Abstract
MepR is a multidrug binding transcription regulator that represses expression of the Staphylococcus aureus multidrug efflux pump gene, mepA, as well as its own gene. MepR is induced by multiple cationic toxins, which are also substrates of MepA. In order to understand the gene regulatory and drug-binding mechanisms of MepR, we carried out biochemical, in vivo and structural studies. The 2.40 Å resolution structure of drug-free MepR reveals the most open MarR family protein conformation to date, which will require a huge conformational change to bind cognate DNA. DNA-binding data show that MepR uses a dual regulatory binding mode as the repressor binds the mepA operator as a dimer of dimers, but binds the mepR operator as a single dimer. Alignment of the six half sites reveals the consensus MepR binding site, 5'-GTTAGAT-3'. ‘Drug’ binding studies show that MepR binds to ethidium and DAPI with comparable affinities (Kd = 2.6 and 4.5 μM, respectively), but with significantly lower affinity to the larger rhodamine 6G (Kd = 62.6 μM). Mapping clinically relevant or in vitro selected MepR mutants onto the MepR structure suggests that their defective repressor phenotypes are due to structural and allosteric defects.
INTRODUCTION
Staphylococcus aureus is a leading cause of multidrug resistant infections, ranging from skin and soft tissue infections to endocarditis, in hospitals as well as community settings (1). Multidrug-resistant organisms employ a variety of mechanisms to negate the toxic effects of various antimicrobials that include alteration of the target or drug and preclusion of their intracellular accumulation by active extrusion by polyspecific or low specificity efflux transporters (2–4). Multidrug resistance efflux pumps (mdr pumps) were originally identified in human cancer cells that showed resistance to fluoroquinolones, but are present in all living organisms (5). These promiscuous membrane-embedded transporters bind to structurally and chemically dissimilar compounds and pump them out of the cell using the proton motive force or ATP hydrolysis (2). The activity of mdr efflux pumps promotes the persistence of pathogens in sanitized environments and allows them to acquire high level target-based resistance.
Efflux pumps are categorized into five different families: the major facilitator super (MFS) family, ATP-binding cassette (ABC) transporters, small multidrug resistance (SMR) transporters, multidrug and toxic compound extrusion (MATE) family and resistance-nodulation-cell division (RND) family (6,7). Amongst the various mdr pumps identified in S. aureus, most belong to the MFS or SMR family (5) but recent genetic and microbiological studies on multidrug resistant S. aureus strains identified the first MATE family multidrug transporter, MepA (8,9). MepA confers resistance to a wide range of compounds, which include various dyes, biocides, fluoroquinolones and the glycylcycline, tigecycline (9,10). Genetic analysis of the open reading frame of mepA reveals that the gene is part of the three-gene cluster, mepRAB, which encodes, MepR, a MarR family transcription regulator, MepA, the MATE-family transporter with 12 transmembrane segments and MepB, a protein of unknown function that belongs to the uncharacterized but highly conserved COG4815 family (Figure 1) (9). Analogous to other described multidrug efflux systems MepR negatively regulates mepR and mepA (and possibly mepB) expression by binding to promoter sequences upstream of the mepR and mepA transcription start sites, respectively (Figure 1). Repression is relieved by MepR binding to one of a multitude of structurally and chemically different monovalent and bivalent cationic, lipophilic drugs (11). The significance of MepR-dependent mepA regulation and its role in multidrug resistance is underscored by the finding of multidrug-resistant clinical isolates of S. aureus, which contain mutations in the mepR gene that result in defective repressor phenotypes (8,12).
Figure 1.
Genetic organization of the mepR, mepA and mepB genes. The open rectangular boxes represent the region encoding the respective genes, which are labelled below, and the shaded rectangles indicate the relevant promoter sequences, which encompass MepR-binding sites. The inverted repeats within the MepR-binding sites are indicated by head-to-head inverted arrows. The thick bent arrows above the rectangles denote the transcription start sites and the thin curved arrows with the negative signs below, indicate the promoters negatively regulated by MepR. The mepB gene does have a MepR-binding site.
MepR is a 139-residue protein that belongs to the MarR family of transcription regulators. Members of the MarR family have been implicated in the regulation of a variety of bacterial adaptive responses including those to oxidative stress and attack by toxins and biocides (13). MarR family proteins also regulate genes that are responsible for catabolism of aromatic compounds and encode virulence determinants (13,14). Analogous to other MarR family proteins, MepR represses transcription from the mepA and mepR promoters by binding to sequences overlapping the –35 and –10 hexamers (11). MepR is induced by binding to most, but not all, MepA substrates (11). Hence, MepR is a cytosolic multidrug sensor and participates in the defence against toxic compounds by regulating the expression of mepA. The DNA-binding site of MepR is characterized by the presence of pseudo palindromes as well as a signature GTTAG motif (11). Though both the mepA and mepR promoters have binding sites for MepR, they differ in location, complexity and multiplicity. The mepA promoter binding site spans a 44 base pair segment that consists of two inverted repeats, one overlapping the –35 element and the other extending into the –10 hexamer. There are two GTTAG motifs. By contrast, the MepR-binding site in the mepR promoter is characterized by a single inverted repeat and one GTTAG motif. This site includes the –10 hexamer and the transcription start site of the gene. An initial biochemical analyses of each MepR–promoter interaction revealed that MepR binds to the mepA promoter sequence with greater affinity than to the mepR promoter and MepR-mediated repression of the mepA promoter is complete relative to the leaky repression of the mepR promoter (11). Upon exposure to MepA substrates, MepR readily dissociates from the mepA promoter, whereas little to no effect was observed on mepR promoter (11). Amongst the various MepA substrates assayed, chlorhexidine, cetrimide, dequalinium, benzalkonium chloride (BAC), pentamidine and tetraphenylphosphonium bromide (TPP+) relieve MepR-dependent repression of mepA (11).
At present, the structural basis of multidrug binding and recognition by MarR family members is quite limited with only the structures of Escherichia coli MarR and Methanobacterium MarR bound to salicylate determined (15,16). It is worth mentioning that the cationic nature of MepR inducers is unusual amongst the MarR family of transcription regulators, as most of the functionally characterized proteins that belong to this family bind to neutral or anionic lipophilic compounds (15,17–20). Regardless, much of our structural understanding of multidrug binding and recognition has been derived from multiple structures of the ligand bound gene regulators, QacR, and TtgR, which belong to TetR family of regulators, and BmrR, a MerR family member (21–23). The structures of QacR and TtgR in complex with various inducers revealed the presence of a voluminous, multifaceted drug-binding pocket that accommodates diverse drugs by their interaction with different parts of the multidrug-binding pocket. The inner face of the drug-binding site is typically lined with an array of aromatic and hydrophobic residues, which are involved in van der Waals, stacking and cation–π interactions with the lipophilic compounds as well as charge neutralizing acidic or basic residues that can interact with the respective positive or negative charge of the drug (21–23). It remains to be seen whether MepR utilizes similar mechanisms to interact with its cationic ligands, specifically the use of multiple negative charges within the cationic drug-binding pocket and the presence of multiple minipockets similar to QacR or a single, more rigidly fixed pocket as observed in BmrR (21,22,24). Moreover, the impressive drug-binding profile of MepR should shed light into a unique model system as well as the multidrug-binding properties of all MarR family proteins.
In order to understand more fully the biochemical and structural mechanisms of multidrug and DNA binding by MepR, we determined the crystal structure of drug-free MepR at 2.4 Å resolution and carried out drug and DNA-binding studies. The structure also provided insight into the loss of repressor function of MepR mutants that have been identified in clinical isolates and in vitro selection.
