Abstract
The human adenovirus type 5 (HAdV-5) E1A 13S oncoprotein is a potent regulator of gene expression and is used extensively as a model for transcriptional activation. It possesses two independent transcriptional activation domains located in the N-terminus/conserved region (CR) 1 and CR3. The protein acetyltransferase p300 was previously identified by its association with the N-terminus/CR1 portion of E1A and this association is required for oncogenic transformation by E1A. We report here that transcriptional activation by 13S E1A is inhibited by co-expression of sub-stoichiometric amounts of the smaller 12S E1A isoform, which lacks CR3. Transcriptional inhibition by E1A 12S maps to the N-terminus and correlates with the ability to bind p300/CBP, suggesting that E1A 12S is sequestering this limiting factor from 13S E1A. This is supported by the observation that the repressive effect of E1A 12S is reversed by expression of exogenous p300 or CBP, but not by a CBP mutant lacking actyltransferase activity. Furthermore, we show that transcriptional activation by 13S E1A is greatly reduced by siRNA knockdown of p300 and that CR3 binds p300 independently of the well-characterized N-terminal/CR1-binding site. Importantly, CR3 is also required to recruit p300 to the adenovirus E4 promoter during infection. These results identify a new functionally significant interaction between E1A CR3 and the p300/CBP acetyltransferases, expanding our understanding of the mechanism by which this potent transcriptional activator functions.
INTRODUCTION
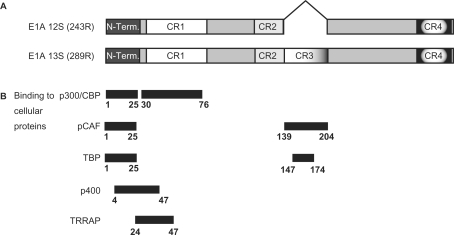
Human adenovirus type 5 (HAdV-5) early region 1A (E1A) is the first viral gene to be transcribed upon infection and plays an essential role in activating transcription (1,2). The 13S and 12S E1A mRNAs encode two major products of 289 residues (R) and 243R, respectively (Figure 1A), and these share identical amino and carboxyl sequences. The only difference between them is the presence of an additional 46 amino acids in the 289R protein that arises as the result of differential splicing of the primary E1A transcript (2). The region unique to the 13S encoded E1A protein coincides with a region that is highly conserved amongst the E1A proteins of different adenovirus serotypes, referred to as conserved region 3 (CR3) (3–5). Of the two major E1A polypeptides, the larger is considered to be primarily responsible for transcriptional activation of gene expression. Indeed, alterations within CR3 generally abolish E1A transactivation (6–10). Interestingly, a synthetic CR3 peptide corresponding to residues 140–188 of E1A was sufficient to transactivate adenovirus early promoters when microinjected into HeLa cells (11). Later work identified an adjacent acidic region spanning residues 189–200, termed Auxiliary Region 1 (AR1) as essential for efficient transactivation of early viral promoters by E1A (12).
Figure 1.
Schematic of E1A isoforms and locations of binding sites for indicated proteins. (A) Schematic representation of E1A 12S and E1A 13S splice isoforms. (B) Binding sites for p300/CBP, pCAF, TBP, p400 and TRRAP on E1A are indicated.
The mechanism by which CR3 of E1A activates transcription has been the subject of intense investigation. Despite this, some aspects of transactivation by E1A remain unclear. CR3 interacts with a wide variety of different transcription factors (13–17), allowing it to strongly activate transcription of many different genes that have no obvious similarities (16). These observations suggested that the interaction of E1A with certain sequence specific transcription factors results in the localization of E1A to target promoters in the infected cell.
Extensive mutational analyses identified a promoter targeting region embedded within CR3 that is located within residues 180–188 (15). This region is not required for transactivation if E1A is artificially targeted to a promoter as a fusion with a heterologous DNA-binding domain (DBD) (18). These residues confer interaction with a number of unrelated sequence specific transcription factors, such as ATF1-3, c-jun, SP1, USF, Oct-4 and CBF/NF-Y (13–17) and several TBP associated factors (TAFs), including TAFII55, TAFII110, TAFII135 and TAFII250 (19–22).
Interestingly, mutations within the promoter targeting region of CR3 exhibit a pronounced dominant negative effect on transcriptional activation by wild-type E1A (23,24). This phenomenon, commonly referred to as squelching, suggested that these particular mutants were sequestering limiting factors necessary for transactivation by wild-type E1A. The first of these factors to be identified was TBP (25). Further studies led to the identification of the Sur2/TRAP150β/Med23 component of the Mediator/TRAP complex as a target of the CR3 domain of E1A (26,27). More recent work has also suggested distinct roles for different proteasome complexes in CR3-dependent transcription (28). Clearly, the unusually strong transcriptional activation function of CR3 results from a complex orchestration of the activities of numerous transcriptional components.
When fused to a heterologous DBD, which directly tethers E1A to a promoter, a second transactivation domain distinct from CR3 was identified within the N-terminus/CR1 portion of E1A (29). This region of E1A interacts with a number of transcriptional regulators, including the p300, CBP (CREB-binding protein) and pCAF acetyltransferases, TBP, TRRAP and p400 (Figure 1B) (30). Paradoxically, this region appears to function primarily as a transcriptional repression domain in the context of the E1A 12S protein, by sequestering limiting factors, such as p300 and CBP from cellular transcription factors (2). Indeed, recent work has shown that expression of E1A 12S induces global changes in histone H3 K18 acetylation, consistent with the sequestration/retargeting of p300/CBP by E1A (31).
p300 and CBP are highly related transcriptional co-activators that are recruited to gene promoters via their association with numerous otherwise unrelated DNA-binding transcription factors (32). Once recruited to target promoters, p300/CBP activate transcription by acetylating histone tails or target lysines within other transcription factors using their intrinsic acetyltransferase activity.
In the present study, we demonstrate direct binding of p300/CBP to CR3, thus identifying a novel second site of interaction in E1A 13S for these acetyltransferases. We also show that p300/CBP and their associated acetyltransferase activities are important factors for transcriptional activation by CR3. In summary, our results identify a new factor required for transcriptional activation by the E1A oncoprotein.
MATERIALS AND METHODS
Cell lines, tissue culture and viruses
U2OS, A549 and HeLa cells were grown in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), streptomycin and penicillin.
HeLa cells were infected with an m.o.i. of 10 with the indicated viruses. dl309 expresses both E1A 13S and 12S, pm975 expresses only E1A 13S and dl520 expresses only E1A 12S. Roughly 5 × 106 cells were infected per virus. Infections were carried out for 1 h in serum-free media, after which 10 ml of complete media was added and the cells were incubated for an additional 16 h.
