Abstract
The 5′-untranslated region of the hepatitis C virus (HCV) RNA contains a highly structured motif called IRES (Internal Ribosome Entry Site) responsible for the cap-independent initiation of the viral RNA translation. At first, the IRES binds to the 40S subunit without any initiation factors so that the initiation AUG codon falls into the P site. Here using an original site-directed cross-linking strategy, we identified 40S subunit components neighboring subdomain IIId, which is critical for HCV IRES binding to the subunit, and apical loop of domain II, which was suggested to contact the 40S subunit from data on cryo-electron microscopy of ribosomal complexes containing the HCV IRES. HCV IRES derivatives that bear a photoactivatable group at nucleotide A275 or at G263 in subdomain IIId cross-link to ribosomal proteins S3a, S14 and S16, and HCV IRES derivatized at the C83 in the apex of domain II cross-link to proteins S14 and S16.
INTRODUCTION
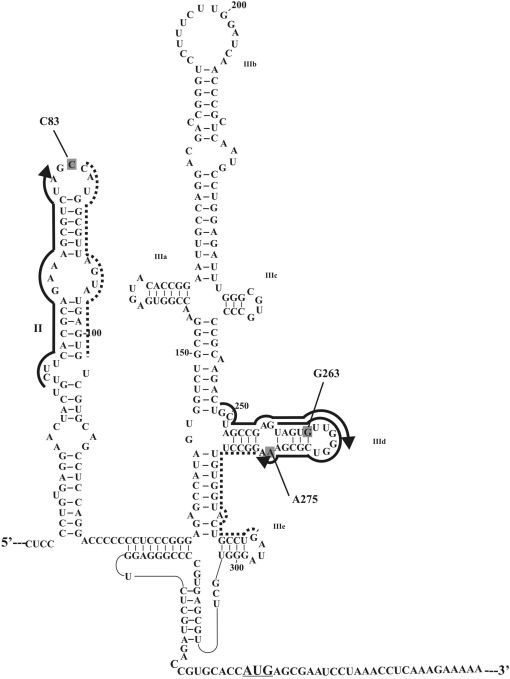
Hepatitis C virus (HCV) is one of the most dangerous human pathogens. It infects about 180 million people worldwide (1) and can lead to cirrhosis, hepatocellular carcinoma and other damages of the liver (2,3). HCV is a flavivirus whose genome consists of one positive 9600 nucleotide-long RNA strand lacking a cap at the 5′-terminus (4). One of the key steps of the HCV life cycle in human host cells is initiation of viral RNA translation. It is mediated by a specific highly structured motif comprising nucleotides 40–372 of the viral RNA; it includes the 5′ untranslated region and the adjacent part of the coding sequence, and is known as IRES (Internal Ribosomal Entry Site) (5–7) (Figure 1). The HCV IRES is responsible for cap-independent internal initiation of translation; it is an alternative to the conventional mechanism generally used by capped eukaryotic cellular mRNAs involving a number of initiation factors that provide association of the mRNA to the cap, leading 40S ribosome binding and scanning to the initiation codon (6). The organization of domains and several RNA motifs in the domains are conserved among different strains of HCV and related viruses of the Flaviridae family (6,8,9). The HCV IRES folds under physiological ionic conditions into a defined 3D structure whose integrity is essential for efficient IRES-mediated translation (10); spatial structures of the HCV IRES domains have already been resolved by X-ray crystallography and NMR (11).
Figure 1.
Structural organization of the HCV IRES (7), the initiation AUG codon is underlined. RNA sequences complementary to deoxy-oligomers used for site-specific modification of the HCV IRES are marked with thick lines with arrows indicating the 5′-phosphates derivatized with alkylating groups. RNA sequences complementary to helper oligomers used together with the alkylating derivatives are marked with dotted lines. Nucleotides of the HCV IRES cross-linked to the oligonucleotide derivatives are shaded.
The sequence of events leading to the formation of 80S initiation complex with IRES competent for the start of viral RNA translation is well known (12–14). The functional roles of individual IRES domains and subdomains have been studied using mutant IRESes (6,12–16), cross-linking with chemically reactive derivatives of oligonucleotides complementary to target RNA sequences (17), and enzymatic and chemical footprinting (15,16,18). According to the generally accepted model, in the first step of the assembly of the 80S initiation complex, the HCV IRES forms a stable binary complex with the 40S subunit where the initiation codon is positioned on the P site without any initiation factors. Binary complex formation crucially depends on the basal portion of domain III (mainly hairpins IIId and IIIe) and on the four-way junction IIIabc (Figure 1). Thereafter, eIF3 and the ternary complex (eIF2•Met-tRNAiMet •GTP) assemble to form a 48S preinitiation complex; binding of eIF3 is mediated by domain IIIb and four-way junction IIIabc. Binding of eIF5 to this complex promotes hydrolysis of eIF2-bound GTP and subsequent release of eIF2•GDP, which is modulated by HCV IRES domain II (14). Finally, the release of the remaining factor eIF3 requires eIF5B and GTP hydrolysis and occurs during joining of the 60S subunit to form the 80S initiation complex.
Knowledge concerning the ribosomal components responsible for binding of HCV IRES is important to understand the mechanism of translation initiation of the viral RNA and could be essential for the development of novel efficient therapies for HCV. However, the structural organization of the IRES-binding site on the 40S subunit is less studied than the functional role of the domains, and the resulting data are rather contradictory. Direct UV-induced cross-linking of HCV IRES to the 40S subunit revealed ribosomal protein S5 as the only target for cross-linking (19). Other investigators using HCV IRES with uridines randomly substituted by 4-thiouridines reported cross-linked proteins S2, S3, S10, S15, S16/S18 and S27 (20). These studies highlighted a set of proteins that could surround the 5′-terminal part of HCV RNA, but did not provide information on the specific ribosomal proteins neighboring particular RNA nucleotides. Based on the cross-linking results and the data on inability of 40S subunits from lower eukaryotes to bind HCV IRES, it was suggested that the IRES-binding site on the 40S subunit is formed by protein sequences specific of higher eukaryotes rather than by the 18S rRNA that is fairly similar between yeast and human (78% identity) [(20) and refs therein].