EXPERIMENTAL PROCEDURE
Bacterial strains, plasmids, media and reagents
The strains and plasmids used in this study are listed in Table 1. LB broth was used for routine cell growth and protein overexpression. The growth media was obtained from DIFCO, whereas the tryptic soy broth and brain heart infusion were from BD Biosciences. Fluoresceinated DNA for the fluorescent polarization assays was purchased from Operon Biotechnologies. All other reagents were obtained from Sigma-Aldrich Chemical Co.
Table 1.
Strains and plasmids used for MepR functional analyses
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| S. aureus strains | ||
| SA-K2916 | SH1000 mepR::lacZ | (11) |
| SA-K2916-R | SA-K2916/pK434 | (11) |
| SA-K2916-R (Q18P) | SA-K2916/pK580 | (8) |
| SA-K2916-R (G97E) | SA-K2916/pK582 | (8) |
| SA-K2916-R (G97W) | SA-K2916/pK491 | This study |
| SA-K2916-R (A103V) | SA-K2916/pK519 | (46) |
| Plasmids | ||
| pALC2073 | S. aureus vector containing a tetracycline-inducible promoter controlling expression of cloned genes; Cmr | (35) |
| pK434 | pALC2073 mepR wt | (9) |
| pK491 | pALC2073 mepR (G97W) | This study |
| pK519 | pALC2073 mepR (A103V) | (46) |
| pK580 | pALC2073 mepR (Q18P) | (8) |
| pK582 | pALC2073 mepR (G97E) | (8) |
aCmr, chloramphenicol resistance selection.
Protein overexpression and purification
The construction of the MepR overexpression plasmid has been described elsewhere (11). Briefly, the mepR gene was cloned under the control of the T7 promoter with a hexa-histidine tag encoded at its 3′-end. Protein overexpression was achieved using the strain BL21 DE3 star. An overnight culture grown in 200 ml LB broth containing 40 μg/ml ampicillin was transferred into 9 l of LB broth supplemented with 40 μg/ml ampicillin and allowed to grow at 37°C until the OD600 reached 0.5–0.6. MepR overexpression was induced by the addition of 1 mM IPTG and incubation at 37°C for 3 h. Cells were harvested by centrifugation and stored at –80°C. The frozen cells were thawed and resuspended in 50 ml of Buffer A (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 2.5% glycerol and 1 mM TCEP) with one protease cocktail inhibitor pellet and 200 μg of DNase I. Cells were lysed by a microfluidizer M-110L (Microfluidics) followed by centrifugation at 15 000 rpm for 30 min to remove the cell debris. The cleared lysate was applied to Ni-NTA resin pre-equilibrated with Buffer A. After washing with 25 column volumes of Buffer A, MepR was eluted with a 0–300 mM imidazole gradient. MepR elutes at ∼100 mM imidazole. Further purification and buffer exchange with Buffer A was achieved by gel-filtration chromatography using a Superdex-200 column. Protein was purified to >95% homogeneity and was concentrated to ∼30 mg/ml using a YM-10 (10 kDa cutoff) centricon. The protein concentration was estimated by a Bradford assay (25) using bovine serum albumin as the standard and confirmed by mass spectrometry and amino acid analysis. Selenomethionine-substituted MepR (SeMet MepR) was overexpressed using the methionine inhibitory pathway (26) and purified as described for native MepR.
Crystallization and data collection
MepR was crystallized at room temperature using the hanging drop vapor diffusion method by mixing equal volumes of purified MepR and the reservoir solution, which contained 14–16% PEG 4000, 0.2 M MgSO4, 10% glycerol and additive NDSB-256. Both native and SeMet-MepR crystals grown in this condition belong to the space group P212121 with cell dimensions: a = 32.2 Å, b = 96.6 Å, c = 110.3 Å. Native and SeMet crystals were flash frozen in liquid nitrogen using 20% glycerol as the cryo-protectant. X-ray intensity data were collected at the Advanced Light Source (ALS) Beamline 8.3.1 at 100K. Intensity data processing and scaling were carried out using MOSFLM and SCALA (27,28). Native intensity data were collected to 2.40 Å resolution and multiple wavelength anomalous dispersion (MAD) data were collected to 2.60 Å. Selected intensity data statistics are given in Table 2.
Table 2.
Selected crystallographic data and statistics
| SeMet | Native | |||
|---|---|---|---|---|
| Data collection and phasing | ||||
| Wavelength (λ) | 0.9797 | 0.9798 | 1.02 | 1.116 |
| Resolution (Å) | 50.0–2.60 | |||
| Rsyma | 0.092 (0.22)b | 0.06 (0.19) | 0.07 (0.21) | 0.046 (0.14) |
| I/σ(I) | 12.3 (4.1) | 16.1 (5.5) | 14.5 (4.7) | 25.6 (8.4) |
| Total reflections (#) | 43794 | 44245 | 43757 | 54688 |
| Unique reflections (#) | 12590 | 12712 | 12621 | 14174 |
| Completeness (%) | 100 (100) | 99.9 (100) | 100 (100) | 100 (99.8) |
| Selenium sites (#) | 10 | |||
| Overall figure of meritc | 0.52 | |||
| Native MepR refinement statistics | ||||
| Resolution range (Å) | 50.0–2.40 | |||
| Rwork/Rfree(%)d | 23.9/28.8 | |||
| Atoms (#) | ||||
| Protein | 2128 | |||
| Sulphate ions | 5 | |||
| Solvent | 196 | |||
| B factors (Å2) | 38.2 | |||
| Rmsd | ||||
| Bond lengths (Å) | 0.006 | |||
| Bond angles (°) | 1.007 | |||
| B for bonded main-chain atoms (Å2) | 1.397 | |||
| Ramachandran analysis | ||||
| Most favoured (%) | 95.9 | |||
| Add. allowed (%) | 4.1 | |||
| Gen. allowed (%) | 0.0 | |||
| Disallowed (%) | 0.0 |
aRsym = ∑∑|Ihkl − Ihkl(j)|/∑Ihkl, where Ihkl(j) is the observed intensity and Ihkl is the final average intensity value.
bValues in parentheses are for the highest resolution shell.
cFigure of Merit = <|ΣP(α)eiα/ΣP(α)|>, where α is the phase and P(α) is the phase probability distribution.
dRwork = Σ||Fobs| − |Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|; where all reflections belong to a test set of 5% randomly selected reflections.
Structure determination and refinement
Ten selenium sites, indicative of two MepR subunits (12 selenium sites in totu) per asymmetric unit, were located and refined with SOLVE using MAD data to 2.6 Å resolution. The resulting figure of merit was 0.52. Solvent flattening in RESOLVE (29,30) resulted in a much improved electron density map. Model building of the MepR dimer was done with ‘O’ (31) and Coot (32) and all refinement was performed using CNS (33). After successive rounds of positional and B-factor refinement and model rebuilding, the initial model contained residues 4–82 and 88–139 of each subunit. This structure was refined against the native data to 2.40 Å resolution. The final model contains residues 3–84 and 87–139 of subunit A and residues 4–81 and 89–139 of subunit B, 196 water molecules and five sulphate ions. Due to poor electron density, the side chains of residues 82, 83, 87 and 88 of subunit A were truncated to alanine. The refined model has a final Rfree of 0.288 and Rwork of 0.239 for all data in the resolution range of 50.0–2.40 Å. Selected data collection, phasing and refinement statistics are given in Table 2. All figures were generated using Pymol (34). The coordinates and structure factors have been deposited in the RCSB with PDB accession code 3ECO.