E3 and E4 reporter plasmid construction
The region spanning the adenovirus E3 promoter was PCR amplified from a dl309 genomic DNA preparation using the following primers:
Forward: 5′-GATCCTCGAGGGCGGCTTTCGTCACAGGG-3′
Reverse: 5′-GCATGAAGCTTTCTGAAATGTCCCGTCCGG-3′
The E4 promoter was amplified using the following primers:
Forward: 5′-GTAAAGCTTCGACACGGCACCAGCTCAATCAG-3′
Reverse: 5′-GATCCTCGAGATTTGAGGAAGTTGTGGGTTTTTTG-3′
The PCR product was then cloned in at the HindIII and XhoI sites of pGL3-Basic (Promega).
E1A and p300/CBP expression vectors
E1A12S and E1A13S cDNAs were cloned into the pcDNA3 mammalian expression vector (Invitrogen) while the N-terminal deletions and the RG2 point mutant of 12S E1A were cloned into pcDNA3.1 (Invitrogen). EGFP fusions of E1A 12S fragments were generated by cloning the specific fragment into the pEGFP-C1 expression vector (Clontech) in-frame with the N-terminal EGFP. GAL4-fusions of CR3 were generated by subcloning CR3 (encoding residues 139–204 of HAdV-5) into the pM vector (Promega) in-frame with the N-terminal GAL4-DBD. GST fusions of CR3 (residues 139–204) and residues 1–82 of E1A were made by subcloning the respective fragments into pGEX-4T1 (GE Healthcare Life Sciences) in-frame with the N-terminal GST tag. Expression vectors for p300, CBP, CBP AT- and pCAF were previously described (33,34).
Transfection and reporter assay
Twenty-four hours prior to transfection, U2OS or HeLa cells were seeded in six-well dishes (Sarsteadt) at a density of 100 000 cells per well in DMEM. U2OS cells were transfected with 1 μg of the reporter plasmid (pGal6-Luc) and 1 μg of the transactivator pM-GAL4-CR3 (unless otherwise noted), which expresses HAdV-5 E1A residues 139–204 fused to the GAL4 DBD, and the indicated amounts of a plasmid expressing E1A 12S or various deletion mutants. HeLa cells were transfected with 0.5 μg of the reporter plasmid (pGL3-E4 or pGL3-E3) and 1.5 μg of E1A 13S plasmid, and 0.1 μg or indicated amounts of a plasmid expressing E1A 12S. In addition 0.2 μg of a β-galactosidase plasmid was transfected with all reporter/driver plasmids for transfection normalization purposes. Transfections were carried out using the Superfect reagent (Qiagen) according to manufacturer's instructions. Luciferase activity was assayed 24 h after transfection for U2OS cells or 48 h after transfection for HeLa cells using the Promega Luciferase Reporter kit according to the manufacturer's instructions. Luciferase activity was normalized to both transfection efficiency using β-galactosidase activity and protein levels. U2OS cells were used for the GAL4-CR3 experiments instead of HeLa cells because of poor activity of the GAL4-CR3 fusion in HeLa cells. While HeLa cells were used for the E3 and E4 reporter experiments because of high background activity of these two reporters in U2OS cells.
siRNA transfection
siRNA for p300 was described previously (35). The custom siRNA was ordered from Ambion and was transfected into either U2OS or HeLa cells using the SilentFect transfection reagent (Bio-Rad) according to manufacturer's instructions. Negative control siRNA #1 was purchased from Ambion (cat #4611). Seventy-two hours after siRNA transfection, the cells were transfected with the indicated plasmid mixes for luciferase assays. Luciferase activity was assayed 24 or 48 h after plasmid transfection.
Immunoprecipitation and GST pulldowns
For immunoprecipitation of wild-type E1A or E1A RG2 mutants, transfected HeLa cells were lysed in high salt NP-40 lysis buffer (0.5% NP-40, 50 mM Tris pH 7.8, 500 mM NaCl) supplemented with protease inhibtor cocktail. Two milligrams of the cell lysate was used for immunoprecipitation with the monoclonal M73 anti-E1A antibody (36). E1A was detected using the M73 monoclonal antibody, while p300 was detected using the RW128 monoclonal antibody (Upstate).
HeLa cells transfected with 10 μg of either E1A 12Sdl1101, E1A 13Sdl1101 or genomic E1A expressing plasmids were fixed with 1.5% formaldehyde for 10 min at room temperature directly in the growth media. Crosslinking was stopped with glycine and incubation at room temperature for an additional 5 min. Cells were then scraped from the dishes, washed with phosphate buffered saline and resuspended in NP-40 lysis buffer (1% NP-40, 50 mM Tris pH 7.8, 150 mM NaCl) supplemented with protease inhibitor cocktail (Sigma). The cell suspension was then sonicated for 30 seconds. One milligram of the cell lysate was used in immunoprecipitation using a mix of the M58 and M73 monoclonal anti-E1A antibodies.
GST pulldowns were performed using purified full-length CBP expressed from a baculovirus with purified GST-CR3 (E1A residues 139–204) and purified GST-1-82 as previously described (37,38).
For GST pulldown of CR3 with endogenous p300, A549 cells were lysed in high salt NP-40 lysis buffer. Four milligrams of A549 lysate were mixed with 50 µg of either GST or GST-CR3 and Glutathione Sepharose 4B and incubated at 4°C. Bound proteins were recovered by centrifugation, washed with high salt NP-40 lysis buffer, eluted in SDS sample buffer and resolved on a gradient 4–12% SDS–PAGE. Associated p300 was detected using RW128 monoclonal antibody.
Chromatin immunoprecipitation (ChIP)
ChIP was carried out essentially as previously described (39). HeLa cells were infected with the indicated adenoviruses at an m.o.i. of 10 and harvested 16 h after infection for ChIP analysis. For immunoprecipitation of E1A, the monoclonal M73 antibody was used. For immunoprecipitation of p300 the monoclonal RW128 anti-p300 antibody was used. Mouse anti-rabbit antibody was used as a negative IgG control (Sigma). The E4 primers used for amplification of the precipitated DNA were:
Forward: 5′-GTAAAGCTTCGACACGGCACCAGCTCAATCAG-3′
Reverse: 5′-GATCTCGAGCATCATCATAATATACCTTATTTTGGATTGAAGCC-3′
PCR reactions were carried out using Taq DNA Polymerase Supermix (Invitrogen) using 0.5% of total ChIP DNA as template according to manufacturer's instructions. The annealing temperature used was 55°C and 35 cycles were run.
