Abstract
From mutants of Escherichia coli unable to utilize fructose via the phosphoenolpyruvate/glycose phosphotransferase system (PTS), further mutants were selected that grow on fructose as the sole carbon source, albeit with relatively low affinity for that hexose (Km for growth ≈8 mM but with Vmax for generation time ≈1 h 10 min); the fructose thus taken into the cells is phosphorylated to fructose 6-phosphate by ATP and a cytosolic fructo(manno)kinase (Mak). The gene effecting the translocation of fructose was identified by Hfr-mediated conjugations and by phage-mediated transduction as specifying an isoform of the membrane-spanning enzyme IIGlc of the PTS, which we designate ptsG-F. Exconjugants that had acquired ptsG+ from Hfr strains used for mapping (designated ptsG-I) grew very poorly on fructose (Vmax ≈7 h 20 min), even though they were rich in Mak activity. A mutant of E. coli also rich in Mak but unable to grow on glucose by virtue of transposon-mediated inactivations both of ptsG and of the genes specifying enzyme IIMan (manXYZ) was restored to growth on glucose by plasmids containing either ptsG-F or ptsG-I, but only the former restored growth on fructose. Sequence analysis showed that the difference between these two forms of ptsG, which was reflected also by differences in the rates at which they translocated mannose and glucose analogs such as methyl α-glucoside and 2-deoxyglucose, resided in a substitution of G in ptsG-I by T in ptsG-F in the first position of codon 12, with consequent replacement of valine by phenylalanine in the deduced amino acid sequence.
Escherichia coli grow readily on fructose as the sole source of carbon. When this sugar is present in the growth medium at concentrations ≤2 mM, it is taken up and phosphorylated predominantly to fructose 1-phosphate (1) via two membrane-associated proteins specified by fruA (2) and fruB (3); at concentrations >2 mM, it also is taken up and phosphorylated (but to fructose 6-phosphate) via the membrane-associated uptake system for mannose, specified by manXYZ (4, 5). Strains of E. coli impaired in both these systems have been shown to mutate further and to take up and phosphorylate fructose to fructose 6-phosphate via derepression of glucitol transport proteins (6). In all of these instances, the simultaneous translocation and phosphorylation of fructose requires the action of the phosphoenolpyruvate/glycose phosphotransferase system (PTS) (7–9).
It has been shown that Salmonella mutants lacking one or more of the cytosolic proteins of the PTS, which consequently are unable to use any of the sugars normally taken up via the PTS, can give rise to further mutants that are able to grow on fructose as the sole carbon source (9). Such mutants also had elevated levels of fructo(manno)kinase (Mak) (10), which implies that fructose entered the cells in an unphosphorylated form and only thereafter received a phosphate group from ATP. But it was not known how fructose was translocated across the cell envelope: the available evidence was interpreted (8) as ruling out involvement of any enzyme II of the PTS as well as of the galactose permease, which is known to be able also to transport sugars other than galactose.
We here report the isolation and properties of fructose-positive E. coli mutants from strains in which the utilization of fructose by PTS-mediated routes was abolished by deletions in the fructose and glucitol operons, as well as by insertion of a chloramphenicol-resistance transposon into manXYZ. That the PTS was not involved in the growth on fructose of such mutants was confirmed by further insertion of a [ΔptsH ptsI crr]KanR cassette (11): this did not alter the fructose-positive phenotype. We show that fructose enters the cells via an isoform of enzyme IIGlc specified by a ptsG gene in which the G at position 34 in the published E. coli nucleotide sequence (12) has been replaced by T; mutants not containing this isoform take up fructose to only a negligible extent. We designate the former as ptsG-F and the latter as ptsG-I.
Materials and Methods
Strains, Plasmids, and Growth Conditions.
The bacterial strains used are listed in Table 1. Bacteria were grown aerobically at 37°, either in liquid culture consisting of LB broth (25 g of LB base per liter of water) or in defined media (13), or on such media solidified with 2% agar.