Fluorescence polarization assay
Fluorescence polarization-based MepR-ligand binding experiments were performed with a Panvera Beacon Fluorescence polarization system (Invitrogen) utilizing the intrinsic fluorescence of fluorescein labelled DNA or MepR inducers, ethidium (Et), rhodamine 6G (R6G) and 4′,6-diamidino-2-phenylindole (DAPI). The polarization (P) of the labelled DNA or drugs increases as a function of protein binding and equilibrium dissociation constants are determined from plots of millipolarization (P × 10–3) against protein concentration. For MepR-DNA-binding studies, 1 nM 5′-fluoresceinated oligodeoxynucleotide of various lengths in 1 ml binding buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl and 2.5% glycerol) was titrated against increasing concentrations of purified MepR and the resulting change in polarization measured. Samples were excited at 490 nm and emission measured at 530 nm. The MepR-drug-binding studies were carried out in a drug-binding buffer composed of 20 mM Tris–HCl pH 7.5, 200 mM NaCl and 2.5% glycerol. The excitation wavelengths were 490, 490 and 330 nm, respectively for R6G, Et and DAPI and the emission measured at wavelengths of 560, 620 and 560 nm, respectively. All data were plotted using Kaleidagraph and the resulting plots were fitted with the equation P = {(Pbound − Pfree)[protein]/(KD + [protein])} + Pfree, where P is the polarization measured at a given protein concentration, Pfree is the initial polarization of the free ligand, Pbound is the maximum polarization of specifically bound ligand and [protein] is the protein concentration. Non-linear least squares analysis was used to determine Pbound, and Kd. The binding constant reported is the average value from three independent experimental measurements.
β-galactosidase assay
An in vivo transcription assay system was employed to determine the functional integrity of various MepR mutant proteins that had been identified in previous studies (8,12). The construction of the tetracycline-inducible MepR overexpression plasmid pALC2073 has been described elsewhere (35). These pALC2073-based constructs were transduced into SA-K2916, which contains a chromosomal mepR::lacZ transcriptional fusion (11). Using a β-galactosidase assay the effect of induction of plasmid-based mepR expression on chromosomal mepR regulated β-galactosidase expression could be determined. Test strains were grown overnight in Trypticase Soy broth, washed with phosphate-buffered saline (pH 7.0), and diluted 1:200 in 100 ml prewarmed Brain–Heart Infusion broth, with or without 50 ng/ml tetracycline, and grown at 37°C with agitation. β-galactosidase activity was measured employing 4-methylumbelliferyl-β-d-galactopyranoside (MUG) as a substrate. Briefly, 0.5 ml culture aliquots were taken at intervals during growth for measurement of OD600. Cells in a second 0.5 ml aliquot were harvested by centrifugation and the pellets snap frozen at –80°C whilst awaiting assay. To assay for β-galactosidase activity, cells were thawed, resuspended in 0.5 ml ABT (100 mM NaCl, 60 mM K2HPO4, 40 mM KH2PO4, 0.1% Triton X-100), and incubated at 37°C for 15 min. Fifty microlitres MUG (4 mg/ml stock) were then added and the mixture incubated at room temperature for an additional hour. The reaction was stopped by the addition of 0.5 ml 0.4 M Na2CO3. Samples were serially diluted in a 1:1 (v:v) mixture of ABT and Na2CO3 in 96-well polystyrene microtiter plates. A range of concentrations of MUG was used to generate a standard curve and β-galactosidase activity (expressed in MUG units; 1 unit = 1 pmol MUG cleaved per min per OD600) was determined by fluorescence using a Bio-Tek FLx800 plate reader (Bio-Tek Instruments, Inc., Winooski, VT, USA). Cumulative chromosomal mepR expression over the course of 10 h experiment was determined by integrating the area beneath expression curves using software from the SigmaPlot 11.0 suite (Systat Software, Inc., Point Richmond, CA, USA).
Size exclusion/gel-filtration chromatography
Size exclusion/gel-filtration chromatography was used to determine the oligomerization states of apo MepR and two MepR–DNA complexes. A Superdex 200 26/60 column (Amersham Pharmacia Biotech) with a mobile phase containing 20 mM Tris–HCl pH 7.5, 200 mM NaCl and 2.5% glycerol was used in the gel-filtration experiments. Blue dextran (Sigma) was used to determine the void volume of the column. The size exclusion column was calibrated using RNase A (Mr 13 700 kDa), carbonic anhydrase (Mr 29 000 kDa), bovine serum albumin (Mr 66 000 kDa) and alcohol dehydrogenase (Mr 150 000 kDa). The KAverage (Kav) was calculated using the equation Kav=(VE – VO)/(VT – VO), where VT, VE and VO are the total column volume, elution volume and void volume of the column, respectively. A standard plot was obtained by graphing the logarithm of the molecular weight (Mr) against the Kav (Kaleidagraph). The Kav of each marker as well as the experimental samples were the average value of two or more experiments.
RESULTS AND DISCUSSION
Structure of MepR
The drug-free or apo structure of MepR was determined to 2.4 Å resolution by multiwavelength anomalous dispersion (MAD) methods using selenomethionine-substituted MepR. The asymmetric unit contains one MepR dimer. The final refined models exhibited excellent stereochemistry with 100% of the observed residues in the most favored (95.9%) and additionally allowed (4.1%) regions of the Ramachandran plot (Table 2).
MepR is predominantly α helical with six α helices and a two-stranded antiparallel β hairpin and an overall topology of α1 (residues 5–26) -α2 (residues 31–42) -α3 (residues 50–57) -α4 (residues 61–74) -β1 (residues 77–81) - W1 (the tip of the Wing, residues 82–84)-β2 (residues 88–93) -α5 (residues 95–118) -α6 (residues 121–139) (Figure 2A and C). Each subunit is composed of two functional domains: a dimerization domain that includes helices α1, α5 and α6, and a DNA-binding domain with a winged helix-turn-helix (wHTH) motif (α3,α4,β1,W1, β2) in the middle of the polypeptide chain (Figure 2B). The DNA-binding domain is connected to the dimerization domain by helices α2 and α5. Unlike most MarR family members, the apo MepR structure lacks a third β strand typically located between α2 and α3. Residues located at the tip of the wing, between β1 and β2, are either missing completely or have had their side chains truncated to alanines. The disorder in this region is likely due to the inherent flexibility of the wing in the absence of DNA as observed in other MarR family members such as OhrR (36,37).
Figure 2.
Structure of MepR. (A) Ribbon diagram of the MepR subunit. The secondary structural elements are labelled and rainbow colored from red (N-terminus) to blue (C-terminus). The N- and C-termini are indicated as N and C, respectively. With the exception of the loops between β1 and β2, all loops are colored grey. (B) Ribbon diagram of the MepR dimer. The secondary structure elements of one subunit is color coded as in Figure 2A. The dyadic mate is colored magenta. The dimerization interface and the winged helix-turn-helix motif (wHTH) are labelled. (C) Primary sequence alignment of MepR with other structurally characterized MarR family members [MepR, B. subtilis OhrR (OhrR-B), X. campestris OhrR (OhrR-X), E. coli MarR, P. aeruginosa MexR and Methanobacterium MarR, MTH313]. The multiple sequence alignment was made with ClustalW (45). The secondary structure elements of MepR are indicated above the alignment, α helices as rectangles and β strands as arrows, and colored as in Figure 2A. Identical residues are colored red and the chemically similar residues are blue. The conserved hydrophobic residues that are involved in dimerization are highlighted in shaded boxes. The magenta asterisks (*) indicate the positions of MepR mutations that had been identified from clinically isolated or in vitro selected multidrug-resistant strains of S. aureus.