RESULTS
E1A 12S inhibits transactivation by E1A 13S and a GAL4-CR3 fusion
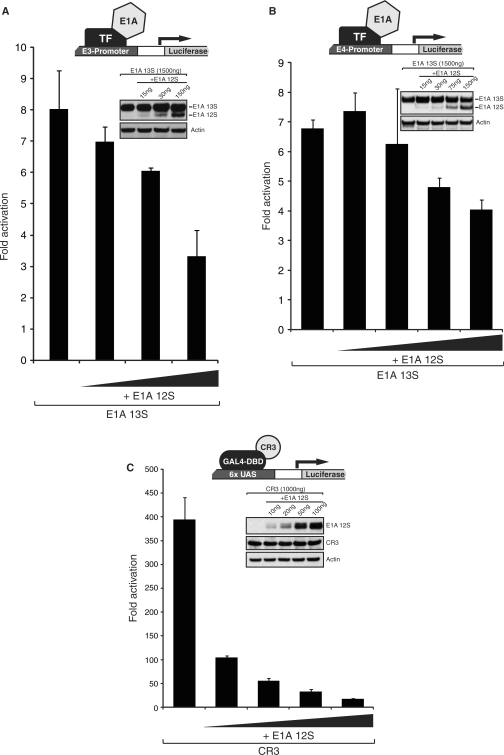
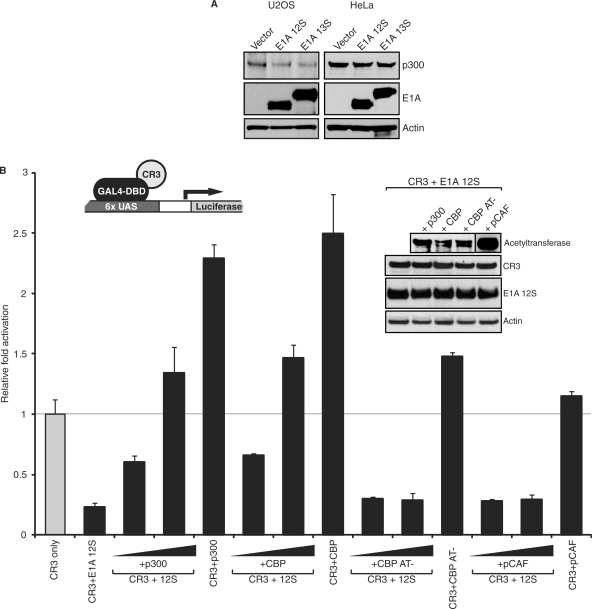
E1A 13S can strongly activate transcription from a luciferase reporter gene under the control of the adenovirus E3 regulatory region (Figure 2A). Intriguingly, co-transfection of limiting amounts of E1A 12S repressed activation of luciferase expression by E1A 13S (Figure 2A). Indeed, activation was reduced by ∼50% when one-tenth the amount of 12S expression vector was present. The effect of E1A 12S on 13S transactivation was not due to a reduction of 13S expression levels as determined by Western blot analysis. We also tested an adenoviral E4 promoter construct (Figure 2B). This reporter showed similar response to the E3 reporter, with robust activation by E1A 13S and a pronounced decrease in activation when sub-stoichiometric amounts of E1A 12S were co-expressed with E1A 13S.
Figure 2.
Repression of E1A 13S- and CR3-mediated transactivation by E1A 12S is dose-dependent and independent of the method by which E1A is targeted to the promoter. (A) HeLa cells were co-transfected with plasmids expressing E1A 13S (1.5 μg) and E1A 12S (as indicated) together with an adenovirus E3-luciferase reporter plasmid (0.5 μg). Luciferase activity was assayed 48 hours after transfection. TF, transcription factor. (B) HeLa cells were co-transfected with plasmids expressing E1A 13S (1.5 μg) and E1A 12S (as indicated) together with an adenovirus E4-luciferase reporter plasmid (0.5 μg). Luciferase activity was assayed 48 h after transfection. (C) U2OS cells were co-transfected with plasmids expressing a GAL4 DNA-binding domain-CR3 (1 μg) fusion and E1A 12S (as indicated) together with a GAL4 responsive luciferase reporter (1 μg). Luciferase activity was assayed 24 h after transfection.
Interference by E1A 12S with E1A 13S-mediated activation suggests that it affects a key factor necessary for 13S-dependent transcriptional activation. E1A 12S could sequester a factor from E1A 13S that is necessary for transcriptional activation. This process is often referred to as ‘transcriptional squelching’ (40) and can be used as an experimental tool to identify limiting factors necessary for transcriptional activation. Alternatively, 12S E1A could induce the degradation of a factor necessary for transcriptional activation by the 13S E1A. 13S E1A contains two independent activation domains, located in the N-terminal/CR1 and CR3 regions, respectively. As the N-terminal/CR1 activation region is also present in the 12S E1A isoform, it is unlikely that sub-stoichiometric amounts of 12S would efficiently compete for limiting factors with the identical region in 13S E1A. It seemed more likely that E1A 12S was sequestering a limiting factor required by the CR3 region, which is unique to the E1A 13S (Figure 1A). To directly determine if E1A 12S could affect CR3-dependent transactivation, luciferase assays were performed with increasing concentrations of E1A 12S in conjunction with a GAL4 DBD fusion of CR3 (amino-acid residues 139–204) and a GAL4 responsive-luciferase reporter. Activation by GAL4-CR3, which is directly recruited to the reporter by the fused GAL4 DBD, was reduced to ∼30% of normal activity even at the lowest levels of E1A 12S co-expression (one-hundredth the amount of GAL4-CR3 plasmid transfected; Figure 2C). Increasing the levels of 12S to 100 ng (one-tenth of GAL4-CR3 transfection) further reduced CR3 activity by up to 90%. Western blot analysis showed that the expression level of GAL4-CR3 was not affected by increasing concentrations of E1A 12S, suggesting that the strong repression resulted from direct interference with CR3 transactivation (Figure 2C).
Taken together, these results suggest that E1A 12S is a potent dose-dependent inhibitor of 13S- and CR3-dependent transactivation. E1A 12S appears to function by sequestering or inducing the degradation of a key transcriptional co-regulator required by CR3. Moreover, this is independent of the mechanism by which the activator is recruited to the promoter, and is not mediated by changes in the expression levels of E1A 13S or GAL4-CR3.
