Abstract
The availability of completely sequenced genomes from eight species of nematodes has provided an opportunity to identify novel cis-regulatory elements in the promoter regions of Caenorhabditis elegans transcripts using comparative genomics. We determined orthologues for C. elegans transcripts in C. briggsae, C. remanei, C. brenneri, C. japonica, Pristionchus pacificus, Brugia malayi and Trichinella spiralis using the WABA alignment algorithm. We pooled the upstream region of each transcript in C. elegans with the upstream regions of its orthologues and identified conserved DNA sequence elements by de novo motif discovery. In total, we discovered 158 017 novel conserved motifs upstream of 3847 C. elegans transcripts for which three or more orthologues were available, and identified 82% of 44 experimentally proven regulatory elements from ORegAnno. We annotated 26% of the motifs as similar to known binding sequences of transcription factors from ORegAnno, TRANSFAC and JASPAR. This is the first catalogue of annotated conserved upstream elements for nematodes and can be used to find putative regulatory elements, improve gene models, discover novel RNA genes, and understand the evolution of transcription factors and their binding sites in phylum Nematoda. The annotated motifs provide novel binding site candidates for both characterized transcription factors and orthologues of characterized mammalian transcription factors.
INTRODUCTION
The binding of transcription factors (TFs) to DNA sequences upstream of a gene is an important element in transcriptional control (1). The genome of the nematode Caenorhabditis elegans is well characterized and almost all of its genes have been identified (2), including 664 genes predicted to encode TFs (3). However, binding sites have been identified for less than 50 of these TFs, and transcriptional regulation is understood for only a few genes. Because regulatory elements are shared among the upstream regions of orthologous (4,5) and coexpressed (6,7) genes, computational methods involving DNA sequence motif discovery among upstream regions of putative co-regulated (orthologous or coexpressed) genes have been used to direct laboratory experiments such as reporter gene and gel shift assays (5,8). Recently, the pace of genome sequence generation has increased and the assembled sequences of eight nematode species have become publicly available. Here, we take advantage of this information and attempt to predict regulatory elements in upstream regions of C. elegans genes by comparing these regions to orthologous regions in other nematode genomes. We hypothesized that most regulatory elements are conserved between many of the eight species, and conversely, that many conserved promoter elements have regulatory function.
To find novel regulatory elements in the C. elegans genome using a comparative genomics approach, we used eight sequenced nematode genomes that were available from either the WormBase (2) or Washington University Genome Sequence Center public FTP servers (Supplementary Table S1). These included the genome sequences or assemblies of C. elegans (9), C. briggsae (10), C. remanei (unpublished), C. brenneri (11), C. japonica (unpublished), Pristionchus pacificus (12), Brugia malayi (13) and Trichinella spiralis (14).
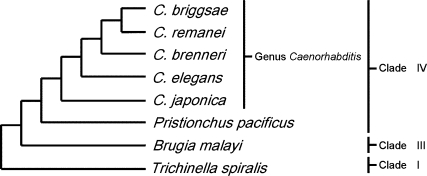
The first five of these species are in the same genus as C. elegans (15) (Figure 1). C. elegans diverged from the other species in genus Caenorhabditis between 18 and 100 million years ago (10,16). P. pacificus is similar to Caenorhabditis species in that it is also a free-living soil bacteriovore, and is classified in the same clade; C. elegans and P. pacificus diverged between 280 and 430 million years ago (12). B. malayi and T. spiralis are mammalian parasites from different clades (17), and are therefore much more remotely related. C. elegans and B. malayi diverged between 350 and 540 million years ago (12), while C. elegans and T. spiralis diverged more than 600 million years ago (14).
Figure 1.
Phylogenetic tree of species. C. briggsae, C. remanei and C. brenneri are all more closely related to each other than they are to C. elegans, while C. japonica is an outgroup within genus Caenorhabditis. Pristionchus pacificus, like C. elegans, is a hermaphroditic bacteriovore and belongs to the same clade of nematodes as C. elegans, but Brugia malayi and Trichinella spiralis are mammalian parasites from other clades in phylum Nematoda. Evolutionary distances are not to scale.