Table 1.
Bacterial strains used in this study
| Strain | Relevant genotype | PtsG isoform | Source (ref.) |
|---|---|---|---|
| BW 6160 | Hfr (PO 118 of Broda 8) zdi57∷Tn10 | I | 21 |
| BW 7622 | Hfr (PO 4 of KL 96) trp∷Tn10 | I | 21 |
| HK 1691 | fruA ΔfruK manXYZ∷CmR rpsL | F | This work |
| HK 1787 | as HK 1691 but Mak+ | F | This work |
| HK 2140 | as HK 1691 and ΔgutA | F | This work |
| HK 2148 | [P1(2200) × 2140]Fru+ | F | This work |
| HK 2190 | as HK 2148 but ptsG∷CmR | — | This work |
| HK 2200 | as HK 1787 and [ΔptsH ptsl crr] KanR | F | This work |
| HK 2225 | as HK 2240 and [ΔptsH ptsl crr] KanR | I | This work |
| HK 2237 | [P1(2148) × 2190]Glc+ | F | This work |
| HK 2240 | [P1(BW 6160) × 2190] Glc+ | I | This work |
A 254-bp deletion, which removed amino acids 139–392 from the FruK protein but maintained the reading frame of the gene, (designated Δfruk in this work), was constructed as described in ref. 14; a 504-bp deletion in the gutA gene(15) (designated ΔgutA in this work) was constructed as described in ref. 16. Procedures used for Hfr-mediated conjugations and phage-mediated transductions are described in ref. 17 and for the uptake of [14C]fructose in ref. 18: this isotopic sugar was obtained from DuPont/NEN.
Manipulations of DNA fragments and agarose gel electrophoresis were performed as described in ref. 19. Genomic DNA was prepared with the aid of a QIAmp Tissue kit (Qiagen, Chatsworth, CA), following the manufacturer's instructions; VENT polymerase (New England Biolabs) was used in PCRs and DNA thus amplified was gel-purified with a QIAquick Gel Extraction kit (Qiagen). Plasmid DNA was isolated and purified with a Wizard Plus SV Miniprep Purification system (Promega); restriction enzymes and T4 DNA ligase were from New England Biolabs and were used as recommended by the supplier.
Mak activity was measured in extracts of cells that had been disrupted by sonic oscillation and clarified by centrifugation for 5 min at 9,000 × g by using the method described in ref. 10.
Sequencing reactions were carried out with the use of the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin–Elmer), as recommended by the manufacturer. Reaction products were purified on Centri-Sep spin columns (Princeton Separations). Samples were analyzed by the Boston University Sequencing Facility on an ABI 377XL DNA sequencer (Perkin–Elmer).
Results
E. coli Mutants Unable To Take up Fructose via the PTS Give Rise to Further Mutants that Grow on Fructose.
In the course of studies of the routes by which fructose enters E. coli cells and is used for growth, we isolated a mutant (HK 1691) in which the normal PTS-linked modes of fructose uptake were abolished by the introduction of a point mutation in the gene specifying the enzyme II for fructose (fruA), a deletion in the gene specifying 1-phosphofructokinase (ΔfruK), and insertion of a chloramphenicol-resistance transposon into the genes specifying the uptake and utilization of mannose (manXYZ∷CmR) (Table 1). When spread on agar plates containing 40 mM fructose as the sole carbon source and incubated at 37° for several days, HK 1691 gave rise to further mutants that grew on this hexose, albeit perceptibly more slowly than did wild-type E. coli. One such fructose-positive pseudorevertant (HK 1787) was isolated for further study.