Cryo-electron microscopic (cryo-EM) visualization of HCV IRES in its complexes with the 40S subunit (21) and the 80S ribosome (22) provided a general idea of the positioning of the IRES on the ribosome; two ribosomal proteins, namely, S5 on the head of the subunit interface and S14 on the platform were mapped close to the apex of domain II of the IRES (22). However, domain II in contrast to subdomains IIId/e and the four-way junction IIIabc is not essential for binding of HCV IRES to 40S subunit (see above). Thus, ribosomal proteins that could contact keystone structural elements of HCV IRES leading to its high affinity for the 40S subunit remained unknown.
Highly sensitive tools to investigate the molecular environments of RNA ligands bound to the ribosome are RNA derivatives bearing a perfluorophenyl azide cross-linker at specific locations. Using a set of short mRNA analogues bearing such groups at the specific locations, the structure of the mRNA-binding channel of the human 80S ribosome has been studied in detail (23–26). The data on ribosomal components neighboring mRNA nucleotides in positions −9 to +12 with respect to the first nucleotide of the P site codon were recently confirmed in a study of 48S initiation complexes using mRNAs bearing 4-thiouridines capable of forming ‘zero-length’ cross-links (27). This additionally demonstrated that perfluorophenyl azide-modified RNAs provide correct and adequate information on the structures of binding sites of various RNA ligands bound to the mammalian ribosome. Recently, we suggested a novel strategy that makes it possible to selective introduce perfluorophenyl azide groups into long structured RNAs (28). This strategy is based on the site-specific alkylation of an RNA with [4-(N-2-chloroethyl-N-methylamino)benzyl]phosphoramide derivatives of oligodeoxyribonucleotides complementary to a sequence adjacent to the target site. After hydrolysis of the phosphoramide bond in the covalent adduct formed, aryl azide moiety is selectively introduced at the liberated benzylamine group. Using HCV IRES derivative bearing the cross-linker at A296 in hairpin IIIe for a site-directed cross-linking study on human ribosomes, we determined the ribosomal components neighboring this keystone subdomain in the binary complex of the IRES with the 40S subunit. This approach makes it possible to monitor the molecular environment of each of structural elements of the IRES on the ribosome at every step of translation initiation of the viral RNA.
In this study, applying the strategy based on site-specific modification, we obtained HCV IRES derivatives bearing a perfluorophenyl azide group at various nucleotides in hairpin IIId or in the apex of domain II (Figure 1), and with the use of these derivatives determined the ribosomal components neighboring these nucleotides in the binary complex of HCV IRES with the human 40S ribosomal subunit. The results obtained show which ribosomal components neighbor the structural elements of HCV IRES critical for its binding to the 40S subunit and which components surround the apex of domain II that is located close to the ribosomal surface based on cryo-EM data.
MATERIALS AND METHODS
Acrylamide, N,N′-methylene-bis-acrylamide and urea were purchased from AppliChem, ribonucleoside triphosphates were from Sigma, Sephadex G-75 from Pharmacia and T7 RNA polymerase from NEB. DNA oligomers complementary to definite sequences of the HCV IRES were synthesized in the Laboratory of Medical Chemistry (Institute of Chemical Biology and Fundamental Medicine SB RAS), [α-32P]GTP (1 mCi/nmol), 4-(N-2-chloroethyl-N-methylamino)benzylamine (ClRCH2NH2) and N-oxysuccinimide ester of 4-azidotetrafluorobenzoic acid were synthesized in the Laboratory of Biotechnology of this Institute.
Preparation of labeled HCV IRES RNA
The fragment corresponding to HCV RNA nucleotides 40–372 was obtained by in vitro transcription of plasmid pXL40-372.NS (29) linearized by BamH1 (‘Fermentas’, Lietuva) as described (17) using [α-32P]GTP. It was isolated and examined as described (28). The specific radioactivity of the RNA transcript was typically about 25 000 cpm/pmol.
Introduction of perfluorophenyl azide groups into the IRES
4-(N-2-chloroethyl-N-methylamino)benzyl-5′-phosphoramides of oligodeoxyribonucleotides were synthesized and purified as described (17). Site-specific modification of the HCV IRES was carried out as described (28) starting with 300 pmol of the RNA transcript. Isolation of the RNA modified with deoxy-oligomers derivatives, determination of the modified RNA nucleotides, hydrolysis of the phosphoramide bond in the deoxyribonucleotide derivatives attached to the RNA, selective introduction of perfluorophenyl azides at the aliphatic amine groups liberated after hydrolysis and purification of the photoactivatable derivatives of HCV IRES were carried out as described (28).
Ribosomes, ribosomal complexes and cross-linking procedures
40S ribosomal subunits with intact rRNA were isolated from unfrozen human placenta as indicated (30). Prior to use, the subunits were re-activated by incubation in binding buffer A (20 mM Tris–HCl, pH 7.5, 100 mM KCl, 2.5 mM MgCl2 and 0.25 mM spermidine) at 37°C for 10 min. Binary complexes of HCV IRES and 40S subunits were obtained by incubating the subunits (2.0 × 10−6 M if not specified otherwise) with IRES (0.5 × 10−6 M if not specified otherwise) in buffer A at 37°C for 10 min. The extent of binding of the 5′-32P-labeled RNAs to 40S subunits was examined by the nitrocellulose filtration assay using alkali pretreated filters as described (31). In the experiments on the determination of cross-linked ribosomal proteins, the binary complexes were formed starting from 10–20 pmol of the derivatized HCV IRES. UV-irradiation of the complexes was carried out according to (25). The distribution of the cross-linked labeled IRES between the rRNA and ribosomal proteins was examined by centrifugation in a 5–20% sucrose density gradient in the presence of SDS and EDTA as described (32).