Superimposition of the individual subunits of the MepR dimer revealed a remarkable degree of flexibility with a root mean square deviation (r.m.s.d.) of 2.2 Å for 127 corresponding Cα atoms (Figure 3A). This degree of conformational asymmetry of subunits within the same dimer has not been observed previously in the unliganded structures of any other MarR family members. Given the variation amongst the MarR family members in the lengths of their cognate DNA-binding sites and their spacer regions as well as the size, charge and hydrophobicity of their different inducers, an inherently flexible molecule is clearly required for the transition between different functional states (14).
Figure 3.
Conformational plasticity of MepR. (A) Superimposition of the two subunits of the MepR dimer with subunit A colored light pink and subunit B, red. Subunits are depicted as ribbons and the location of the N- and C-termini are labelled N and C, respectively. (B) Superimposition of the MepR dimer (light pink) and DNA-bound OhrR dimer (cyan), using the entire polypeptide chain. The wHTH motifs of MepR and DNA-bound OhrR are colored red and yellow, respectively. The midpoints of the recognition helices of MepR and OhrR are shown as spheres and their intersubunit distances provided above by the arrows. (C) Superimposition of the subunits of selected MarR family members (MepR, apoOhrR, Xc OhrROxi, DNA-bound Bs OhrR, E. coli MarR, Methanobacterium MarR, and MexR). The wHTH motifs of each protein are colored light pink and labelled. The N- and C-termini of MepR are colored black and cyan, respectively, and labelled, whereas the N- and C-termini of all the other proteins are colored green and red and labelled N (green) and C (red), respectively. On the right, the overlays are rotated 180° along the y-axis.
The dimer interface of MepR
The MepR dimer has the typical ‘safety triangle’ shape of the MarR family with its dimerization domain in the apex of the triangle, whilst the two-winged helix-turn-helix (wHTH) motifs are located at the base (Figure 2B). In this arrangement, a wide cleft is created between the two DNA-binding domains. The dimer interface is formed by the burial of hydrophobic residues located on the internal faces of helices α1, α5 and α6 of one subunit along with similar residues from their dyadic mates (α1′, α5′ and α6′, where ′ denotes the second subunit). Specifically, Tyr5, Leu8, Phe9, Ile12 and Met16 in the N-terminal half of α1 interact with Met111′, Leu115′ and Leu119′ of α5′ as well as Met127′, Leu131′ and Leu135′ of α6′. The same interactions are made by their dyadic mates. These hydrophobic interactions are masked from the solvent by polar interactions between Asn130 and Glu123′, Gln118′ and Lys133, and Gln126 and Gln126′. The extensive interaction between the subunits leads to the burial of 3475 Å2 accessible surface area (ASA) in the dimer interface. Although the sequence identity of these residues varies amongst the MarR family members, the hydrophobic nature of the residues is highly conserved (Figure 2C).
Conformational incompatibility for DNA binding
The wHTH motif of MepR is composed of helices α3 and α4 (the recognition helix) and the following antiparallel β hairpin formed by β1, β2 and a four-residue connecting loop, i.e. the wing (W1) (38,39) (Figure 2B and C). Most of the intradomain contacts of the wHTH motif are limited to the hydrophobic interactions between the family wide, highly conserved non-polar residues located on the internal face of helices α2, α3, α4, β1, β2 and the N-terminal half of helix α5. With few exceptions, the recognition helix makes base-specific contacts in the major groove, whereas the wing interacts specifically with the bases in the adjacent minor groove or with the phosphate backbone in a non-specific fashion (38,39). To date only one crystal structure of a MarR family member–DNA complex has been reported, that of B. subtilis OhrR bound to the ohrA operator sequence (36). That structure revealed that the duplex DNA runs through a cleft between the DNA-binding domains with the 2-fold related recognition helices of the OhrR dimer interacting with pseudo palindromic sequences of consecutive major grooves. The wings are extended on either side of the recognition helices making specific and non-specific contacts with the bases in the minor groove (36). A third DNA-binding element, the helix-helix (HH) motif, formed by helices α1 and α2, interacts with the DNA backbone in the spacer region that connects the pseudo palindromes (36). As observed for most apo MarR family member structures, drug-free MepR is not configured for DNA binding. First, the distance between the midpoints of the recognition helices [R69 (Cα) – R69′ (Cα)] is ∼43 Å (Figure 3B) which is incompatible with binding consecutive major grooves of B-DNA (∼34 Å). Second, the superimposition of MepR and DNA-bound OhrR reveals the outward displacement of all DNA-binding elements of MepR such that in addition to the increased distance between the recognition helices, the distance between the HH motifs of MepR is 7 Å longer than those of DNA-bound OhrR (Figure 3B). Third, the corresponding winged helices of MepR and DNA-bound OhrR, although equally distanced in the two dimers, take quite different orientations whereby the wings of MepR are nearly perpendicular to those of OhrR (Figure 3B).
MepR is captured in an extended form incompatible with B-DNA binding. Indeed, comparison with all other structurally characterized MarR family members reveals that MepR takes the most extended structure (Supplementary Table 1). Only reduced apo Bs OhrR (35 Å) and oxidized Xc OhrR (35 Å) show significant widening between their recognition helices (Supplementary Table 1). By contrast, the midpoints of the recognition helices of most MarR family members range from 23 to 32 Å. Of these, only apo HucR and one of the four different dimer conformers of MexR (MexR-CD), which shows a midpoint distance of 29 Å, could be modelled successfully onto B-DNA. Interestingly, we were also able to place the recently determined MarRM-salicylate complex, which has a center-to-center distance between recognition helices of 30 Å, onto a B-DNA site (data not shown) (40,41). The 29–30 Å distances between the midpoints of the recognition helices are close to that measured between the recognition helices of Bs OhrR bound to DNA (31 Å). By contrast to MepR, some MarR family members, including apo MarRM and one dimer of apo MexR (MexR AB) display quite short center-to-center distances of 23 and 24 Å, respectively. Thus, MarR family members display a huge range of motion allowing the DNA-binding domains to span at least 20 Å. Of course, the center-to-center distance is not the only factor for DNA-binding as the proper conformation and orientation of DNA-binding elements are necessary as well. However, it would appear that the center-to-center distance of the recognition helices of MarR family members must be ∼30 Å for DNA binding. Clearly, the observed conformation of apo MepR is not suited for high affinity DNA binding and requires significant changes to effect such binding to the mepA and mepR operator sites.
Conformational flexibility of MepR and its structural homologues
The superimposition of the subunits of MepR revealed a remarkable degree of conformational plasticity with an r.m.s.d. of 2.2 Å. Inspection of the overlay shows that most of the structural differences lie in the dimerization domain of each subunit (Figure 3A and B). Such structural asymmetry within a dimer is unusual amongst the structurally characterized MarR family members, i.e. their dyadic subunits typically take very similar conformations. By contrast and as seen with other MarR family members, the DNA-binding domains of MepR (residues 50–93) and other MarR family members exhibit a high degree of structural similarity with r.m.s.d. ranging from 0.9 to 1.0 Å (Figure 3C). Given the 2.2 Å difference between the two subunits of MepR, it might be expected that the dimerization domains of MepR and other MarR family members display significant differences in the relative positions of their α1, α5 and α6 helices. Indeed, the largest positional differences reside in the N-terminus of α1, the C-terminal half of α5, and the entirety of helix α6 (Figure 3C). Conformational plasticity of these helices has been observed previously in the structures of OhrR, MexR and EmrR (37,40,42) and we would suggest that the intrinsic flexibility of these structural elements provides the impetus for the structural transitions that occur between the different functional states of each protein viz., the apo, DNA bound and induced states. Consistent with this supposition, the structures of OhrR in its different functional states, i.e. reduced, DNA-free, DNA-bound and oxidized (induced), reveal that key structural and functional rearrangements are found in the loop between α1 and α2, helix α5, the loop between α5 and α6, and α6. The end result is a rigid body movement of the DNA-binding domains (36,37).