That the general soluble proteins of the PTS (enzyme I, HPr, and enzyme IIAGlc) were not involved in this mode of fructose utilization was confirmed with a derivative (HK 2200) that, through insertion of a [ΔptsH ptsI crr]KanR transposon (11), also lacked these proteins. Like its parent organism, this strain grew in liquid culture with fructose as the sole carbon source at rates strongly dependent on the substrate concentration in the medium: half-maximal rate of growth was not achieved until the fructose concentration exceeded 8 mM, with a doubling time at infinite substrate concentration (Vmax for generation time) calculated to be ≈1 h 10 m. It had been observed previously (9, 20) that growth on fructose in the absence of PTS function was associated with derepression of Mak: we also found that the specific activity of this enzyme in sonic extracts of HK 1787 and HK 2200 was 15–20 times greater than that in similar extracts of the fructose-negative strain HK 1691. As was expected from the postulated role of Mak, the phage-mediated introduction into strain HK 1787 of a pfkA mutation, which prevented the ATP-dependent conversion of fructose 6-phosphate to fructose 1,6-bisphosphate, abolished growth on fructose.
However, for fructose utilization to occur via this ATP-linked cytosolic enzyme, some agent must facilitate the entry of fructose into the cells. Neither the location of the gene for this translocating agent nor of that for Mak have yet been reported. To locate them, an F− mutant (HK 2140) was constructed that carried fruA ΔfruK and manXYZ∷CmR and in which also the gene specifying the enzyme II for glucitol transport (15) was deleted (ΔgutA). A culture of this strain was infected with bacteriophage grown on HK 2200, and transductants were selected on medium containing 40 mM fructose as the sole carbon source. The Mak activity in sonic extracts of such transductants was found to be 17–25 times higher than that of the donor strain, which showed that it was the gene specifying this activity that had been transferred by the phage; it also indicated that the agent that facilitated the entry of fructose was already present in the recipient cells. One such transductant (HK 2148) was used for further study.
Location of the Gene Facilitating Fructose Uptake.
To determine where on the chromosome of E. coli genes might lie that would affect growth on fructose, the F− streptomycin-resistant strain HK 2148 was crossed with a variety of Hfr strains, each of which was streptomycin-sensitive and contained the tetracycline-resistance transposon Tn10 integrated at known sites (21) and which therefore enabled exconjugants to be selected, on plates containing tetracycline and streptomycin, after transfer of relatively small regions of the genome. Recombinants that had virtually lost the ability to grow on fructose were found in only two regions of the E. coli linkage map (22). Those derived from crosses in which the Hfr strain BW 6160 [point of origin (PO ≈ minute 8, clockwise)] was the donor had only low Mak activity, whereas those that had received DNA from the Hfr strain BW 7622 (PO [≈ minute 45, counterclockwise) had retained the high Mak activity of HK 2148. Analysis of the former type will be reported in a later paper.
The virtual loss of ability to grow on fructose despite high Mak activity suggested that ptsG, located at minute 24.9 (22), or a gene close to it, might be involved in fructose translocation. Evidence to this effect was provided by the introduction into HK 2148 of ptsG∷CmR, a chloramphenicol-resistance transposon integrated into ptsG. Because that recipient already contained a chloramphenicol-resistance transposon integrated into manXYZ, this introduction required two steps: initially, strain HK 2148 was infected with phage carrying pyrC∷Tn10, which introduces a requirement for uracil and is known to be ≈10% cotransducible with ptsG; uracil+ transductants were selected therefrom after infection with phage carrying ptsG∷CmR. Whereas 40 of 45 such transductants grew well on glucose and on fructose, five had simultaneously lost the ability to grow on either hexose; one of these (HK 2190) was used for further study. This finding suggested that the isoform of ptsG+ into which the CmR transposon had been introduced was involved in the transport of both sugars.