Of the eight nematode genomes, only C. elegans has been extensively characterized in terms of gene location, expression, and function. Given this, we first identified orthologues for C. elegans protein-coding genes in the other seven genomes using WABA (Supplementary Figure S2) (18). Although genes have been predicted for some of the species, and orthologues from C. elegans to C. briggsae and C. remanei have been inferred, we chose to use a single consistent orthologue prediction method for all species. We included alternative transcripts for C. elegans genes because such transcripts frequently have different translation start sites (ATG) and transcripts with the same ATG can have different predicted orthologues if the coding exons vary widely.
We then assembled sets of orthologous upstream sequence regions (Supplementary Figure S2). To do this, we pooled the upstream region of each C. elegans transcript with that of its predicted orthologues, extending each upstream region to the next protein-coding sequence, to a maximum of 1500 base pairs (bp). We used the Gibbs sampler MotifSampler (19) to find conserved DNA sequence motifs in each set of upstream region sequences. All motifs were loaded into the C. elegans cisRED database (20) and are publicly available via the database web interface at www.cisred.org. We used 44 experimentally proven transcription factor binding sites (TFBSs) from ORegAnno (21), found in 28 of the upstream regions, to validate the motif discovery process. Lastly, we compared motif sequences to TF-binding sequences from TRANSFAC (22), JASPAR (23) and ORegAnno, and annotated a motif as similar to a binding sequence if the comparison was statistically significant.
METHODS
Orthologue identification
Genome sequences were obtained from the WormBase and Washington University FTP servers (Supplementary Table S1). WS170 was used because the cisRED web interface makes extensive use of the UCSC Genome Browser and that was the version of the C. elegans genome at UCSC as of May 2008. WABA (18) was used to find one or more orthologous sequences in each of the other genomes for each of the 23 212 chromosomal protein-coding transcripts in WormPep. Only single alignments from WABA that aligned beginning at the ATG of the C. elegans sequence (i.e. ‘high-quality orthologues’) were retained.
Orthologous upstream sequence regions
The upstream region of each C. elegans WormPep transcript was combined with the upstream regions of its orthologues in the other nematode genomes to form an orthologous upstream sequence region set. Only transcripts that had at least three out of a possible seven high-quality orthologues were used. Of the 192 curated C. elegans TFBSs in ORegAnno, 83% were within 1500 bp of the ATG. The remaining TFBSs were sparsely distributed up to 9-kbp upstream and up to 9-kbp downstream of the ATG; the region further upstream than 1500 bp was not enriched for TFBSs. Half of C. elegans transcripts had another gene within 1500 bp of the ATG. The upstream sequence used was defined as 1500 bp upstream of the ATG (including the 5′ UTR, if present) or up to the end of the nearest protein-coding transcript, WABA match or end of contig. The 1500 bp excluded masked repeats and undefined sequence (Ns), and was limited to a maximum total length of 3000 bp. A minimum of 100 bp was required for C. elegans to avoid transcripts whose upstream region was too short to analyse efficiently. We excluded 59 C. elegans transcripts for this reason; of these the closest upstream transcript was on the same strand for 28 and on the opposite strand for 31.
Motif discovery
We applied the motif discovery algorithms MEME (24), CONSENSUS (25) and MotifSampler (19) to the upstream sets and compared their relative performance in detecting a set of experimentally discovered TFBSs obtained from ORegAnno. Of the three methods, only MotifSampler could detect the positive controls with greater than 25% sensitivity and combining the results of two or more methods did not improve the sensitivity. Consequently, we used only MotifSampler to detect motifs in the orthologous upstream sets. For each orthologous upstream sequence region set, a background sequence set was generated that contained randomly selected upstream sequences from each species in the same proportions as the foreground sequences. A third-order Markov background model was then generated from each background sequence set.