Isolation and identification of ribosomal proteins cross-linked to HCV IRES derivatives
To isolate ribosomal proteins, the irradiated complexes were treated under conditions for dissociation of the subunits into the rRNA and proteins and exhaustively hydrolyzed with RNases A and T1 according to (28). The proteins were separated from oligoribonucleotides by gel-filtration and analyzed by 1D and 2D PAGE as described (28). To analyze cross-linked proteins by immunoprecipitation, we used rabbit antibodies against proteins S2 and S3, goat antibodies against proteins S3a and S13/S16 and sheep antibodies against protein S14 obtained as described (33). These antibodies did not cross-react with other 40S proteins. Irradiated binary complexes of the 32P-labeled IRES derivatives and 40S subunits were supplied with 2-mercaptoethanol, SDS and EDTA to final concentrations of 0.2%, 0.1% and 10 mM, respectively, together with 100 pmol of unlabeled IRES. The mixture was incubated at 37°C for 10 min to dissociate the subunits into rRNA and proteins and then diluted with TBS (20 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.4% Triton X-100). The antibodies were immobilized on protein-G-Sepharose (Fluka) by stirring 20 μl of the sorbent suspension with 5–20 μl of a solution containing 50 mM Tris–HCl, pH 9.0, 4 M urea and 0.3 mg antibodies for 2 h at 0°C. The supernatant was discarded and the sorbent was washed three times with 100 μl TBS. To prevent unspecific binding of RNAs and cross-linked proteins to protein-G-Sepharose, the washed sorbent with immobilized antibodies was supplied with 20 μl of a solution containing 1 A260 unit of total Escherichia coli tRNA and bovine serum albumin (final concentration of 0.5 mg/ml); the mixture was incubated 1 h at 0°C with stirring. The supernatant was discarded and the sorbent was mixed with a solution containing cross-linked proteins and incubated for 3 h at 0°C. The sorbent with bound cross-linked proteins was pelleted by low-speed centrifugation, washed five times with TBS buffer (100 μl each time) and their radioactivity was measured by Cherenkoff counting. The values obtained with antibodies in a control experiment with unmodified IRES were subtracted from the values obtained with IRES derivatives.
RESULTS
HCV IRES derivatives
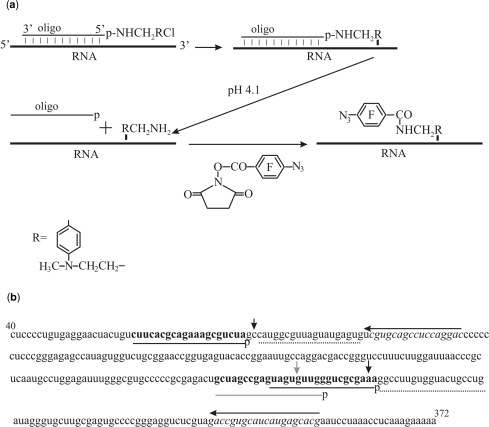
To introduce photoactivatable perfluorophenyl azide groups into hairpin IIId or the apex of domain II of the IRES (Figure 1), an approach based on the site-specific alkylation of RNA with phosphoramide derivatives of deoxyribonucleotides described previously (28) and schematically shown in Figure 2a was applied. To increase the yield of IRES alkylation with the derivatives of deoxy-oligomers complementary to sequences 259–276 and 62–81, helper oligomers were used that facilitate binding of oligomers bearing the alkylating group to the structured RNA [see (34) and Refs. therein] (Figures 1 and 2b). Helpers were not used in the experiments with the derivative of oligomer complementary to sequence 248–267 since the yield of HCV IRES alkylation was sufficiently high. The covalent adducts resulting from IRES alkylation with the oligonucleotide derivatives were separated from unmodified RNA by denaturing PAGE on an 8% gel (the respective electrophoregrams are not shown). Generally, about 50% of the RNA was converted into the covalent adduct in the course of alkylation.
Figure 2.
Scheme of site-specific introduction of a photoactivatable group into specific RNA sites based on the site-specific alkylation of the RNA with [4-(N-2-chloroethyl-N-methylamino)benzyl]-phosphoramides of oligodeoxyribonucleotides. (a) Order of chemical reactions. (b) Nucleotide sequence of HCV IRES. Target sequences for the deoxy-oligomer derivatives are given in bold, the respective complementary oligodeoxyribonucleotides are shown with either gray or black lines under the sequence, and the letters ‘p’ indicate the terminal 5′-phosphates derivatized with alkylating groups. Helper oligomers are shown by dotted lines. Lines with arrows above the sequence indicate primers used for reverse transcription (arrows show the direction of primer extension). RNA sequences complementary to the primers are given in italics. Either gray or black vertical arrows show cross-linked nucleotides.
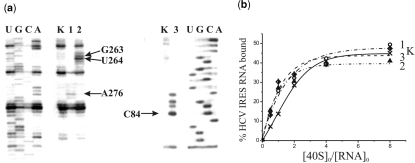
To identify modified IRES nucleotides, reverse transcription was used that makes it possible to detect modified nucleotides by stop or pause of primer extension. The modification site is generally assumed to be the nucleotide 5′ of the primer extension stop site. Therefore, stops at A276 (Figure 3a, left panel, lane 1), U264 (left panel, lane 2) and C84 (right panel, lane 3) correspond to modified A275, G263 and C83, respectively (Figures 1 and 2b). With HCV IRES alkylated with the derivative of oligomer complementary to sequence 248–267, a weaker stop at G263 was also observed (Figure 3a, left panel, lane 2). This signal could not be assigned to the primer extension stop caused by the modification of U262 since uridine is unable to react with aromatic 2-chloroethyl amines at nearly neutral pH (35). Thus, the signal at nucleotide in position 263 was assigned to a pause of reverse transcription that could occur at guanine alkylated on the N7 atom, which is not involved in Watson–Crick base pairing. One could expect that in the heteroduplex of the HCV IRES with the derivative of oligomer complementary to sequence 248–267 the alkylating group is located closer to nucleotides G265–267 rather than to G263 (Figure 1). The unusual target of alkylation in this case might be due to peculiarities of the spatial structure of the IRES and also to the low reactivity of guanines in oligoG fragments towards alkylation with aromatic 2-chloroethyl amines (36). Weaker stops with the derivative of oligomer complementary to sequence 62–81 probably correspond to minor alkylation sites adjacent to C83 (taking into account the discussion above, these are G82 and A81).