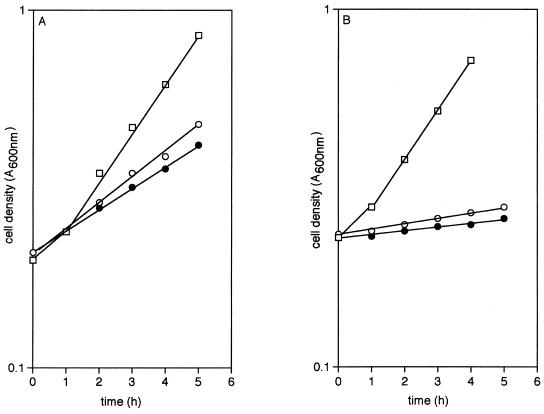
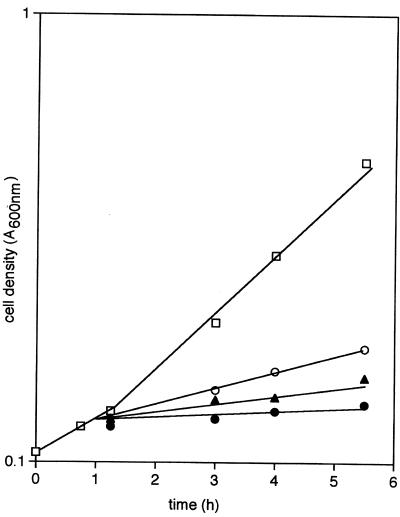
Confirmation of this hypothesis was obtained in a number of ways. When phage grown on a ptsG+ donor that was also fructose-positive was used to infect HK 2190 and transductants were selected on glucose, all were found to be able to grow also on fructose; in contrast, infection of HK 2190 with phage grown on a ptsG+ donor that was unable to take up fructose yielded glucose-positive transductants that grow only poorly on fructose. Because both types of transductant were also devoid of the PTS-dependent mannose permease specified by manXYZ, the growth of the fructose-positive strain on mannose (Fig. 1A) and the virtual inability of the fructose-negative transductant to do so (Fig. 1B) confirms (23, 24) that E. coli, unlike Salmonella typhimurium (25), can admit both of these sugars via the PtsG protein but also suggests that only one isoform of PtsG (which we designate ptsG-F) does so readily.
Figure 1.
The growth of strains containing ptsG–F (A) and ptsG –I (B), but otherwise isogenic, on 10 mM glucose (□), 10 mM mannose (○), or 20 mM fructose (●).
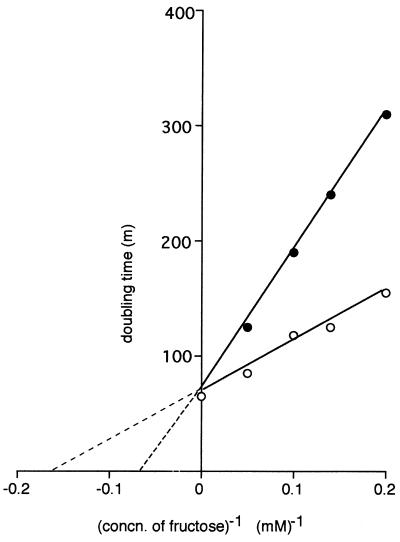
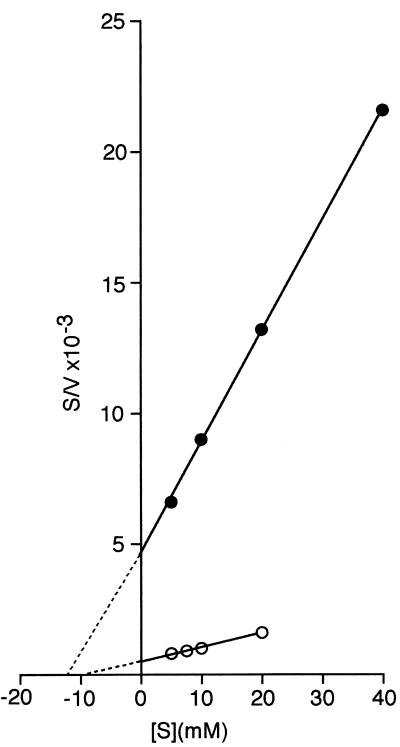
After introduction of the [ΔptsH ptsI crr]KanR cassette into such a fructose-negative transductant (yielding strain HK 2225), a direct estimate could be made of the kinetics of facilitated fructose diffusion in the presence of Mak but in the absence of the cytoplasmic proteins of the PTS (Fig. 2). It is evident that, although fructose can diffuse into either strain, the rates at which this process occurs differ greatly. The doubling times at infinite fructose concentration (Vmax for generation time) are ≈65 min for strain HK 2200 and ≈400 min for strain HK 2225; however, there is no significant difference in the concentrations of fructose at which half-maximal rates (Km for growth) are achieved.