The N-terminal/CR1 portion of E1A 12S inhibits CR3 transactivation
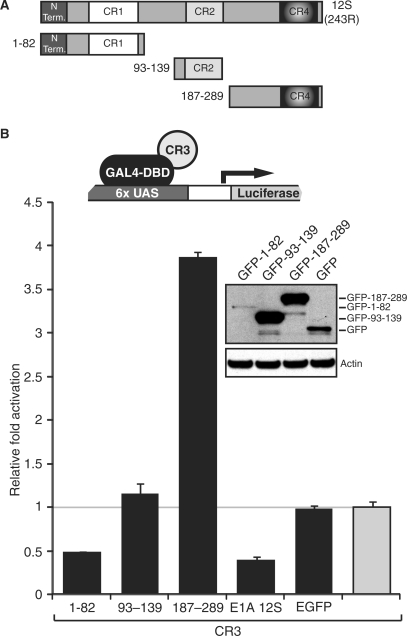
In order to determine which region of E1A 12S mediates the silencing effect, a set of GFP fusion constructs that collectively encompass most of the E1A 12S protein (Figure 3A) were co-expressed with the GAL4-CR3 fusion protein in the presence of a GAL4-luciferase reporter plasmid (Figure 3B). Neither the CR2 region (residues 93–139), nor the C-terminal region (residues 187–289) of E1A were capable of silencing CR3 transactivation. Indeed, the C-terminal region, which tightly binds CtBP, had the opposite effect, inducing the reporter 4-fold. This result is consistent with our previous observations that the interaction of CtBP with CR3 limits CR3-dependent activation (41). The only region of E1A capable of silencing CR3-mediated transactivation mapped to the N-terminal 82 residues of E1A. Interestingly, this region is the only portion of 12S E1A that functions as a transcriptional activation domain when fused to a heterologous DBD (29). This suggests that the N-terminus of E1A and CR3 may be binding a common factor(s). Sequestration or degradation of this factor(s) by E1A 12S presumably causes the observed reduction in transcription by E1A 13S.
Figure 3.
Repression of CR3 transactivation by E1A 12S maps to the N-terminal/CR1 region. (A) Schematic representation of E1A 12S and the locations of fragments used as GFP fusions in the squelching assay. (B) U2OS cells were co-transfected with 0.1 μg of the indicated GFP-E1A fragment fusions, GAL4-CR3 (1 μg) and a GAL4-luciferase reporter (1 μg). Luciferase activity was assayed 24 h after transfection and the results were plotted versus GAL4-CR3 (shown in grey), which was set to 1.
E1A 12S mutants that lose p300/CBP binding no longer inhibit CR3 transactivation
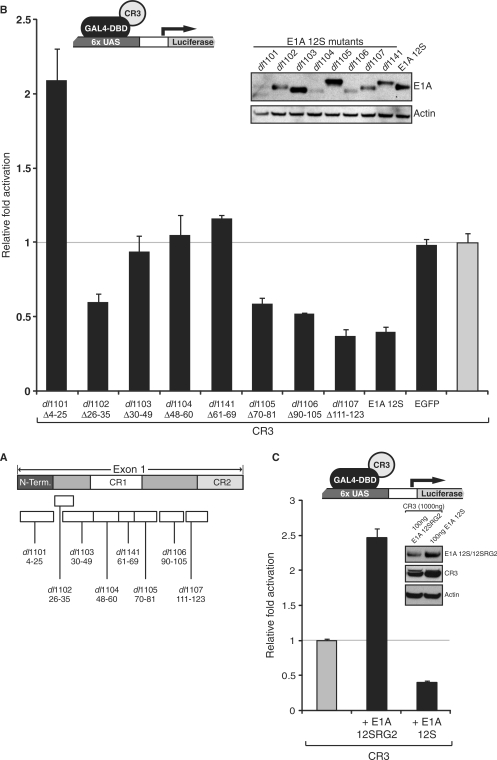
The N-terminus of E1A binds a large number of proteins involved in transcriptional regulation, including p300/CBP, pCAF, TBP, p400 and others (Figure 1B) (30,42). To further map the region within the N-terminus of E1A that was responsible for the squelching phenotype and to potentially ascertain which factor is involved in repression, a series of E1A 12S deletion mutants (Figure 4A) were tested for their ability to repress GAL4-CR3 mediated transactivation (Figure 4B). Since their initial construction (43), these mutants have been studied extensively and are well characterized for binding to various factors involved in transcriptional control, including p300/CBP (44), pCAF (45) (data not shown), TBP (46) and p400 (47) (summarized in Table 1, Figure 1B). E1A 12S deletion mutants lacking residues 4–25, 30–49, 48–60 and 61–69 (mutants dl1101, dl1103, dl1104 and dl1141, respectively) lost their ability to repress CR3-mediated transactivation. The inability of mutants to repress CR3 activation correlated perfectly with loss of binding to p300/CBP (44). Although many of these E1A mutants are impaired for interaction with multiple transcriptional regulators, loss of function does not match with the binding profile of any other target protein (Table 1, Figure 1B). This suggests that sequestration of p300/CBP by the N-terminal/CR1 portion of 12S E1A may be the sole mechanism of squelching CR3-mediated transactivation. The E1A 12SRG2 point mutant, which also does not bind p300/CBP in vivo, was tested in the squelching assay and was also unable to repress CR3-dependent activation (Figure 4C). Indeed, 12SRG2 behaved similarly to the E1A 12Sdl1101 mutant, enhancing CR3-mediated activation, presumably by sequestering CtBP-associated transcriptional repressors. These results suggest that p300/CBP is most likely the sole factor sequestered by the N-terminus of E1A 12S and that it is involved in transactivation by CR3.
Figure 4.
Repression of CR3 transactivation by E1A 12S maps to the same regions that bind p300/CBP. (A) Schematic representation of E1A 12S deletion mutants within exon 1 used in the study. (B) U2OS cells were co-transfected with 0.1 μg of the indicated E1A 12S deletion mutants together with GAL4-CR3 (1 μg) and a GAL4-luciferase reporter (1 μg). Luciferase activity was assayed 24 h after transfection and the results were plotted versus GAL4-CR3 (shown in grey), which was set to 1. (C) U2OS cells were co-transfected with GAL4-CR3 fusion (1 μg), GAL4-luciferase reporter (1 μg) and 0.1 μg of E1A 12SRG2 or wild-type E1A 12S. Luciferase activity was assayed 24 h after transfection and the results were plotted versus GAL4-CR3 (shown in grey), which was set to 1.
Table 1.
E1A 12S mutants binding profile for p300, p400, TBP and pCAF
| E1A mutant | Squelching of CR3 | Interaction with |
|||
|---|---|---|---|---|---|
| p300 | p400 | TBP | pCAF | ||
| dl1101 | − | − | − | − | − |
| dl1102 | ++++ | ++++ | − | + | ++ |
| dl1103 | − | − | − | ++++ | +++ |
| dl1104 | − | − | ++++ | +++ | ++++ |
| dl1141 | − | − | ++++ | ++++ | ++ |
| dl1105 | ++++ | ++++ | ++++ | ++++ | ++ |
Binding properties of E1A 12S N-terminal deletion mutants to p300, p400, TBP and pCAF.
Expression of exogenous p300 or CBP restores transactivation by CR3 when E1A 12S is present
The extensive mutational analysis presented above indicates that reduction of transcriptional activity by E1A 12S relies on binding to p300; although, other factors cannot be fully excluded from playing a role. We initially determined if E1A 12S had an effect on the steady state level of p300 by Western blot analysis in both U2OS and HeLa cells (Figure 5A). E1A 12S expressing cells showed similar levels of endogenous p300 expression as compared to vector transfected cells or cells expressing E1A 13S. This indicates that the mechanism by which E1A 12S blocks transcriptional activation by E1A 13S is not mediated by degradation of p300.