DNA binding by MepR
MepR autoregulates its own expression and mepA expression independently by binding to similar but not identical gene-specific operator sequences (11). Biochemical analysis of the sequences upstream of mepR and the sequences in the intergenic region of mepR-mepA has identified two MepR binding sites (11). In that study, a differential response to drugs on these promoters was noted as MepR readily dissociated from the mepA promoter upon the addition of drugs but far less so from the mepR operator (11). Footprinting studies of the mepA and mepR promoters with MepR identified a 43 bp binding site in the mepA promoter and 26 bp site in the mepR promoter (11). Analysis of these binding sites revealed pseudo palindromic sequences that overlapped crucial promoter elements as well as a GTTAG signature motif. The mepA promoter possesses two of these motifs whilst the mepR promoter has only one (11). Based on these observations, it was proposed that a differential affinity for mepA and mepR operator sequences could be due to the plurality of these MepR-binding elements in the former promoter region.
In order to determine the minimal sequence required for MepR binding, fluorescent polarization-based DNA-binding assays were carried out with various lengths of oligodeoxy-nucleotides, which encompassed the proposed MepR-binding sites in the mepA and mepR promoters. Results from the titration experiments indicate that MepR binds to a 44 bp duplex covering the ‘full’ MepR footprint of the mepA promoter with a dissociation constant (Kd) of 6.3 nM (Figure 4A). Interestingly, truncation of downstream sequences had little effect on MepR binding whereby the dissociation constant for a 26 bp oligodeoxynucleotide, which does not include 18 bp from the 3′-end of the 44 bp site, was only ∼2.4-fold lower (Kd = 15.3 nM) than that of the complete mepA site (Figure 4B). Removal of four additional base pairs further impairs MepR binding to the mepA site another 3.3-fold (Kd = 46.4 nM) but binding remains relatively tight with only a cumulative ∼7-fold loss in affinity as compared to the 44 bp site (Figure 4C and F). By contrast, deletion of eight base pairs from the 5′-end of the 44mer causes a greater than 30-fold reduction (Kd = 186.9 nM) in the affinity of MepR for the mepA-binding site (Figure 4D). DNA-binding experiments with the 26 bp MepR-binding site from mepR promoter also showed high-affinity binding (Kd = 24.3 nM) that essentially matches the affinity of the tested 26-bp fragment from the mepA promoter and is only ∼4-fold lower than that of the full 44 bp mepA site (Figure 4B and E). Thus, the previously observed differential response to drugs, i.e. the slower release of MepR from mepR operator site, does not appear to correlate well with MepR–DNA-binding affinity and suggests that MepR binds to mepA and mepR-binding sites somewhat differently, whereby subsequent drug binding to MepR when bound to mepR promoter is less effective in induction than drug binding to MepR bound to mepA-binding site.
Figure 4.
Binding isotherms of MepR and selected ligands. (A through D) MepR-binding isotherms to various lengths of cognate DNA from the mepA promoter. (A) MepR binding to a 44-bp site encompassing the entire mepA promoter DNase I footprint. (B) MepR binding to a 26-bp site of the mepA promoter that has been truncated from the 3′-end. (C) MepR binding to a 22-bp site of the mepA promoter that has been truncated from the 3′-end. (D) MepR binding to a 31-bp site of the mepA promoter that has been truncated from the 5′-end. (E) MepR-binding isotherms to a 26-bp DNA binding site from the mepR promoter. (F) Nucleotide sequence of the mepA promoter. The –10 and –35 elements of the promoter are shown in bold and labelled and the transcription start site (TSS) is indicated as a bent arrow. The pseudo-inverted repeats of the promoter are shown by horizontal arrows and the signature GTTAG motifs are underlined. The boundaries of the 22, 26 and 31-bp sequences used in the binding studies are indicated by blue, red and purple rectangles, respectively. G–I, MepR-binding isotherms for three ‘drugs’. (G) The MepR-Et-binding isotherm. (H) The MepR-DAPI-binding isotherm. (I) The MepR-R6G-binding isotherm. The change in polarization, shown in red circles and indicated in millipolarization units (mP), was plotted against the MepR dimer concentration indicated inside each plot. The binding constants are shown in each plot. The chemical structure of each drug is shown below the respective binding curve.
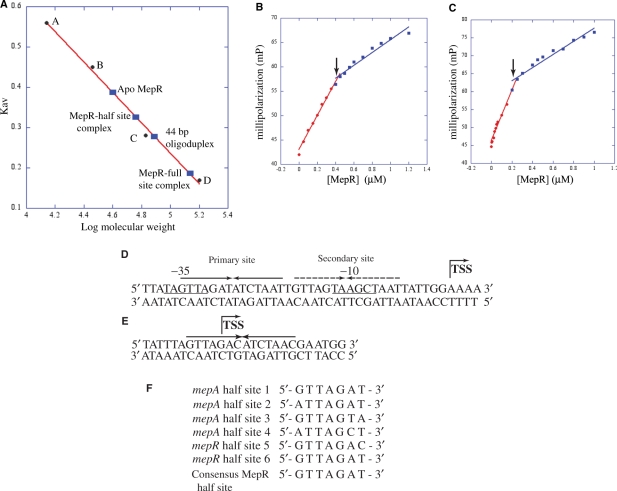
The different lengths of the MepR-binding sites in the mepA and mepR promoters suggest that MepR binds to these sequences with different stoichiometries. To determine the number of MepR dimers that bind each site, size exclusion chromatography was utilized. The elution volumes of MepR, MepR complexed with the ‘half site’ (27 bp DNA) and ‘full site’ (44 bp DNA) sequences from the mepA promoter were determined and the molecular weights for each species calculated (Figure 5A, Table 3). As expected, purified MepR eluted at a volume, which corresponds closely to the calculated molecular weight of a dimer (37.6 versus 33.7 kDa). The elution volume for MepR bound to the half site indicates a molecular mass of 62.8 kDa, which is near but noticeably larger than the calculated 50.3 kDa molecular weight of a MepR dimer bound to the 27 bp half site (Figure 5A). The discrepancy between the calculated and experimental values is the likely result of the non-spherical shape of the MepR–DNA complex. Gel-filtration experiments with MepR complexed to the full site give a molecular weight of 147.2 kDa. This value would suggest strongly that more than one MepR dimer bind to the 44 bp promoter sequences (calculated Mr = 60.8 kDa) (Table 3). Given the differences in DNA and protein shapes, a thin rigid rod of DNA bound to a triangle-shaped protein very likely runs anomalously through the column. Consistent with this, the elution volume of the 44 bp duplex DNA comprising the mepA full site alone indicates a molecular weight of ∼75 kDa, which is significantly greater than its calculated molecular weight of ∼29 kDa. In light of this information, the added values of the calculated molecular weight of the anomalously migrating individual components, 75 and 37 kDa for the 44 bp duplex and MepR dimer, respectively, correlates well with the experimentally determined molecular weight (147 kDa) of two MepR dimers complexed to the 44 bp site (2 × 37 kDa + 75 kDa = 149 kDa) (Table 3
Figure 5.