Figure 2.
Growth of strains HK 2200 (○) and HK 2225 (●) on various concentrations of fructose. Both strains carried ΔgutA [ΔptsH ptsI crr]KanR fruA ΔfruK and manXYZ∷CmR, were rich in Mak activity, and differed only in their ability to effect the facilitated diffusion of fructose. [S]−1, Fructose concentration (mM)−1; v−1, doubling time (min).
Although these findings indicate that isoforms of PtsG determine whether fructose enters the cells, it is conceivable that the observed phenotypes are associated not with ptsG but with a gene located close to it. Several lines of evidence argue against this view.
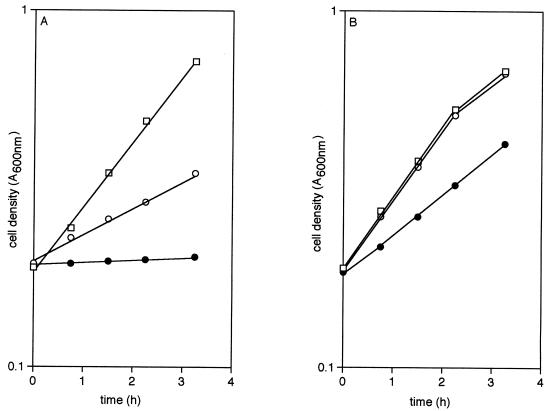
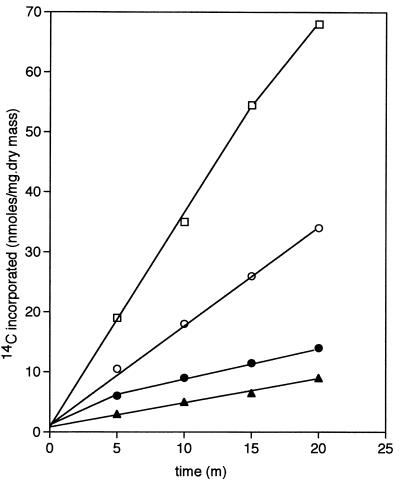
Mutants carrying that form of PtsG that is able to effect the diffusion of fructose (ptsG-F) and those able to transport glucose but not fructose (ptsG-I), but which are otherwise isogenic, differ in their response to analogs of glucose. Addition of 5 mM methyl α-glucoside (αMG) or 5 mM 2-deoxyglucose (2DG) to cultures of ptsG-F strains growing on 20 mM glycerol produced rapid inhibition of growth. The former analog caused a virtual cessation of growth, whereas the latter increased the doubling time from 84 min to 230 min (Fig. 3A). In contrast, growth on glycerol of cultures of strains containing ptsG-I was virtually unaffected by the addition of 2DG and was only slightly inhibited by αMG (Fig. 3B), which indicated that, during growth on glycerol, the analogs were not taken up by this isoform of PtsG and consequently did not manifest the known toxic effects of accumulated hexose phosphates (for review, see ref. 1). This hypothesis was confirmed by the behavior of ptsG-F and ptsG-I mutants in which the PTS had been rendered inoperative by insertion of a [ΔptsH ptsI crr]KanR cassette. Such mutants now were unaffected by the analogs when growing on glycerol, which showed that the PTS was required for the hexose phosphates to be formed and accumulated. However, both the uptake of [14C]fructose by washed suspensions of such fructose-positive PTS− mutants that had been grown on fructose (Fig. 4), as well as the growth of such mutants on fructose (Fig. 5), were powerfully inhibited by either analog and also by glucose, which supported the view that the analogs and glucose probably entered the cells by the same route as did fructose; in the absence of PTS function, metabolism of glucose did not occur, and this hexose and its analogs acted as competitive inhibitors of fructose transport. Indeed, even as little as 1 μM αMG powerfully inhibited the utilization of 40 mM fructose by strains carrying the [ΔptsH ptsI crr]KanR-transposon (Fig. 6).