Figure 5.
Overexpression of p300 or CBP restores E1A 12S-repressed CR3 transactivation. (A) U2OS or HeLa cell extracts from cells transfected with empty vector, E1A 12S expression plasmid or E1A 13S expression plasmid were assayed for levels of endogenous p300 and E1A. (B) U2OS cells were co-transfected with plasmids expressing GAL4-CR3 (600 ng) and 600 ng of vectors expressing the indicated acetyltransferases, together with a GAL4-luciferase reporter plasmid (600 ng) and 60 ng of E1A 12S as indicated. Luciferase activity was assayed 24 h after transfection and the data was plotted versus CR3 alone (shown in grey), which was set to 1. Overexpressed p300, CBP, CBP AT- and pCAF were detected using anti-HA antibody for p300 and anti-FLAG M2 for CBP, CBP AT- and pCAF.
To determine if sequestration of p300/CBP was responsible for the repressive effects observed, we tested whether expression of exogenous p300 would restore transactivation by the GAL4-CR3 fusion protein in the presence of E1A 12S. Expression of increasing levels of p300, or the closely related acetyltransferase CBP, restored CR3 transactivation in the presence of E1A 12S (Figure 5B). This was dependent on p300/CBP acetyltransferase activity, as an acetyltransferase-defective CBP mutant protein was unable to restore transactivation. Furthermore, the observed effect was specific to p300/CBP, because the pCAF acetyltransferase was unable to restore CR3 transactivation (Figure 5B). These results conclusively show that p300/CBP is indeed the key factor sequestered by E1A 12S, as supplementation by it alone was sufficient to restore activity. Furthermore, these results suggest that p300/CBP is a limiting factor in CR3-mediated transactivation, as CR3-dependent transcriptional activity was enhanced by overexpression of p300/CBP (Figure 5B).
Depletion of p300 strongly impairs transactivation by CR3 and E1A 13S
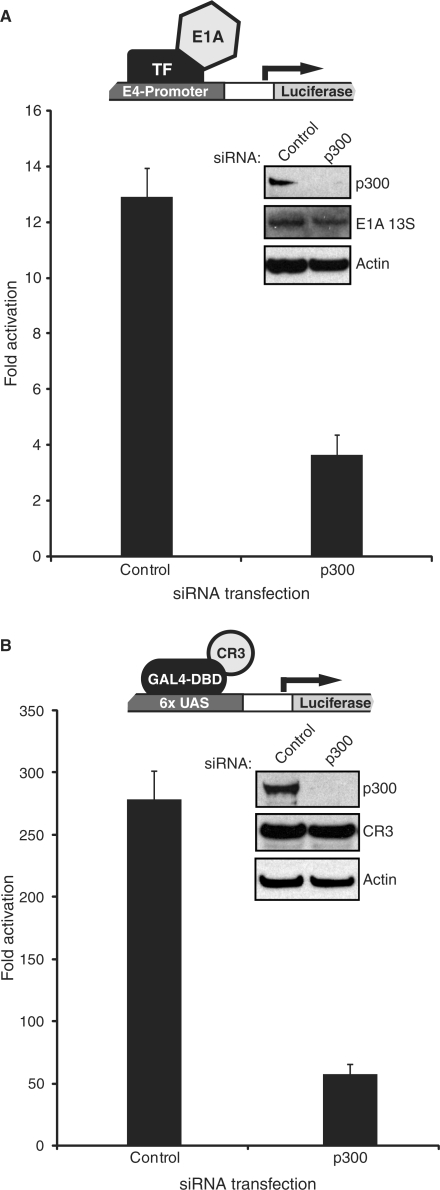
To definitively determine if p300 played a role in CR3-mediated transactivation, p300 was knocked down in human HeLa cervical cancer cells (for the E4/E1A 13S assay) or human U2OS osteosarcoma cells (for the GAL4-CR3 assay) using siRNA previously shown to be specific to p300 mRNA (35). p300 levels were greatly reduced in cells transfected with siRNA specific for p300, but not in those that were transfected with a negative control siRNA (Figure 6). Knockdown of p300 had a dramatic effect on transactivation by both 13S and the GAL4-CR3 fusion, without affecting the expression levels of either activator (Figure 6). This effect was not due to a general reduction of transactivation or loss in viability in cells in which p300 was knocked down because a reporter not regulated by E1A or CR3 was largely unaffected by the knockdown (data not shown), in agreement with what was previously observed with this siRNA (35). This result conclusively demonstrates that p300 is required for transactivation by CR3.
Figure 6.
Knockdown of p300 by siRNA impairs activation by E1A 13S and CR3. (A) HeLa cells were transfected with siRNA for p300 or a negative control siRNA and subsequently with 1 μg of plasmid expressing E1A 13S and 1 μg of E4-luciferase reporter. Luciferase activity was assayed 5 days after the initial siRNA transfection. (B) U2OS cells were transfected with siRNA for p300 or a negative control siRNA and subsequently with 1 μg of plasmid expressing the GAL4-CR3 fusion and 1 μg of GAL4-luciferase reporter. Luciferase activity was assayed 5 days after the initial siRNA transfection.
CR3 binds p300/CBP independently of the N-terminus and CR1
Numerous reports have indicated that p300/CBP stably binds to E1A via the N-terminus/CR1 region (2). The data presented here indicates that p300/CBP is also required for CR3-dependent transactivation, suggesting that CR3 may represent a previously unidentified second independent site of interaction with p300/CBP.