Figure 3.
Identification of sites of cross-linking of oligonucleotide derivatives to the HCV IRES by reverse transcription and binding properties of the HCV IRES derivatives. (a) Extension of [5′-32P]-labeled primers complementary to the HCV IRES sequences 331–350 (left panel) and 103–120 (right panel). Lanes 1, 2 and 3, primer extension on HCV IRES alkylated with derivatives of deoxy-oligomers complementary to the sequences 259–276, 248–267 and 62–81, respectively. Lanes K, primer extension with control HCV IRES incubated under conditions of alkylation but without oligomer derivatives. Lanes U, G, C, A, sequencing of HCV IRES. Arrows indicate positions of the reverse transcription stops caused by the cross-links. (b) Isotherms of binding of control unmodified HCV IRES (K) and its derivatives containing a perfluorophenyl azide group at A275, G263 or C83 to 40S subunits (1, 2 and 3, respectively). The initial concentration of the HCV IRES or its derivatives was 1.0 × 10−7 M. Relative error was about 10%.
To introduce a photoactivatable moiety on modified RNA nucleotides, the phosphoramide bond in the covalent adducts obtained from 32P-labeled IRES was hydrolyzed under mild acidic conditions (pH 4.1) that resulted in the release of the aliphatic amino group −RCH2NH2 linked to the RNA. The modified IRES RNAs were purified by denaturing 8% PAGE and the −RCH2NH2 moieties were selectively derivatized by treatment with the N-oxysuccinimide ester of 4-azidotetrafluorobenzoic acid. HCV IRES derivatives bearing photoactivatable groups at nucleotides A275, G263 or C83 were thus obtained (Figures 1 and 2b). These derivatives were used in the cross-linking experiments with human 40S ribosomal subunits in parallel with control HCV IRES (K) treated under identical conditions but without addition of alkylating derivatives of oligodeoxyribonucleotides.
Binding and cross-linking of photoactivatable HCV IRES derivatives to 40S subunits
Filter retention assays revealed that the affinities of HCV IRES derivatives bearing the cross-linker at A275, G263 or C83 to 40S subunits were similar to each other and did not significantly differ from that of unmodified IRES (Figure 3b) as was shown previously with HCV IRES similarly derivatized at the nucleotides A296 and G87 (28). For the cross-linking experiments, saturating concentrations of 40S subunits were used. To obtain the cross-links, the binary complexes of the IRES derivatives and the 40S subunits were irradiated with mild UV light (>280 nm). Subsequent analyses of the distribution of the radioactive label between the rRNA and the ribosomal proteins after centrifugation in sucrose density gradients containing SDS and EDTA did not reveal radioactivity in the fractions containing 18S rRNA (the respective sedimentation profiles are not presented). Radioactivity was found only in the top fractions containing proteins and IRES not cross-linked to ribosomal components. Therefore, none of the HCV IRES derivatives cross-linked to the rRNA.
Identification of ribosomal proteins cross-linked to the photoactivatable HCV IRES derivatives
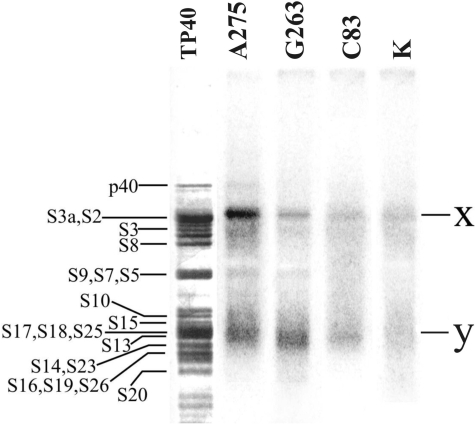
Ribosomal proteins cross-linked to derivatives of the IRES were first resolved by 1D SDS–PAGE. Prior to analysis, the RNA cross-linked to proteins was exhaustively hydrolyzed with RNases A and T1; ribosomal proteins were separated from labeled oligoribonucleotides resulting from hydrolysis by gel-filtration under strongly denaturing conditions (6 M guanidine chloride) to decrease possible effects related to unspecific binding of oligoribonucleotides to the proteins. The results presented in Figure 4 show that all IRES derivatives cross-linked mainly to two protein groups corresponding to the bands ‘x’ in the upper and ‘y’ in the lower parts of the gel, the intensities of the bands depending on the location of the cross-linker in the IRES. With the IRES derivatized at C83, the yield of cross-linking was significantly lower than with the two other derivatives (Figure 4). It should be also noted that in the lane corresponding to the control RNA (lane K), weak diffuse bands appeared at positions similar to those of ‘x’ and ‘y’. These bands clearly originated from a small portion of unspecific complexes of ribosomal proteins with labeled oligoribonucleotides resulting from hydrolysis of the IRES and remaining in the protein fraction after gel-filtration (see above), and not separated from the proteins even under conditions of SDS–PAGE.
Figure 4.
Autoradiogram of the 40S ribosomal proteins cross-linked to the 32P-labeled derivatives of HCV IRES by 1D PAGE in the presence of SDS. Nucleotides bearing the cross-linker are indicated. Lane K, proteins isolated from an irradiated control complex of unmodified HCV IRES with 40S subunits. Lane TP40, silver stained gel; positions of the 40S ribosomal proteins are indicated (43,44).