Figure 3.
Effect of glucose analogs on the growth, on glycerol, of strains containing ptsG-F (A) and ptsG-I (B) but otherwise isogenic. After overnight growth on 20 mM glycerol, cells were suspended in 20 mM glycerol medium, to which was added water (□) 2DG to 5 mM (○) or αMG to 5 mM (●).
Figure 4.
Incorporation of 14C from [14C]fructose by a mutant carrying ptsG-F and [ΔptsH ptsI crr]KanR. Cells were grown overnight on 20 mM fructose, harvested, and washed. Suspensions (A600 nm = 1) were incubated with 5 mM [14C]fructose to which was added water (□), 1 mM 2DG (○), 1 mM αMG (●), or 5 mM glucose (▴). Samples were taken at known times, the cells were filtered and washed, and their radioactivity was assayed.
Figure 5.
Effect of glucose analogs on the growth, on fructose, of a mutant carrying ptsG-F and [ΔptsH ptsI crr]KanR. Cells were grown overnight on 20 mM fructose and resuspended in four flasks of the same medium (□). After 1 h 15 min, 2DG (○), glucose (▴), or αMG (●) to final concentrations of 1 mM were added.
Figure 6.
Hanes plot of the effect of αMG on the growth, on various concentrations of fructose, of a mutant carrying ptsG-F and [ΔptsH ptsI crr]KanR. Cells were grown overnight on 20 mM fructose, harvested, washed, and resuspended in flasks of the same medium but containing fructose at 5 mM, 7.2 mM, 10 mM, or 20 mM (○), and 5 mM, 10 mM, 20 mM, or 40 mM (●). αMG to a final concentration of 1 μM was added to flasks indicated by ●.
Direct evidence that it is PtsG and not a protein specified by an adjacent gene that effects fructose uptake was provided by the introduction into the [ptsG∷CmR] strain HK 2190 of the ptsG-F+ and ptsG-I+ genes cloned into a plasmid. These genes had been amplified by PCR from genomic DNA prepared from otherwise isogenic donor strains and the resultant fragments, ≈1.4 kb in length and covering only the structural regions, were ligated into the pGSB7 plasmid (26), replacing the ptsG in this plasmid with the two amplified isoforms. Both types of transformant grew readily on glucose but only that containing ptsG-F also grew on fructose. Because only the structural genes had been cloned and transferred to the previously PtsG-negative recipient, this shows that the facilitated diffusion of fructose is effected solely by the PtsG-F isoform of the glucose carrier.
Sequence Analysis.
DNA prepared from the chromosomal ptsG genes of the fructose-positive strain HK 2237 and from the fructose-negative strain HK 2240 and amplified by PCR was sequenced completely. The only difference detected between the sequence of ptsG-F and that of ptsG-I [the latter of which was identical to the Blattner sequence (12)] was a G/T substitution of the 34th nucleotide, which is the first position of codon 12 and which alters the deduced amino acid sequence of the membrane-spanning protein from valine in PtsG-I to phenylalanine in PtsG-F (V12F). This finding was confirmed by sequencing the DNA that had been cloned into the pGSB7 plasmid: again, the only difference between the two isoforms of ptsG that was detected was in nucleotide 34. It was further confirmed that position 34 of the nucleotide sequence of ptsG in all of the Hfr strains used for mapping (22) was occupied by G and that these strains therefore contained the ptsG-I isoform.