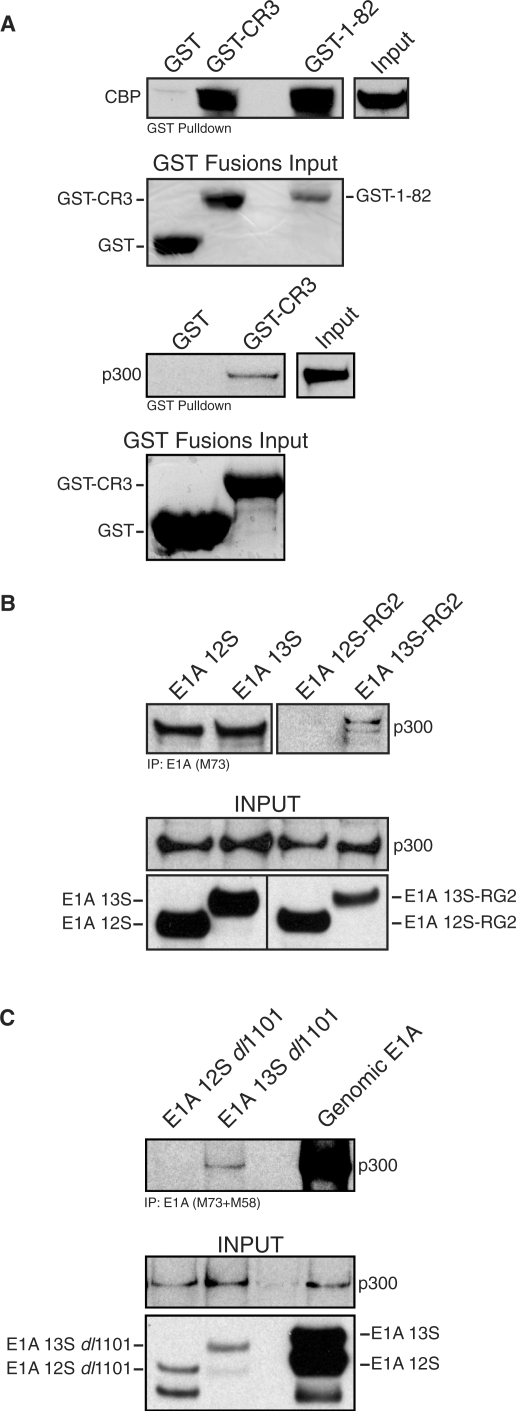
To determine whether CBP binds directly to CR3, purified recombinant full-length CBP and purified GST fused CR3 (amino-acids 139–204) were used in a GST pulldown experiment (Figure 7A). Recombinant GST-E1A residues 1–82 was used as a positive control, as it contains the high affinity p300/CBP-binding region (48). A direct interaction between CR3 and CBP was readily detectable under these in vitro conditions, whereas GST alone did not bind to CBP. The interaction observed between CR3 and CBP was somewhat weaker than that with the N-terminus of E1A. This data indicates that recombinant CR3 can directly and independently bind CBP in vitro. Interestingly, addition of HeLa total cellular extract to the pulldown assay reduced the interaction between CR3 and CBP by ∼50% (Supplementary Figure S1), suggesting that cellular factors present in the lysate may be competing with E1A for CBP.
Figure 7.
p300/CBP bind directly to E1A CR3. (A) Top panel: Full-length, purified FLAG-tagged CBP was incubated with either purified GST, GST-CR3 or GST-E1A-1-82. Protein complexes were then bound to Glutathione Sepharose 4B beads, washed and resolved on 4–12% gradient SDS–PAGE. Associated CBP was detected with anti-FLAG M5 monoclonal antibody, and input levels of GST-fusion proteins were determined by Ponceau S staining of the same membrane. Bottom panel: A549 total cell extract was incubated with either purified GST or GST-CR3. Protein complexes were then bound to Glutathione Sepharose 4B beads, washed and resolved on 4–12% gradient SDS–PAGE. The blot was probed for p300 using RW128 anti-p300 antibody and input levels of GST-fusion proteins was determined using Ponceau S staining of the same membrane. (B) HeLa cells were transfected with plasmids expressing either wild-type E1A 12S, 13S, 12SRG2 or 13SRG2 together with a plasmid expressing HA-tagged p300. Immunoprecipitations were carried out using M73 anti-E1A antibody and the proteins were resolved on a 4–12% gradient SDS–PAGE and western blots performed for p300 using the RW128 anti-p300 antibody. Input levels are shown. (C) HeLa cells were transfected with plasmids expressing either wild-type genomic E1A, E1A 12Sdl1101 or E1A 13Sdl1101, and immunoprecipitated with a mix of M73 and M58 anti-E1A antibodies. The immunoprecipitated proteins were resolved on a 3–8% gradient SDS–PAGE and Western blots performed for p300 using the RW128 anti-p300 antibody. Input levels are shown.
To determine whether p300 was capable of interacting with purified GST-CR3 in vitro, total A549 cell extract was mixed with purified GST or GST-CR3. GST-CR3 was able to pull-down endogenous p300 from A549 cell extract (Figure 7A).
We tested whether CR3 could independently bind p300 in vivo in the context of full-length E1A by co-immunoprecipitation (Figure 7B). E1A 12S and 13S co-precipitated comparable amounts of HA tagged p300. As reported previously, incorporation of the RG2 point mutation abrogated binding of p300 with E1A 12S in vivo (49,50). In contrast, E1A 13SRG2 differed from 12SRG2 as it was able to co-precipitate detectable amounts of p300 (Figure 7B). This result indicates that, in the absence of the N-terminal-binding site, CR3 can independently interact with p300 in vivo. Additional co-immunoprecipitations using the deletion mutant dl1101, which does not bind p300/CBP as it lacks residues 4–25, also reproducibly detected p300 (Figure 7C). Clearly, the observed interaction with the two E1A 13S mutants is not mediated by residual binding at the N-terminus, because the same mutants in the E1A 12S protein do not show any binding.
Taken together, these results show a direct physical interaction between p300/CBP and CR3 of E1A 13S in vitro and in vivo.
p300 is recruited to the adenovirus E4 promoter during infection
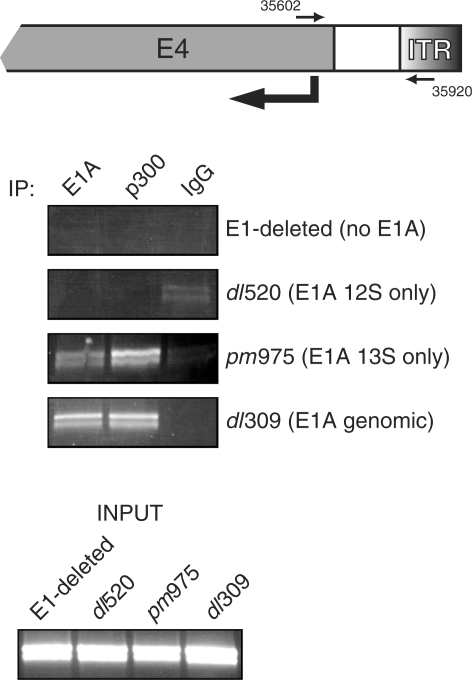
During viral infection, E1A facilitates transcriptional initiation at the viral early promoters (42). To determine whether p300 is recruited to the adenovirus E4 promoter during viral infection, we performed chromatin immunoprecipitations with E1A and p300 antibodies (Figure 8) on infected HeLa cells. In the context of a viral genome, E1A and p300 were found occupying the viral E4 promoter only in the presence of the E1A 13S isoform (pm975 and dl309 viruses), but not when the cells were infected with an E1A 12S only virus (dl520).