Stoichiometry of MepR binding to the conserved pseudo palindromes located in the mepA and mepR promoters. (A) Size exclusion chromatography of MepR and MepR–DNA complexes. The linear fit of the elution values of four standard proteins [(A), ribonuclease A (Mr, 13.7 kDa); (B), carbonic anhydrase (Mr, 29 kDa); (C), bovine serum albumin (Mr, 66 kDa); (D), alcohol dehydrogenase (Mr, 150 kDa)] is shown as a red line. The Kav of MepR and MepR bound to the 44 and 27 bp high-affinity binding sites of the mepA promoter was plotted on the graph (solid blue rectangles) and labelled. (B and C) Fluorescence polarization-based determination of MepR–mepA-binding stoichiometry. (B) Binding to the 44 bp mepA operator site. (C) Binding to the 26 bp mepA operator site. The linear fit for the high affinity (specific) and low affinity (non-specific) binding are color coded in red and blue, respectively. The inflection points that denote the breaking point of high and low-affinity binding are indicated by black arrows. (D) The sequence of the MepR-binding sites on the mepA promoter. The 44-bp sequence comprising the MepR footprint of the mepA promoter is shown with the –10 and –35 hexamers of the promoter underlined and labelled. The primary binding site of MepR is indicated by solid inverted arrows, whereas the secondary site is shown as broken inverted arrows. The transcription start site (TSS) is marked by a bent arrow. (E) The sequence of the MepR-binding sites on the mepR promoter. The pseudo palindrome is shown as solid inverted arrows and the transcription start is indicated by the bent arrow. (F) Alignment of the heptanucleotide sequences of the MepR binding half sites of the mepA and mepR promoters. The half sites from the MepR primary (MepR sites 1 and 2) and secondary sites (sites 3 and 4) of the mepA operator and from the MepR-binding site of the mepR promoter (sites 5 and 6) are aligned from 5′→3′ direction and the consensus sequence derived from the sequence similarity amongst the six sequences is shown at the bottom of the alignment.
Table 3.
Data from size exclusion/gel filtration experiments for MepR and MepR bound to various lengths of cognate DNA
| MepR/MepR–DNA complexes | Oligomerization state | Predicted molecular weight (kDa) | Experimentally determined molecular weight (kDa) |
|---|---|---|---|
| Apo MepR | Dimer | 33.7 | 37.6 |
| Tetramer | 67.4 | ||
| MepR-27 bp | Dimer—27 bp | 50.3 | 63.1 |
| Dimer of dimers—27 bp | 84.0 | ||
| MepR-44 bp | Dimer—44 bp | 60.8 | 147.2 |
| Dimer of dimers—44 bp | 94.5 | ||
| 44 bp oligoduplex | – | 27.0 | 75.0 |
The gel-filtration elution properties of MepR and DNA prompted us to carry out a fluorescent polarization-based stoichiometry assay in which the DNA-binding site was added in 20-fold molar excess of its Kd and titrated with protein. Polarization was then measured at each protein dimer titration point and the MepR concentration at the inflection point of the resulting curve used to deduce the stoichiometry. The results from the 44 bp duplexes showed that the inflection point, i.e. the point at which all high-affinity (specific) sites are bound, occurs at 400 nM MepR dimer in a binding experiment that contains 200 nM dsDNA, thus demonstrating that two MepR dimers bind specifically to the full site (Figure 5B). Analogous experiments with the 27 bp duplex showed that MepR binds to the smaller site as a dimer as the inflection point occurs at 200 nM MepR dimer (Figure 5C). The different stoichiometries of MepR for the full and half sites of cognate DNA indicate that MepR employs different mechanisms in its regulation of mepA and mepR expression.
The mechanism with which MepR binds the mepA operator will require the crystal structures of appropriate repressor–DNA complexes. However, insight into the mepA-binding mode of MepR can be gained by reference to the crystal structure of B. subtilis OhrR–ohrA complex, in which the ohrA-binding site contains the 12 bp inverted repeat (5′-ATTGTA•TACAAT-3′) that is flanked on either side by A-T rich sequences (36). The OhrR recognition helices make symmetric contacts with the palindromic sequences whilst the wings make specific as well as non-specific contacts with the upstream and downstream A-T rich minor grooves. Inspection of mepA promoter sequences reveals a similar arrangement of binding elements and leads to the proposal that the nearly perfect repeat, 5′-GTTAGAT•ATCTAAT-3′, which spans from –35 to –22 of the mepA promoter and contains the GTTAG signature motif, is the ‘specificity’ element for the recognition helices, whilst the flanking A-T rich region provides the interacting surface for the wings of the wHTH motif (Figure 5D). In addition, a second putative MepR-binding site, 5′-GTTAGTA•AGCTAAT-3′, that exhibits strong sequence conservation to the other MepR-binding element is found between –20 and –7 and overlaps the –10 hexamer of the mepA promoter (Figure 5D).
On the basis of these observations and our binding data, we propose that the palindrome, which overlaps the –35 hexamer, is the primary, higher affinity MepR-binding site and the pseudopalindrome that encompasses the –10 hexamer is a lower affinity second site. Consistent with this, MepR binds with high affinity to the 26 bp fragment that contains the entire primary site as a single dimer, whereas two dimers interact with the full site, which is comprised of both the primary and the secondary binding site (Figures 4A,B, 5A and B) (Table. 3). Removal of downstream sequences, which keeps the primary palindrome intact, has little effect on the binding affinity (Figure 4B, C and F). However, destruction of the left half of primary site, as seen in the mepA 31 bp fragment, lowers the binding affinity by >30-fold (Figure 4D and F) indicating that the secondary site is significantly weaker than the proposed primary site. Similar to the mepA sites, the MepR-binding site in the mepR promoter is a nearly perfect palindrome, 5′-GTTAGAC•ATCTAAC-3′, that includes the transcription start site and is located between –4 and +10 sequence with a laterally located upstream A-T rich sequence (Figures 4E and 5E). Thus, alignment of the sequences of the six heptanucleotide half sites of the mepA and mepR promoters reveals the consensus half site sequence 5′-GTTAGAT-3′ (Figure 5F).
Given the nature of the specificity elements, location, and the multiplicity of binding site, the tighter repression from the mepA promoter by MepR can be explained by the presence of both high- and low-affinity binding sites that overlaps crucial promoter elements. Thus, when MepR is bound to the mepA promoter, perhaps cooperatively as a dimer of dimers, the –10 and –35 hexamer binding elements of RNA polymerase (RNAP) are occluded, thereby mediating effective repression. By contrast, the weaker interactions between MepR and its binding site in the mepR promoter as well as the locus of the MepR binding site results in the blockage of only the –10 box from RNAP. RNAP binding at the –35 element would result in transcription from the mepR promoter by dissociation of MepR as part of its normal on and off DNA-binding equilibrium (11). Although still unclear, the poorer ability of drugs to induce MepR from the mepR promoter suggests the intriguing possibility that the differences between the mepR and consensus sequences, for example substitution of the central TpA step by CpA, alter the MepR conformation such that when bound to mepR, drug binding by the repressor is diminished as suggested by earlier drug induced DNA ‘knock off’ experiments (11), or conformational changes in MepR induced by weaker inducer binding is insufficient to promote complete induction. Moreover, cooperative binding by MepR dimers to the mepA operator could also play a role in the rapid induction of this promoter by drug. Additional structural and biochemical studies should clarify these observations.