To identify candidate cross-linked proteins, we took into account our previous data (25,26) allowing evaluation of the effect of oligoribonucleotides cross-linked to ribosomal proteins based on their electrophoretic mobilities. Moreover, exhaustive hydrolysis of HCV IRES with RNases A and T1 should produce fragments ApA*pApGp, G*pUp and C*pCp/C*p (asterisks indicate modified nucleotides) cross-linked to proteins in the case of the HCV IRES derivatized at the A275, G263 and C83, respectively (Figures 1 and 2b). These fragments somewhat decrease the electrophoretic mobility of the cross-linked proteins (the greater the molecular mass of the protein, the lower the effect of the cross-linked oligomer). Considering all these factors, one could suggest that bands ‘x’ might correspond to cross-linked proteins S2/S3a and less probably to protein S6, and bands ‘y’ to cross-linked proteins S14/S16/S19/S23/S26. With the IRES derivatized at A275 and at G263, a minor band in the middle of the electrophoregram was also detected; its position with respect to proteins S9, S7 and S5 was very similar to that of cross-linked S5 in the experiments with the IRES derivatized at A296 (28). With IRES modified at A275 an additional weak band could be seen at position corresponding to a single protein p40, which was shown earlier to cross-link to A296 in subdomain IIIe (28).
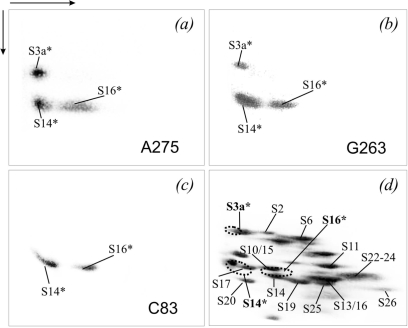
Cross-linked protein(s) were more precisely identified resolving them after exhaustive hydrolysis of HCV IRES with RNAses A and T1 by ‘basic-SDS’ 2D PAGE (separation in the first dimension was at pH 8.3, and in the second dimension at pH 6.75 in the presence of SDS), the results are presented in Figure 5. Cross-linked proteins were identified considering that in the electrophoretic system used, oligoribonucleotide fragments cross-linked to proteins decrease their mobilities in the first dimension but barely affect their migration in the second dimension. Consequently, the radioactive spot of the cross-linked protein should be somewhat shifted to the left of the corresponding stained spot of the unmodified protein. The extents of the shifts were evaluated based on data of our previous studies on cross-linking of short mRNA analogues to ribosomes (25,26). The results showed that HCV IRES derivatized at the A275 and the G263 cross-linked to the same three groups of proteins, namely, to S3/S3a (less probably, S2), S10/S15/S14 and S13/S16 (much less probably, S25), the distribution of cross-links between these groups depended on the location of the cross-linker in the HCV IRES (Figure 5a and b). With C83-modified IRES, only two spots corresponding to cross-linked proteins S10/S15/S14 and S13/S16 were detected (Figure 5c). Given the results of the 1D gels (Figure 4), more exact identification of the cross-linked proteins could be performed. In the group S13/S16/S25, only protein S16 was actually cross-linked since S13 and S25 were not among the candidate cross-linked proteins (see above). For the same reason, proteins S10 and S15 could be excluded from the group S10/S15/S14 and the only cross-linked protein was S14. Similarly, protein S3a could be identified as the only cross-linked protein in the group S3/S3a. Thus, combining the data of 1D and 2D PAGE made it possible to conclude that all HCV IRES derivatives used here cross-linked mainly to proteins S14 and S16, and the derivatives bearing the modifying group at A275 or G263 also cross-linked to protein S3a (Figure 5d).
Figure 5.
Analysis by 2D PAGE of proteins cross-linked to 32P-labeled HCV IRES derivatives in the binary complexes with the 40S subunits. (a–c), The autoradiograms correspond to the experiments with HCV IRES derivatives containing the cross-linker at nucleotide A275, G263 or C83 (marked in the respective panels). (d), Coomassie stained gel corresponding to (b) (as example); the positions of the proteins are indicated (44, 45). The locations of the radioactive spots corresponding to the cross-linked proteins are indicated on the stained gel by dotted lines. The cross-linked proteins are highlighted by an asterisk (*), and are also in bold in (d).
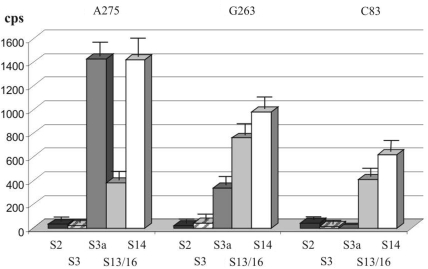
The identity of the cross-linked proteins S3a, S14 and S16 was confirmed by immunoprecipitation using specific antibodies. In parallel, immunoprecipitation with antibodies against proteins S2 and S3 whose modification was practically excluded on the basis of the comparison of the results of 1D and 2D PAGE analysis was carried out as control. The results shown in Figure 6 are in a good accordance with the data of 1D and 2D PAGE separations discussed above (Figure 5) and confirm cross-linking of the HCV IRES derivatives to ribosomal proteins S3a, S14 and S16.
Figure 6.
Analysis of 40S proteins cross-linked to 32P-labeled HCV IRES derivatives (nucleotides bearing the cross-linker are indicated at the top) by immunoprecipitation using antibodies against mammalian 40S ribosomal proteins (indicated at the bottom).
DISCUSSION
Choice of sequences within the HCV IRES for the site-specific introduction of cross-linker and binding properties of the derivatized IRES
The criteria for the choice of derivatives of oligonucleotides for introduction of cross-linker into the HCV IRES were based on our earlier data on alkylation of the IRES with the derivatives of oligodeoxyribonucleotides complementary to its specific sequences and testing abilities of the resulting covalent adducts to bind to the 40S subunits (17). The oligos were 20-mers that was enough to enable them to form relatively stable heteroduplexes specifically with the target sequences, and contained an alkylating group at the 5′-termini. This group generally modifies RNA nucleotides near the 5′-end of the oligomer bound to the RNA, the more accessible is target RNA sequence, the higher is yield of the covalent adduct (34). To increase the yields of the covalent adducts, we used helper oligomers to disrupt the IRES secondary structure in the target region and thus to facilitate formation of the heteroduplexes with alkylating derivatives. We found that affinity of 40S subunits for the HCV IRES covalent adducts bearing deoxy oligomers complementary to sequences in hairpin IIId and IIIe was drastically lower than that for the unmodified IRES (17). In contrast, attachment of a deoxy oligomer complementary to the apical loop of domain II barely affected the affinity of the IRES for the 40S subunits (17). These results confirmed earlier data (12–16,18) that HCV IRES subdomains IIId and IIIe are the main determinants for binding to 40S subunits and that domain II does not contribute to the binding. Based on all these, we chose target RNA sequences for site-specific introduction of the cross-linker (Figure 1). Two of them were in hairpin IIId [hairpin IIIe has been studied in our previous report (28)] and third one was mainly in the stem of domain II to introduce cross-linker to the middle of the apical loop [since IRES derivative with a photoactivatable group on the G87 used in our previous study (28) did not cross-linked to the 40S subunit].