Discussion
Although early work (8) had suggested that enzymes II of the PTS do not effect the transport of their substrates in the absence of phosphorylation, there is a substantial body of evidence that this view is not correct. In particular, mutants in which glucose transport had been uncoupled from phosphorylation can take up glucose in the absence of the cytoplasmic proteins of the PTS (27); moreover, it has been known since 1976 that galP mgl strains of E. coli, which are devoid of the genes for the active transport of galactose, can still grow on this hexose, which now enters the cells by facilitated diffusion through the PtsG protein (28). This finding has been confirmed with mutants of S. typhimurium in which the PTS had been rendered nonfunctional (29). It had been suggested that non-PTS systems with altered activities might be responsible for such a putative PTS-mediated process (8), but this explanation can be ruled out for the mutants of E. coli in which altered forms of PtsG have been shown to effect the entry of mannitol (26), ribose (30), and (in the present work) fructose, mannose, αMG, and 2-DG: in each of these, the sites of the mutations were mapped and shown unequivocally to lie within the ptsG gene that encodes the membrane-spanning protein of the glucose permease. However, unlike the other mutations in the ptsG of E. coli that have been shown to be capable of transporting substrates other than glucose or its analogs (26, 27, 30, 31), the isoforms of ptsG that affect fructose transport are altered in a deduced amino acid located only 10 residues from the amino terminus of the PtsG protein, which is presumed (31) to be located entirely within the cytosol.
It is likely that this change affects the abundance rather than the specificity of the membrane-spanning protein. The data recorded in Fig. 2 indicate that even strains devoid of PTS activity and containing the PtsG-I isoform can take up fructose, albeit much less readily than similar strains containing PtsG-F; moreover, it has long been known that enzymes II for hexitols with structure analogous to fructose, such as glucitol (6) or mannitol (M. L. Becker and H.L.K., unpublished results), can effect fructose uptake if these permeases are overproduced. It thus may be that the V12F mutation in ptsG leads to increased production of the PtsG protein or to production of a PtsG-protein with improved transport activity: this would be in accord with the observation (Fig. 3) that, when growing on glycerol, strains carrying PtsG-F take up glucose analogs to a greater extent than do strains carrying PtsG-I.
Powerful evidence in support of this view also has been provided by the isolation of a variety of different mutants of E. coli from continuous cultures subjected simultaneously to glucose and oxygen limitation (32). Remarkably, the predominant type of mutant elicited by this selection procedure was one manifesting the identical V12F change in ptsG described in the present work. Furthermore, this mutation was also shown to result in a greatly increased rate of glucose transport compared with that observed with wild-type ptsG strains. But it remains to be elucidated why the replacement of valine by phenylalanine so close to the amino terminus of the glucose permease should have such remarkable consequences.
Acknowledgments
We are deeply grateful to Drs. M. K. Berlyn, G. M. Church, A. Danchin, B. Erni, N. J. Gay, G. R. Jacobson, and B. L. Wanner for generous gifts of E. coli strains and plasmids used in this work.
