introduction to Combinatorial Chemistry.” Ketcha, D. M.; Taylor, R. T. Miami University, Oxford, Ohio, July 27−August 1, 2003. The workshop was sponsored by the Center for Workshops in the Chemical Sciences (CWCS), Georgia State University, Smith, J. C., Director (award number: DUE-0341138).
- Scott, W. L.; Schonegg, R. A.; Cwi, L. Vessel Handling System Useful for Combinatorial Chemistry. U.S. Patent 5,785,927, July 28, 1998.
- Leads Metal Products, PO Box 441186, Indianapolis, IN, 46244−1186 (larry@leadsmetal.com).
- Dothager R. S.; Putt K. S.; Allen B. J.; Leslie B. J.; Nesterenko V.; Hergenrother P. J. J. Am. Chem. Soc. 2005, 127, 8686–8696. [DOI] [PubMed] [Google Scholar]
- a For access to the IUPUI—Distributed Drug Discovery (D3) database, register for a free read-download account with Collaborative Drug Discovery (CDD) at https://www.collaborativedrug.com/register/iupui-d3 (accessed November 2, 2008) by completing the “Sign up for IUPUI—Distributed Drug Discovery (D3)” information. (b) A tutorial for use of the IUPUI—Distributed Drug Discovery (D3) database on CDD is provided: (i) in the Supporting Information for the following two articles,4,5 (ii) from the D3 database on the CDD website, and (iii) on the IUPUI Department of Chemistry and Chemical Biology website [http://chem.iupui.edu/ (accessed November 2, 2008)] under the faculty & staff directory for O’Donnell or Scott. The tutorials available at ii and iii will be updated periodically.