Binding experiments showed that introduction of an aryl azide cross-linker at nucleotides C83, G263 and A275 almost did not affect affinity of the HCV IRES to the 40S subunits (Figure 3b). This implies that HCV IRES modified at these nucleotides retained its functional activity in the binding to the 40S subunit. This conclusion could be done on the IRES modified at nucleotides G263 or A275 in subdomain IIId critical for the binding to the 40S subunit as well as on the IRES modified at the C83 in domain II, which is not essential for the binding since apical hairpins of domain II and subdomain IIId adopt similar folds (37). Besides, apical hairpin of domain II does not interact with other subdomains of the IRES (37) so modification in the loop should not affect the IRES spatial structure.
Ribosomal components neighboring the HCV IRES on the 40S subunit
Two proteins cross-linked to HCV IRES derivatives, S3a and S16, were found previously to also cross-link using the derivative bearing perfluorophenyl azide at A296 in subdomain IIIe (28). Besides, this latter derivative as mentioned above, cross-linked to proteins S5 and p40 but much more strongly than the HCV IRES derivatives used here that bore the modifying group in subdomain IIId suggesting that these proteins are closer to hairpin IIIe than to IIId. On the other hand, cross-linking to protein S14 was not found with HCV IRES bearing the modifying group in hairpin IIIe (28), which implies that this protein likely neighbors subdomain IIId and the apical loop of domain II but not subdomain IIIe. We failed to find any cross-links of HCV IRES derivatized at the G87 (in the upper part of the stem of domain II) with the 40S subunit in the binary complex (28), in contrast to the derivative bearing the same cross-linker at C83 (in the apical loop of domain II) used here (Figure 1). This is a notable result since a distance between the C83 and the G87 is within the length of the cross-linker (14 Å) used in this study (28). Therefore, we have a basis to suggest that a distance between the C83 and the 40S subunit does fall within the range of cross-linker action while the distance from the G87 to the subunit does not. This means that the apical loop of domain II in the binary complex is located closer to the 40S subunit than the stem of this domain.
The results obtained in the present study and in our previous report (28) significantly differ from data of two earlier reports on the determination of the 40S subunit components neighboring the IRES as a whole during initiation of the viral RNA translation discussed in the Introduction (19,20). In the article by Otto et al. (20), proteins S2, S3, S10, S15, S16/S18 and S27 were reported to be cross-linked, of which S3, S2 and S27 were the major targets. It is now clear that a significant part of this set of proteins, namely, proteins S2, S3 and S15, belongs to the environment of the mRNA at the decoding site and downstream of it. This is based on data on cross-linking of these proteins in 80S ribosomal complexes to unstructured mRNA analogues with perfluorophenyl azide-modified nucleotides in positions +4 to +12 (25,26) or with 4-thiouridines in positions +4 to +7 (38) with respect to the first nucleotide of the P site codon. Recently these results were confirmed with 4-thiouridine-containing mRNAs in the 48S/80S initiation complexes, in which the modified nucleotide in position +11 cross-linked to proteins S3 and S2, and in positions +4/+5 cross-linked to protein S15 (27). On the cryo-EM images of the 40S subunit (39), locations of proteins S2 and S3 are similar to those of their prokaryotic counterparts S5p and S3p in the 30S subunit that form the mRNA entry site, and S15 lies on the intersubunit side of the small subunit. All these point to unlikely neighborhood of the mentioned proteins and the 5′-untranslated part of HCV IRES. Thus, we have all reasons to suggest that proteins S2, S3 and S15 found in the report (20) cross-linked not to the 5′-untranslated region of the HCV IRES but to the part of the coding sequence domain IV according to the structure presented in (8) that could contain 4-thiouridines in positions +2, +10 and +13. Consequently, only proteins S10, S16/S18 and S27 can be considered as cross-linked to the 5′-untranslated region of the HCV RNA in the binary complex. This set overlaps the set found here only by protein S16. One possible explanation for this discrepancy is that proteins S10 and S27 could be cross-linked to 4-thiouridines located in IRES positions far from C83, G263, A275 or A296 that bore perfluorophenyl azide cross-linkers. As for the data presented in (19), they cannot be compared directly with our results because in the former experiments, mixtures of HCV IRES and HeLa cytoplasmic extracts but not the purified binary complexes were used for irradiation. This did not make it possible to specify which steps of HCV RNA translation were monitored.
Our results reported here and previously (28) provide a view of the molecular environment of subdomains IIId/e, the main determinants for HCV IRES binding to the 40S subunit, and of the apical loop of domain II, which was suggested to locate close to the head of the subunit near protein S5 and the mRNA exit site on the basis of cryo-EM data (22). According to our results, protein S14 neighbors both sides of the stem of subdomain IIId and the apical loop of domain II, protein S16 is located close to both hairpins IIId/e and to the apical loop of domain II, protein S3a neighbors hairpins IIId/e, and proteins S5 and p40 are mainly close to subdomain IIIe. Remarkably, no cross-links to the 18S rRNA were detected confirming the earlier suggestion that ribosomal proteins but not the 18S rRNA play the main role in binding of the HCV IRES to the 40S subunit (20).