Abbreviations
- PTS
phosphoenolpyruvate/glycose phosphotransferase system
- αMG
methyl α-glucoside
- 2DG
2-deoxyglucose
References
- 1.Ferenci T, Kornberg H L. Biochem J. 1973;132:341–347. doi: 10.1042/bj1320341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior T I, Kornberg H L. J Gen Microbiol. 1988;134:2757–2768. doi: 10.1099/00221287-134-10-2757. [DOI] [PubMed] [Google Scholar]
- 3.Reizer J, Reizer A, Kornberg H L, Saier M H. FEMS Microbiol Lett. 1994;118:159–162. doi: 10.1111/j.1574-6968.1994.tb06819.x. [DOI] [PubMed] [Google Scholar]
- 4.Ferenci T, Kornberg H L. Proc R Soc London Ser B. 1974;187:105–119. doi: 10.1098/rspb.1974.0065. [DOI] [PubMed] [Google Scholar]
- 5.Jones-Mortimer M C, Kornberg H L. Proc R Soc London Ser B. 1974;187:121–131. doi: 10.1098/rspb.1974.0066. [DOI] [PubMed] [Google Scholar]
- 6.Jones-Mortimer M C, Kornberg H L. J Gen Microbiol. 1976;96:383–391. doi: 10.1099/00221287-96-2-383. [DOI] [PubMed] [Google Scholar]
- 7.Kundig W, Ghosh S, Roseman S. Proc Natl Acad Sci USA. 1964;52:1064–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postma P W, Stock J B. J Bacteriol. 1980;141:476–484. doi: 10.1128/jb.141.2.476-484.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saier M H, Scott Young W, Roseman S. J Biol Chem. 1971;246:5838–5840. [PubMed] [Google Scholar]
- 10.Sebastian J, Asensio C. Arch Biochem Biophys. 1972;151:227–233. doi: 10.1016/0003-9861(72)90492-4. [DOI] [PubMed] [Google Scholar]
- 11.Levy S, Zeng G-Q, Danchin A. Gene. 1990;86:27–33. doi: 10.1016/0378-1119(90)90110-d. [DOI] [PubMed] [Google Scholar]
- 12.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 13.Ashworth J M, Kornberg H L. Proc R Soc London Ser B. 1966;65:179–188. doi: 10.1098/rspb.1966.0063. [DOI] [PubMed] [Google Scholar]
- 14.Gay N J. J Bacteriol. 1984;158:820–825. doi: 10.1128/jb.158.3.820-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada M, Saier M H. J Bacteriol. 1987;169:2990–2994. doi: 10.1128/jb.169.7.2990-2994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Link A J, Phillips D R, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 18.Amaral D, Kornberg H L. J Gen Microbiol. 1975;90:157–168. doi: 10.1099/00221287-90-1-157. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Aulkemeyer P, Ebner R, Heilenmann G, Jahreis K, Schmid K, Wrieden S, Lengeler J W. Mol Microbiol. 1991;5:2913–2922. doi: 10.1111/j.1365-2958.1991.tb01851.x. [DOI] [PubMed] [Google Scholar]
- 21.Wanner B. J Mol Biol. 1986;191:39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- 22.Berlyn M K. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis S J, Epstein W. J Bacteriol. 1975;122:1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornberg H L, Jones-Mortimer M C. FEBS Lett. 1975;51:1–4. doi: 10.1016/0014-5793(75)80840-4. [DOI] [PubMed] [Google Scholar]
- 25.Postma P W, Roseman S. Biochim Biophys Acta. 1976;457:213–257. doi: 10.1016/0304-4157(76)90001-0. [DOI] [PubMed] [Google Scholar]
- 26.Begley G S, Warner K A, Arents J C, Postma P W, Jacobson G R. J Bacteriol. 1996;178:940–942. doi: 10.1128/jb.178.3.940-942.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruijter G J G, van Meurs G, Verwey M A, Postma P W, van Dam K. J Bacteriol. 1992;174:2843–2850. doi: 10.1128/jb.174.9.2843-2850.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg H L, Riordan C. J Gen Microbiol. 1976;94:75–89. doi: 10.1099/00221287-94-1-75. [DOI] [PubMed] [Google Scholar]
- 29.Postma P W. FEBS Lett. 1976;61:49–53. doi: 10.1016/0014-5793(76)80169-x. [DOI] [PubMed] [Google Scholar]
- 30.Oh H, Park Y, Park C. J Biol Chem. 1999;274:14006–14011. doi: 10.1074/jbc.274.20.14006. [DOI] [PubMed] [Google Scholar]
- 31.Buhr A, Daniels G, Erni B. J Biol Chem. 1992;267:3847–3851. [PubMed] [Google Scholar]
- 32.Manché K, Notley-McRobb L, Ferenci T. Genetics. 1999;153:5–12. doi: 10.1093/genetics/153.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]