MotifSampler was run using the following parameters: -p 0.3 -s 1 -n 25 -r 30. The ‘r’ parameter specifies 30 iterations on each sequence set; we used the score assigned to each motif by MotifSampler to retain the top 30% of motifs from each sequence set. Motif discovery was performed using target widths of 6, 8, 10, 12 and 14 bp because 86% of C. elegans TFBSs in ORegAnno are in this width range. Motifs that overlapped consistently on all sequences on which they were found were merged into one motif. Motifs for which MotifSampler returned multiple instances on the C. elegans sequence were separated and matched with the most conserved instance of that motif on each orthologous sequence. Motifs that occurred on the orthologous sequences but not on the C. elegans sequence were discarded. Each motif in the cisRED database is an aligned collection of sequences containing one sequence from the C. elegans upstream region and not more than one sequence from each orthologous upstream region.
Validation
Experimentally proven TFBSs from ORegAnno (21) were used as positive controls for motif discovery. ORegAnno contains 192 TFBSs for C. elegans, of which 44 were found in 28 of the upstream regions of this analysis. An experimentally proven TFBS from ORegAnno was considered to be discovered when the predicted motif overlapped at least 50% of the site. The average information content (IC) of each motif was calculated as described by Hertz and Stormo (25).
Annotation to show similarity to known TFBSs
Binding sequences for characterized TFs were obtained from TRANSFAC (version 9.2) (22), JASPAR (version 4) (23) and ORegAnno. Each TF in these databases was associated with a set of between 1 and 179 sequences that had been experimentally shown to bind that TF.
The C. elegans sequence of each motif was compared with each database TF and scored as follows. The score between the C. elegans sequence and a single binding sequence was the number of mismatches between the two sequences divided by the width of the binding sequence. We required a minimum overlap of 5 bp between the motif and the binding sequence; flanking genomic sequence was included as needed. We retained the minimum score with respect to relative strand orientation and position of the two sequences, and the minimum such score over all of the TF's binding sequences.
We assigned a P-value to the retained score for each motif-TF pair based on the background score distribution of that TF, which we generated by scoring 1000 randomly chosen C. elegans upstream sequences that were not covered by motifs. Motifs were annotated as similar to a binding site if the P-value of the motif-TF score was below a threshold as follows: ORegAnno binding sites: P-value threshold = 0.00015; TRANSFAC-binding sites: P-value threshold = 0.00001 and JASPAR-binding sites: P-value threshold = 0.0001.
RESULTS
Orthologue identification
For each of the 23 212 C. elegans chromosomal protein-coding transcripts, we used the WABA algorithm (18) to identify putative orthologues in the other seven genomes. WABA is similar to BLAST and was originally designed for use in nematodes (10,26). We found WABA to be particularly useful for our purposes because it finds putative orthologues for protein-coding DNA sequences from an annotated genome to a newly assembled, unannotated genome without intermediate gene prediction and translation steps.
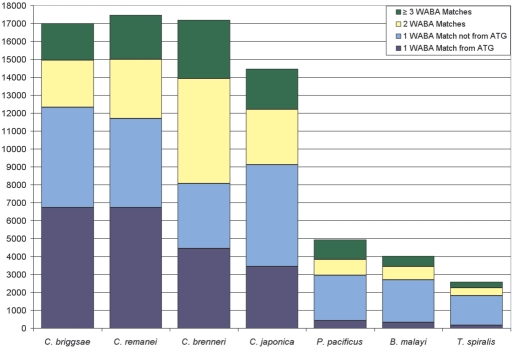
WABA and InParanoid results were concordant. In order to determine whether WABA results were reliable compared to protein-level orthologue determination, we compared its output to the InParanoid database (27). We found that InParanoid identified 12 197 one-to-one orthologues between C. elegans and C. briggsae genes, while WABA identified single orthologues for 12 326 C. elegans transcripts (Figure 2). Of these 12 326, InParanoid also had identified single orthologues for 11 231 (91% of 12 326 and 92% of 12 197). Of the 11 231 C. elegans transcripts with both a single WABA orthologue and a single InParanoid orthologue, the WABA orthologue overlapped the InParanoid orthologue for 11 104 (98.9%), and the start site of the WABA orthologue was within 750 bp of that of the InParanoid orthologue for 9645 (86%).