Figure 8.
p300 is recruited to the adenovirus E4 promoter only in the presence of E1A 13S. Top panel. A schematic representation of the right end of the adenoviral genome with the locations of primer hybridization indicated, numbering refers to the adenovirus type 2 genome. Bottom panel. HeLa cells were infected with the indicated adenoviruses and chromatin immunoprecipitation was carried out 16 h after infection with M73 anti-E1A antibody, RW128 anti-p300 antibody and mouse anti-rabbit antibody as a negative control. Immunoprecipitated DNA was then subjected to PCR analysis using E4-specific primers and the product was resolved on a 2% agarose gel.
DISCUSSION
In this report, we used the transcriptional squelching ability of E1A 12S protein to identify p300/CBP as a co-factor involved in transcriptional regulation by the E1A 13S protein. This conclusion was confirmed using siRNA-mediated knockdown of p300, which demonstrated a dramatic and specific effect on both CR3- and E1A 13S-mediated transactivation (Figure 6). Taken together, these results indicate for the first time that p300/CBP are critical factors for CR3-dependent transcriptional activation.
Prior to this report, p300/CBP interaction had only been mapped to the N-terminal/CR1 region of E1A. Indeed, p300 was identified through its interaction with this region of E1A (51). However, we now show that direct binding of CBP to GST-CR3 can be readily detected with purified proteins (Figure 7). Furthermore, independent binding of p300 to CR3 can be revealed if mutants of E1A 13S that lose high-affinity binding to the N-terminus are used in co-precipitation experiments (Figure 7).
Although both the E1A N-terminus and CR3 function as independent transactivation domains, their individual roles were not clear. This has been further complicated by the original definition of the N-terminal activation domain as a transcriptional repression domain (2). However, our data may provide some insight into the need for two separate activation regions. Specifically, the N-terminal activation domain in the E1A 12S protein may play a role in regulating transactivation by E1A 13S. Effectively, the relative abundance of E1A 12S and 13S may play a role in fine-tuning the overall transcription program carried out by E1A. Our data suggests that E1A 12S does this by sequestering p300/CBP from CR3. Although there are other common targets between the N-terminus of E1A and CR3, such as TBP and the proteasome (28,50,52), our data indicate that p300/CBP is the limiting factor as supplementation with excess p300 or CBP relieves the repressive effects of E1A 12S on CR3-dependent transactivation (Figure 5). Thus, the relative abundance of E1A 12S with respect to E1A 13S could regulate the effects of E1A 13S on transcription.
ChIP analysis reveals that only E1A 13S is efficiently recruited to the E4 promoter region on the viral genomic DNA during infection (Figure 8). Furthermore, occupancy of p300 at this promoter requires E1A 13S, consistent with a role for p300 in E1A-mediated transcriptional activation. The current model of transactivation by E1A 13S is based on recruitment of E1A to the viral promoter via specific interaction of the promoter targeting region of CR3 with sequence-specific factors such as ATF-2. Thus, it is not surprising to see that E1A 12S is not recruited and that it does not recruit p300 to the E4 promoter (Figure 8), as it lacks the promoter targeting domain. We have previously observed that E1A 13S is capable of being recruited to a GAL4-ZNF217 repression complex and activating it (41). Interestingly in that study, although 12S could interact with CtBP and therefore be recruited to ZNF217-occupied promoter region, it was not capable of activating transcription. Together these data suggest that transcriptional activation by E1A is primarily a function of the 13S splice variant. Although the E1A 12S isoform contains an intrinsic activation domain localized at the N-terminus, this is not sufficient to lead to strong activation, at least in the context of the reporter assays we utilized. Indeed, the data presented here suggest that it acts as a repressor, potentially regulating the function of E1A 13S. This agrees with numerous other studies that the E1A 12S protein can function as a general repressor of sequence-specific transcriptional activators (2) and is consistent with a recent report that E1A 12S causes a 3-fold reduction in total cellular histone H3 lysine 18 acetylation, which is a marker of active transcription (31). Our observation that p300/CBP acetyltransferases associate differently with E1A 13S than with the 12S isoform suggests that it may have different effects on global histone acetylation patterns. Unlike E1A 12S, E1A 13S may induce hyperacetylation of histones at specific target promoters. In the context of genomic E1A, where all isoforms of the protein are expressed, the differing functions of 12S and 13S on histone acetylation may fine-tune the reprogramming of the epigenetic code in the infected cell in order to efficiently initiate the cell cycle. This potentially presents an interesting area for future investigation.
In conclusion, this study sheds further light on the mechanism of transcriptional activation by the CR3 region of the E1A oncoprotein. For the first time, we identify a second independent and direct interaction of p300/CBP with E1A 13S and demonstrate a role for p300/CBP in transactivation by CR3. Our work also suggests that E1A 12S may play a role in limiting E1A 13S-dependent activation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by a grant from the Canadian Institutes of Health Research to J.S.M. (Grant number MOP-75647). P.P. was supported by a CIHR Strategic Training Program in Cancer Research and Technology Transfer Fellowship. J.N.G.A. was supported by an Ontario Graduate Scholarship in Science and Technology. Funding for open access charge: CIHR grant MOP-75647.
Conflict of interest statement. None declared.