The sets of ribosomal proteins cross-linked to perfluorophenyl azide-derivatized HCV IRES partially overlapped the sets of proteins cross-linked in the 80S ribosomal complexes to mRNA analogues that bore similar cross-linkers at nucleotides in positions −9 to +12 (25,26). Only protein S14 cross-linked both to HCV IRES derivatives and to mRNA analogues bearing cross-linkers in positions −6 to −9 (25). Protein S14 was also cross-linked to mRNAs bearing 4-thiouridine in the mentioned positions; besides, protein S5 was cross-linked to 4-thiouridines in positions −3/−4 (27). Proteins S5 and S14 are homologous to prokaryotic S7p and S11p, respectively that are known as constituents of the 30S ribosomal exit site of the mRNA (40). Thus, the HCV IRES site has minor overlap with the mRNA-binding channel and is located near the mRNA exit site.
Location of HCV IRES on the 40S subunit
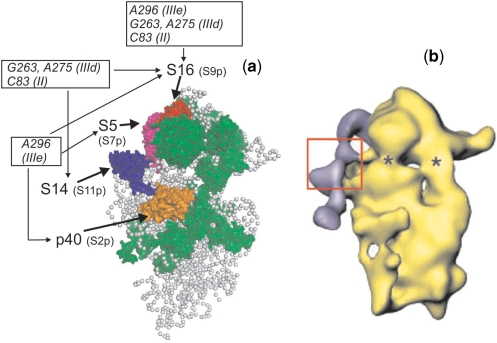
We applied our cross-linking results to the cryo-EM model of the mammalian 40S ribosomal subunit (39) (Figure 7a) and compared the location of the IRES cross-linked to the proteins with its position on the 40S subunit in the binary complex deduced from cryo-EM data (21) (Figure 7b). Cryo-EM data on the 80S ribosomal complex with HCV IRES (22) were not considered since this complex reflects other steps of initiation of viral RNA translation. It should be noted that comparison of our results with cryo-EM-derived data has its limitations since only positions of the conserved parts of ribosomal proteins could be mapped on the models derived from cryo-EM data, while the IRES likely interacts with higher eukaryote-specific sequences of the proteins (20) (mentioned in the Introduction section). Nevertheless, the cross-linking results as a whole are in good agreement with the data obtained by cryo-EM reconstitution of the binary complex of HCV IRES with the 40S subunit (compare Figure 7a and b). Clearly, all the cross-linked proteins are located mainly on the solvent side of the 40S subunit opposite the beak around the cleft between the head and the body, near a site homologous to the mRNA exit site in the 30S subunit and overlapping the E site. The only pronounced dissimilarity between our cross-linking results and the cryo-EM data is that protein S5 appears to neighbor subdomain IIIe (28) rather than domain II (21). This in fact is not a real discrepancy because the absence of cross-link cannot indicate lack of interaction [for a detailed discussion, see (28)], and our results do not exclude possible interaction of protein S5 with domain II of HCV IRES.
Figure 7.
Structural model of the mammalian 40S ribosomal subunit (a) and of the complex of the 40S subunit with HCV IRES (b) obtained from cryo-EM data. View from the solvent side. (a) Model of the 40S subunit of the elongating ribosome [adapted from (39), PDB accession number 2ZKQ]. Locations of ribosomal proteins cross-linked to HCV IRES nucleotides belonging to domain II and subdomains IIId (this study) and IIIe (17) are shown (p40 in orange, S5 in purple, S14 in dark blue and S16 in red), prokaryotic homologues of these proteins are indicated in brackets. All other proteins that have prokaryotic counterparts are shown in green and proteins specific for eukaryotes are not shown; gray balls represent the 18S rRNA. (b) Surface representation of the 40S ribosomal subunit in complex with the HCV IRES [adapted from (21)]. The cryo-EM map is in yellow. The difference map corresponding to the HCV IRES is superimposed and presented in purple. The suggested location of hairpins IIId/e is boxed. Asterics show sites of conformational changes in the 40S subunit caused by HCV IRES binding.
Protein S3a neighboring the HCV IRES according to our data has no prokaryotic counterparts, and therefore its position on the eukaryotic 40S subunit remains unknown. It is conceivable that this protein is located in the same region of the subunit as p40, S5, S14 and S16, namely on the solvent side of the head far from the beak. On the cryo-EM map of the mammalian 40S ribosomal subunit, density corresponding to eukaryote-specific proteins appears close to proteins S14, S5 and p40 (39). A protein density located at the platform near proteins S14 and p40 most likely conforms position of S3a since earlier immunoelectron microscopy studies showed that antibodies against S3a bound at the mentioned region (41).
Thus, our cross-linking results provide significant novel information on the organization of the HCV IRES-binding site on the 40S ribosomal subunit in the binary complex, which forms during the first step of translation initiation of the viral RNA, highlighting the key role played by the ribosomal proteins p40, S3a, S5, S14 and S16 in the organization of this site. The binding sites of various IRESes on the 40S subunit could be significantly different. For instance, in contrast to the HCV IRES, Cricket paralysis virus (CrPV) IRES is located entirely in the mRNA-binding cleft, reaching into the P and A sites (42). Nevertheless, both IRES-binding sites share some extent of similarity: both CrPV IRES and HCV IRES neighbor protein S5 and moreover, the CrPV IRES is located close to S16 as seen upon comparing the models presented in (21,39,42). It is reasonable to suggest also that proteins play a key role in the formation of the 40S ribosomal-binding site not only for HCV IRES but also for other IRESes. However, we cannot extend this suggestion to the later steps of initiation of translation of viral RNAs since in the 80S ribosomal complex with HCV IRES, close proximity of the IRES with 18S rRNA helices was detected (22).
FUNDING
This work was supported by the Russian Foundation for Basic Research (grant #08-04-00508 to G.K.) and the Presidium of Russian Academy of Sciences (Program on Molecular and Cell Biology). The Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully thank Anne-Lise Haenni for critical reading of this manuscript.