Analyses of MepR mutants
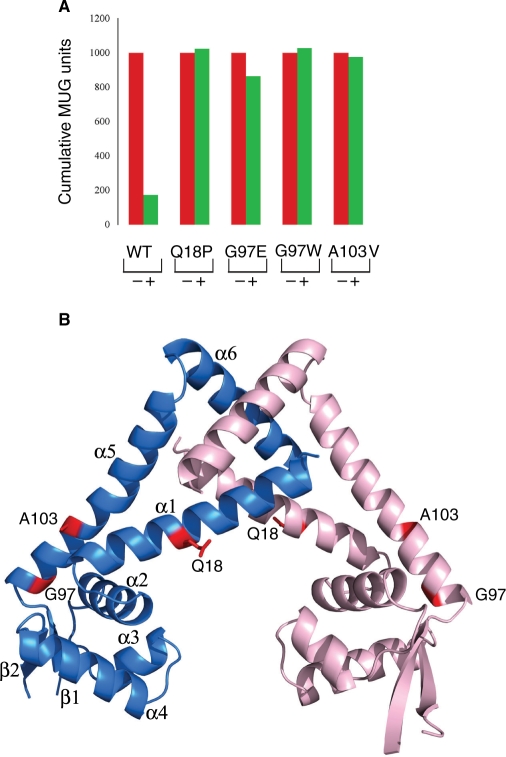
Several MepR mutations have been found in multidrug-resistant S. aureus clinical isolates and by in vitro selection. One mutation, which results in the replacement of MepR residue A103 by valine (A103V), was obtained from a multidrug-resistant strain found in blood samples of a patient (12), whereas three others, G97E, G97W and Q18P, were derived from bacteria that were exposed to a variety of drugs in vitro in either a single step or gradient exposure (8). The functional integrity of these mutants was tested by their ability to repress the expression of β-galactosidase from a mepR-lacZ fusion. The expression of β-galactosidase from a mepR-lacZ fusion was measured before and after the induction of MepR expression from a plasmid carrying the mepR gene under a tetracycline inducible promoter. Upon induction, the wild-type MepR represses gene expression from the mepR promoter by greater than 5-fold, whereas the four mutant proteins displayed a 90% or greater reduction in their repressor function (Figure 6A).
Figure 6.
Functional characterization of clinically isolated or in vitro selected MepR mutants. (A) Histograms of MUG units obtained from β-galactosidase assays for various MepR mutations. Measurements were made in the presence (+) and absence (–) of tetracycline, the addition of which causes MepR production. (B) Structural mapping of MepR mutations. The side chains of the mutated residues are shown as red sticks and labelled. One subunit is colored pink and the other blue. The secondary structure elements of one subunit are labelled.
In order to deduce a possible structural explanation for the observed phenotypes of these mutants, each was mapped onto the MepR structure (Figure 6B). With the exception of residue G97, the side chains of residues Q18 and A103 are surface exposed and located on the external face of helices α1 and α5, respectively (Figure 6B). By contrast, the ‘side chain’ of G97 points towards the hydrophobic core of the wHTH motif and is positioned in the vicinity of multiple hydrophobic residues (L37, Y41, L76, I77, L93, I98, L100 and V101) from α2, α4, β1, β2 and α5. This glycine is highly conserved amongst the members of MarR family (Figure 2C). Model building reveals that introduction of the bulky indole side chain of tryptophan at position 97 would result in steric clash with the side chains of residue L76, I77 and L93 as well as the main chain of I77. Substitution of G97 by a glutamate would place the carboxylate side chain in a hydrophobic environment, which is thermodynamically unfavorable and would likely disrupt the hydrophobic core that is required to maintain the structural integrity of wHTH motif. Interestingly, the analogous G104D mutation in E. coli MarR displayed a transdominant negative phenotype with respect to its repressor function and had reduced solubility, properties consistent with the compromise of the hydrophobic core (43). When the G97E and G97W mutant proteins were overexpressed, more than 95% of each protein ended in the ‘insoluble’ fraction indicating these substitutions disrupt the native MepR structure and its thermodynamic stability (Supplementary Data Figure 1).
The functional defect that arises from the Q18P substitution is likely tied to its structural effects. Proline strongly disfavors the α helical conformation and when centrally located in a helix, almost always kinks that structure. Glutamine 18 is indeed located in the middle of helix α1 and its substitution by proline and any subsequent helix distortion or breaking could affect dimerization by altering the position of the N-terminus of α1 or DNA binding by altering the location of the C-terminal half of α1 or both (Figure 6B). In addition, this change might impair DNA binding as the analogous position in Bs OhrR, residue K27, is part of the HH motif and engages in DNA phosphate backbone contacts (36). Thus, the Q18P substitution could disrupt DNA binding in multiple ways and result in defective repression by MepR. Interestingly, the proline substitution at Q18 is well tolerated, as the overexpression and solubility of this protein are comparable to that of wild-type MepR (Supplementary Data Figure 1).
Perhaps the most intriguing defective repressor is the A103V-substituted MepR as the alanine to valine mutation is a relatively conservative substitution and the side chain is solvent exposed, thus ruling out any obvious steric clash effects (Figure 6B). Regardless, the effect of substituting alanine to valine is possibly conformational simply because alanine prefers the α helical conformation, whilst valine prefers the β-strand structure. The A→V change could weaken the α-helical propensity of α5 as a whole or more likely cause a local conformational deformation that locks in or favors the induced state. Any role of the A→V substitution on solubility and stability of the protein was ruled out, as most of the overexpressed A103V protein remains in the soluble fraction of the cell lysate (Supplementary Data Figure 1). Although a seemingly harmless substitution, the structures of oxidized OhrR and DNA-bound OhrR underscore the critical importance of the conformational plasticity of helix α5, as this helix must be reformed upon the reduction of organic hydroperoxide oxidised OhrR in order to transmit this information to the wHTH motifs to effect cognate DNA binding and repression (36,37). Thus, a seemingly ‘innocuous’ change could have a profound effect on structure and function.
MepR–‘drug’ interactions
MepR is induced by binding to a variety of lipophilic monovalent and bivalent cationic biocides, drugs, dyes, antiseptics and disinfectants (11). The binding affinities of MepR for several inducers namely, Et, DAPI, and R6G were determined using the intrinsic fluorescence polarization of each compound. MepR binds to Et with the highest affinity (Kd = 2.6 μM) but also interacts with DAPI, a bivalent cationic compound, with a nearly equal equilibrium dissociation constant of 4.5 μM (Figure 4G and H). By contrast, MepR binds R6G with relatively poor affinity (Kd value of 62.6 μM) (Figure 4I). Thus, it would appear that the larger R6G (accessible surface area = 754.5 Å2) does not fit the multidrug-binding pocket of MepR as well as the smaller aromatic monovalent cation, Et (ASA = 508.7 Å2) or even the bivalent diamidine DAPI (ASA = 486.6 Å2).
Our current knowledge of inducer binding by MarR family members is mostly derived from the structures of MarR from E. coli and Methanobacterium bound to salicylate, DNA-bound and organic hydroperoxide (OHP) oxidised OhrR, the latter of which is the induced state, and MexR bound to the effector peptide ArmR (15,16,37,40,44). The crystal structure of oxidised OhrR displays drastic conformational changes upon OHP-oxidation that involve the reactive OHP sensor cysteine and aromatic and aliphatic residues from α1, α2, α5 and α6 (37). In OhrR, the only MarR family member for which the apo, DNA-bound and induced structures are available presently, the cascade of structural changes that occurs upon OHP induction originate in the dimerization interface and the structural components, which connect the dimerization and DNA-binding domains. These latter movements in turn lead to the movement of wHTH motifs as rigid bodies to positions incompatible with high-affinity DNA binding. Intriguingly, the salicylate bound structures of MarR from E. coli and Methanobacterium reveal distinct drug-binding modes and stoichiometries (15,16). Amongst the four different salicylate-binding sites, ‘site 1’ of the Methanobacterium MarR was proposed to be the physiologically relevant drug binding site, mostly due to the major structural rearrangements that occur as a result of salicylate binding to this particular site. The salicylate in site 1 interacts primarily by hydrophobic and charged residues from helix α1 of one subunit and α2, α3 and α5 of the second subunit.