Supplementary Material
Footnotes
Correspondence may also be addressed to Dr Joe S. Mymryk. Tel: +1 519 685 8600. Ext.: 53016; Fax: +1 519 685 8616; Email: jmymryk@uwo.ca
REFERENCES
- 1.Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 2.Bayley ST, Mymryk JS. Adenovirus E1A proteins and transformation (Review) Int. J. Oncology. 1994;5:425–444. doi: 10.3892/ijo.5.3.425. [DOI] [PubMed] [Google Scholar]
- 3.Kimelman D, Miller JS, Porter D, Roberts BE. E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J. Virol. 1985;53:399–409. doi: 10.1128/jvi.53.2.399-409.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avvakumov N, Wheeler R, D'Halluin JC, Mymryk JS. Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J. Virol. 2002;76:7968–7975. doi: 10.1128/JVI.76.16.7968-7975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avvakumov N, Kajon AE, Hoeben RC, Mymryk JS. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology. 2004;329:477–492. doi: 10.1016/j.virol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Glenn GM, Ricciardi RP. Adenovirus 5 early region 1A host range mutants hr3, hr4, and hr5 contain point mutations which generate single amino acid substitutions. J. Virol. 1985;56:66–74. doi: 10.1128/jvi.56.1.66-74.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lillie JW, Green M, Green MR. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell. 1986;46:1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- 8.Moran E, Zerler B, Harrison TM, Mathew MB. Identification of separate domains in the adenovirus E1A gene for immortalization activity and the activation of virus early genes. Mol. Cell Biol. 1986;6:3470–3480. doi: 10.1128/mcb.6.10.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran E, Grodzicker T, Roberts RJ, Mathews MB, Zerler B. Lytic and transforming functions of individual products of the adenovirus E1A gene. J. Virol. 1986;57:765–775. doi: 10.1128/jvi.57.3.765-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran E, Mathews MB. Multiple functional domains in the adenovirus E1A gene. Cell. 1987;48:177–178. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- 11.Lillie JW, Loewenstein PM, Green MR, Green M. Functional domains of adenovirus type 5 E1a proteins. Cell. 1987;50:1091–1100. doi: 10.1016/0092-8674(87)90175-9. [DOI] [PubMed] [Google Scholar]
- 12.Strom AC, Ohlsson P, Akusjarvi G. AR1 is an integral part of the adenovirus type 2 E1A-CR3 transactivation domain. J. Virol. 1998;72:5978–5983. doi: 10.1128/jvi.72.7.5978-5983.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholer HR, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 14.Agoff SN, Wu B. CBF mediates adenovirus Ela trans-activation by interaction at the C-terminal promoter targeting domain of conserved region 3. Oncogene. 1994;9:3707–3711. [PubMed] [Google Scholar]
- 15.Liu F, Green MR. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 16.Liu F, Green MR. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 17.Chatton B, Bocco JL, Gaire M, Hauss C, Reimund B, Goetz J, Kedinger C. Transcriptional activation by the adenovirus larger E1a product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1a. Mol. Cell Biol. 1993;13:561–570. doi: 10.1128/mcb.13.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin KJ, Lillie JW, Green MR. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990;346:147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- 19.Geisberg JV, Chen JL, Ricciardi RP. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol. Cell Biol. 1995;15:6283–6290. doi: 10.1128/mcb.15.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazzarelli JM, Mengus G, Davidson I, Ricciardi RP. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAF(II)135. J. Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzarelli JM, Atkins GB, Geisberg JV, Ricciardi RP. The viral oncoproteins Ad5 E1A, HPV16 E7 and SV40 TAg bind a common region of the TBP-associated factor-110. Oncogene. 1995;11:1859–1864. [PubMed] [Google Scholar]
- 22.Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 23.Glenn GM, Ricciardi RP. An adenovirus type 5 E1A protein with a single amino acid substitution blocks wild-type E1A transactivation. Mol. Cell Biol. 1987;7:1004–1011. doi: 10.1128/mcb.7.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster LC, Ricciardi RP. Trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol. Cell Biol. 1991;11:4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee WS, Kao CC, Bryant GO, Liu X, Berk AJ. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 26.Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein [see comments] Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Berk AJ. In vivo association of adenovirus large E1A protein with the human mediator complex in adenovirus-infected and -transformed cells. J. Virol. 2002;76:9186–9193. doi: 10.1128/JVI.76.18.9186-9193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasti M, Grand RJ, Yousef AF, Shuen M, Mymryk JS, Gallimore PH, Turnell AS. Roles for APIS and the 20S proteasome in adenovirus E1A-dependent transcription. EMBO J. 2006;25:2710–2722. doi: 10.1038/sj.emboj.7601169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bondesson M, Mannervik M, Akusjarvi G, Svensson C. An adenovirus E1A transcriptional repressor domain functions as an activator when tethered to a promoter. Nucleic Acids Res. 1994;22:3053–3060. doi: 10.1093/nar/22.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frisch SM, Mymryk JS. Adenovirus-5 e1a: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 31.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 33.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 34.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 35.Bres V, Tagami H, Peloponese JM, Loret E, Jeang KT, Nakatani Y, Emiliani S, Benkirane M, Kiernan RE. Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J. 2002;21:6811–6819. doi: 10.1093/emboj/cdf669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harlow E, Franza B.R., Jr., Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown K, Chen Y, Underhill TM, Mymryk JS, Torchia J. The Coactivator p/CIP/SRC-3 Facilitates Retinoic Acid Receptor Signaling via Recruitment of GCN5. J. Biol. Chem. 2003;278:39402–39412. doi: 10.1074/jbc.M307832200. [DOI] [PubMed] [Google Scholar]
- 38.Shuen M, Avvakumov N, Walfish PG, Brandl CJ, Mymryk JS. The adenovirus E1A protein targets the SAGA but not the ADA transcriptional regulatory complex through multiple independent domains. J. Biol. Chem. 2002;277:30844–30851. doi: 10.1074/jbc.M201877200. [DOI] [PubMed] [Google Scholar]
- 39.Boyd KE, Farnham PJ. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell Biol. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 41.Bruton R, Pelka P, Mapp KL, Fonseca G, Torchia J, Turnell AS, Mymryk JS, Grand RJ. Identification of a second CtBP binding site in adenovirus 5 E1A conserved region 3. J. Virol. 2008;82:8476–8486. doi: 10.1128/JVI.00248-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelka P, Ablack JN, Fonseca GJ, Yousef AF, Mymryk JS. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 2008;82:7252–7263. doi: 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jelsma TN, Howe JA, Evelegh CM, Cunniff NF, Skiadopoulos MH, Floroff MR, Denman JE, Bayley ST. Use of deletion and point mutants spanning the coding region of the adenovirus 5 E1A gene to define a domain that is essential for transcriptional activation. Virology. 1988;163:494–502. doi: 10.1016/0042-6822(88)90290-5. [DOI] [PubMed] [Google Scholar]
- 44.Egan C, Jelsma TN, Howe JA, Bayley ST, Ferguson B, Branton PE. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol. Cell Biol. 1988;8:3955–3959. doi: 10.1128/mcb.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang SE, Hearing P. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene. 2003;22:2836–2841. doi: 10.1038/sj.onc.1206376. [DOI] [PubMed] [Google Scholar]
- 46.Song CZ, Loewenstein PM, Toth K, Tang Q, Nishikawa A, Green M. The adenovirus E1A repression domain disrupts the interaction between the TATA binding protein and the TATA box in a manner reversible by TFIIB. Mol. Cell Biol. 1997;17:2186–2193. doi: 10.1128/mcb.17.4.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbeau D, Charbonneau R, Whalen SG, Bayley ST, Branton PE. Functional interactions within adenovirus E1A protein complexes. Oncogene. 1994;9:359–373. [PubMed] [Google Scholar]
- 48.Shuen M, Avvakumov N, Torchia J, Mymryk JS. The E1A proteins of all six human adenovirus subgroups target the p300/CBP acetyltransferases and the SAGA transcriptional regulatory complex. Virology. 2003;316:75–83. doi: 10.1016/j.virol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Wang H.-G, Rikitake Y, Carter MC, Yaciuk P, Abraham SE, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasti M, Grand RJ, Mymryk JS, Gallimore PH, Turnell AS. Recruitment of CBP/p300, TATA-binding protein, and S8 to distinct regions at the N terminus of adenovirus E1A. J. Virol. 2005;79:5594–5605. doi: 10.1128/JVI.79.9.5594-5605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A- associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 52.Geisberg JV, Lee WS, Berk AJ, Ricciardi RP. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc. Natl Acad. Sci. USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.