REFERENCES
- 1.Brocard M, Paulous S, Komarova AV, Deveaux V, Kean KM. Evidence that PTB does not stimulate HCV IRES-driven translation. Virus Genes. 2007;35:5–15. doi: 10.1007/s11262-006-0038-z. [DOI] [PubMed] [Google Scholar]
- 2.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 3.Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg S. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 2001;313:451–464. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]
- 5.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen C.UT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellen C.UT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 8.Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 50 border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pestova TV, Hellen CU. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- 10.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 11.Pisarev AV, Shirokikh NE, Hellen C.UT. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C.R. Biologies. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl Acad. Sci. USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malygin AA, Graifer DM, Laletina ES, Shatsky IN, Karpova GG. An approach to identify the functionally important RNA sites by complementary addressed modification. Molecular Biology. 2003;37:873–879. (translated from Molekulyarnaya Biologiya 37, 1027–1034) [Google Scholar]
- 18.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40S subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 20.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spahn C.MT, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 22.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Demeshkina N, Repkova M, Ven’yaminova A, Graifer D, Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P and E sites, respectively: a cross-linking study with mRNA analogues carrying aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demeshkina N, Laletina E, Meschaninova M, Ven’yaminova A, Graifer D, Karpova G. Positioning of mRNA codons with respect to 18S rRNA at the P and E sites of human ribosome. Biochim. Biophys. Acta. 2003;1627:39–46. doi: 10.1016/s0167-4781(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 25.Graifer D, Molotkov M, Styazhkina V, Demeshkina N, Bulygin K, Eremina A, Ivanov A, Laletina E, Ven'yaminova A, Karpova G. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004;32:3282–3293. doi: 10.1093/nar/gkh657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molotkov MV, Graifer DM, Popugaeva EA, Bulygin KN, Meschaninova MI, Ven’yaminova AG, Karpova GG. mRNA 3′ of the A site bound codon is located close to protein S3 on the human 80S ribosome. RNA Biology. 2006;3:122–129. doi: 10.4161/rna.3.3.3584. [DOI] [PubMed] [Google Scholar]
- 27.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen C.UT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laletina E, Graifer D, Malygin A, Ivanov A, Shatsky I, Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinolds JE, Kaminsky A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. Embo J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matasova NB, Myltseva SV, Zenkova MA, Graifer DM, Vladimirov SN, Karpova GG. Isolation of ribosomal subunits containing intact rRNA from human placenta. Estimation of functional activity of 80S ribosomes. Analyt. Biochem. 1991;198:219–223. doi: 10.1016/0003-2697(91)90416-q. [DOI] [PubMed] [Google Scholar]
- 31.Graifer DM, Malygin AA, Matasova NB, Mundus DA, Zenkova MA, Karpova GG. Studying functional significance of the sequence 980-1061 in the central domain of human 18S rRNA using complementary DNA probes. Biochim. Biophys. Acta. 1997;1350:335–344. doi: 10.1016/s0167-4781(96)00176-5. [DOI] [PubMed] [Google Scholar]
- 32.Malygin AA, Graifer DM, Bulygin KN, Zenkova MA, Yamkovoy VI, Stahl J, Karpova GG. Arrangement of mRNA at the decoding site of human ribosomes. 18S rRNA nucleotides and ribosomal proteins cross-linked to oligouridylate derivatives with alkylating groups at either the 3′ or the 5′-termini. Eur. J. Biochem. 1994;226:715–723. doi: 10.1111/j.1432-1033.1994.tb20100.x. [DOI] [PubMed] [Google Scholar]
- 33.Noll F, Theise H, Bielka H. Studies on proteins of animal ribosomes. XVIII. Reaction of ribosomes and ribosomal proteins with antibodies against ribosomal proteins. Acta Biol. Med. Germ. 1974;33:547–553. [PubMed] [Google Scholar]
- 34.Bulygin K, Malygin A, Karpova G, Westermann P. Site-specific modification of 4.5S RNA apical domain by complementary oligodeoxynucleotides carrying an alkylating group. Eur. J. Biochem. 1998;251:175–180. doi: 10.1046/j.1432-1327.1998.2510175.x. [DOI] [PubMed] [Google Scholar]
- 35.Ross W.CJ. Biological Alkylating Agents. London: Butterworths and Co., Ltd.; 1962. , pp. 51–63. [Google Scholar]
- 36.Vlassov VV, Skobeltsyna LM. Studying macrostructure of tRNAPhe (E.coli) by chemical modifications. Bioorgan. Khim. 1978;4:550–561. [Google Scholar]
- 37.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 38.Bulygin K, Chavatte L, Frolova L, Karpova G, Favre A. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30 and S15. Biochemistry. 2005;44:2153–2162. doi: 10.1021/bi0487802. [DOI] [PubMed] [Google Scholar]
- 39.Chandramouli P, Topf M, Menetret J.-F, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the Mammalian 80S Ribosome at 8.7 Å Resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yusupova G.Zh., Yusupov MM, Cate J.HD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 41.Lutsch G, Stahl J, Kargel H.-J, Noll F, Bielka H. Immunoelectron microscopic studies on the location of ribosomal proteins on the surface of the 40S ribosomal subunit from rat liver. Eur. J. Cell. Biol. 1990;51:140–150. [PubMed] [Google Scholar]
- 42.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Collatz E, Ulbrich N, Tsurugi K, Lightfoot HN, MacKinlay W, Lin A, Wool I. Isolation of eukaryotic ribosomal proteins. J. Biol. Chem. 1977;252:9071–9080. [PubMed] [Google Scholar]
- 44.Malygin AA, Shaulo DD, Karpova GG. Proteins S7, S10, S16 and S19 of the human 40S ribosomal subunit are most resistant to dissociation by salt. Biochim. Biophys. Acta. 2000;1494:213–216. doi: 10.1016/s0167-4781(00)00252-9. [DOI] [PubMed] [Google Scholar]
- 45.Madjar J.-J, Arpin M, Buisson M, Reboud J.-P. Spot position of rat liver ribosomal proteins by four different two-dimensional electrophoresis in polyacrylamide gel. Mol. Gen. Genet. 1979;171:121–134. doi: 10.1007/BF00269998. [DOI] [PubMed] [Google Scholar]