The recently determined structure of the MexR dimer bound to one molecule of the peptide inducer, ArmR, also reveals a similarly located but slightly larger inducer binding pocket situated between the dimerization and DNA-binding domains of MexR (44). The ArmR-binding site in MexR is composed of four hydrophobic pockets with symmetrical contributions from both subunits. MexR interacts with the C-terminal tail of ArmR, which is rich in aromatic residues largely by using hydrophobic and polar residues from helix α1, α2 and α5 of both subunits (44). Inspection of the corresponding region of MepR reveals that this area too is rich in aromatic and hydrophobic residues (L24, F27, I29, L37, L40, L100, F104 and F109) that are surrounded by a small number of acidic and polar residues (D21, E32 and H36). These acidic residues could serve as charge complements to the binding of lipophilic, cationic drugs. Thus, it is possible that the multidrug-binding pocket of MepR is located similarly to the OHP, salicylate and ArmR-binding sites of OhrR, MarRM and MexR and that this is the canonical ligand binding site of the MarR family. Germane structural studies are underway to test this conjecture.
FUNDING
This work was supported in part by funds from the National Institutes of Health grant (AI048593 to R.G.B.), the Robert A. Welch Foundation grant (G-0040 to R.G.B.) and Veteran Administration Research Funds (to G.W.K.). Funding for open access charge: R.A. Welch Foundation grant G-0040 (to R.G.B).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the beamline scientists at ALS BL 8.3.1 for their great help with MAD data collection. We also acknowledge the Advanced Light Source supported by the Director, Office of science, Office of Basic Energy Sciences, Material Sciences Division, of the US Department of Energy under contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. We thank Dr H. P. Bächinger at Shriners Hospital for Sick Children at Portland, Oregon for determining the absolute values of our protein concentrations by amino acid analysis.
REFERENCES
- 1.Bal AM, Gould IM. Antibiotic resistance in Staphylococcus aureus and its relevance in therapy. Expert Opin. Pharmacother. 2005;6:2257–2269. doi: 10.1517/14656566.6.13.2257. [DOI] [PubMed] [Google Scholar]
- 2.Kohler T, Pechere JC, Plesiat P. Bacterial antibiotic efflux systems of medical importance. Cell. Mol. Life Sci. (CMLS) 1999;V56:771–778. doi: 10.1007/s000180050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 4.Spratt BG. Resistance to antibiotics mediated by target alterations. Science. 1994;64:388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 5.Rice LB. Antimicrobial resistance in gram-positive bacteria. Am. J. Infect. Control. 2006;34:S11–S19. doi: 10.1016/j.ajic.2006.05.220. [DOI] [PubMed] [Google Scholar]
- 6.Lynch AS. Efflux systems in bacterial pathogens: an opportunity for therapeutic intervention? An industry view. Biochem. Pharmacol. 2006;71:949–956. doi: 10.1016/j.bcp.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Kaur P. Multidrug resistance: can different keys open the same lock? Drug Resist. Updat. 2002;5:61–64. doi: 10.1016/s1368-7646(02)00020-1. [DOI] [PubMed] [Google Scholar]
- 8.Huet A, Raygada J, Mendiratta K, Seo S, Kaatz G. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology. 2008;154:3144–3153. doi: 10.1099/mic.0.2008/021188-0. [DOI] [PubMed] [Google Scholar]
- 9.Kaatz GW, McAleese F, Seo SM. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005;49:1857–1864. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAleese F, Petersen P, Ruzin A, Dunman PM, Murphy E, Projan SJ, Bradford PA. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to Tigecycline. Antimicrob. Agents Chemother. 2005;49:1865–1871. doi: 10.1128/AAC.49.5.1865-1871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaatz GW, DeMarco CE, Seo SM. MepR, a repressor of the Staphylococcus aureus MATE family multidrug efflux pump MepA, is a substrate-responsive regulatory protein. Antimicrob. Agents Chemother. 2006;50:1276–1281. doi: 10.1128/AAC.50.4.1276-1281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TAA, Kaatz GW. Efflux-related resistance to Norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51:3235–3239. doi: 10.1128/AAC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- 15.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Mol. Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 16.Saridakis V, Shahinas D, Xu X, Christendat D. Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from methanobacterium thermoautotrophicum. J. Mol. Biol. 2008;377:655–667. doi: 10.1016/j.jmb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Galan B, Kolb A, Sanz JM, Garcia JL, Prieto MA. Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res. 2003;31:6598–6609. doi: 10.1093/nar/gkg851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooun A, Tomashek JJ, Lewis K. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J. Bacteriol. 1999;181:5131–5133. doi: 10.1128/jb.181.16.5131-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson SP, Grove A. Negative cooperativity of uric acid binding to the transcriptional regulator HucR from Deinococcus radiodurans. J. Mol. Biol. 2005;350:617–630. doi: 10.1016/j.jmb.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Xiong A, Gottman A, Park C, Baetens M, Pandza S, Matin A. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance Operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 2000;44:2905–2907. doi: 10.1128/aac.44.10.2905-2907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural mechanisms of QacR induction and multidrug recognition. Science. 2001;294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- 22.Heldwein EEZ, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 23.Alguel Y, Meng C, Tern W, Krell T, Ramos JL, Gallegos M.-T, Zhang X. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J. Mol. Biol. 2007;369:829–840. doi: 10.1016/j.jmb.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 2002;45:885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Doublié S. Methods in Enzymol. Vol. 276. Academic Press; 1997. Preparation of seleomethionyl proteins for phase determination. pp. 523–530. [PubMed] [Google Scholar]
- 27.Collaborative Computational Project. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Leslie A. The integration of macromolecular diffraction data. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger T. Reciprocal-space solvent flattening. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC, Berendzen J. Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and MAD methods. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1999;55:501–505. doi: 10.1107/S0907444998012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A: Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J.-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL. The PyMol Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 35.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. Evaluation of a Tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 2001;69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Gajiwala KS, Burley SK. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 39.Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- 40.Lim D, Poole K, Strynadka NCJ. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux Operon of Pseudomonas aeruginosa. J. Biol. Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 41.Bordelon T, Wilkinson SP, Grove A, Newcomer ME. The crystal structure of the transcriptional regulator HucR from Deinococcus radiodurans reveals a repressor preconfigured for DNA binding. J. Mol. Biol. 2006;360:168–177. doi: 10.1016/j.jmb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Miyazono K, Tsujimura M, Kawarabayasi Y, Tanokura M. Crystal structure of an archaeal homologue of multidrug resistance repressor protein, EmrR, from hyperthermophilic archaea Sulfolobus tokodaii strain 7. Proteins: Struct. Funct. Bioinform. 2007;67:1138–1146. doi: 10.1002/prot.21327. [DOI] [PubMed] [Google Scholar]
- 43.Alekshun MN, Kim YS, Levy SB. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 44.Wilke MS, Heller M, Creagh LA, Haynes CA, McIntosh LP, Poole K, Strynadka NCJ. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc. Natl Acad. Sci. 2008;105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TA, Kaatz GW. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51:3235–3239. doi: 10.1128/AAC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.