Figure 2.
Number of WABA matches for 23 212 chromosomal C. elegans WormPep transcripts. The number of C. elegans transcripts with exactly one match starting from the ATG (‘high-quality orthologues’) is shown at the bottom, in dark blue. The number of remaining C. elegans transcripts with exactly one match is shown in light blue. The number of C. elegans transcripts with two matches in the comparison genome is shown in yellow, and the number of C. elegans transcripts with three or more matches is shown in green.
C. brenneri had two matches for many C. elegans transcripts. All four species from genus Caenorhabditis had at least one match for 14 000–18 000 of the C. elegans transcripts (Figure 2). C. briggsae and C. remanei both had single matches for about 12 000 C. elegans transcripts and two matches for approximately 3000 additional transcripts. However, for C. brenneri, a disproportionately small number of C. elegans WormPep sequences had one match and a large number had two matches. The result was that far fewer C. elegans transcripts had suitable orthologues in C. brenneri (<4500) than in the other two Caenorhabditis species (>6000), even though all three species are the same evolutionary distance from C. elegans. As expected, the three more distant nematode species (P. pacificus, B. malayi and T. spiralis) had far fewer WABA-predicted orthologues than the more closely related nematodes.
Because the analysis described in this paper involved regions directly upstream of ATGs, it was important to accurately identify the N-terminal of each orthologue. Therefore, only high-quality orthologues, i.e. single WABA matches that started at the ATG of the C. elegans transcript, were used for the next step of the analysis.
Orthologous upstream sequence regions
Orthologous upstream sequence region sets were formed by pooling the upstream region of each C. elegans transcript with that of its orthologues from the other genomes. Only transcripts with at least three out of a possible seven high-quality orthologues were retained. The resulting collection contained upstream sets for 3847 C. elegans transcripts, but was somewhat redundant due to both transcripts from the same gene that shared the same ATG and transcripts on bidirectional promoters that shared the same upstream region; 3544 different transcript upstream regions and 3458 genes were represented. Taking orthologous sequences into account, the collection contained 3551 unique upstream sets. WABA identified a unique region of each unannotated genome as an orthologue 96% of the time. Only 141 transcripts had orthologues that overlapped those of another transcript. These may be a result of a gene duplication event that occurred in C. elegans after it diverged from the other species.
Bidirectional promoters were highly conserved among nematodes. We identified 132 C. elegans bidirectional promoters shorter than 1500 bp, of which 25 (19%) were perfectly conserved among all species for which orthologues were found and another 89 (67%) were conserved among orthologues from other species in genus Caenorhabditis. Only 10 (8%) bidirectional promoters were not conserved in any of the species. We also noted that 5 (4%) of the transcript pairs on bidirectional promoters had similar or identical protein-coding sequences and as a result had the same orthologues.
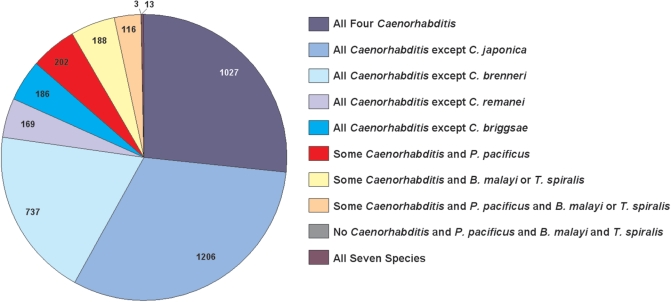
Most transcripts only had orthologues in other species of genus Caenorhabditis; only 14% had orthologues in P. pacificus, B. malayi or T. spiralis. There were 1027 (27%) C. elegans transcripts with orthologues in all four of the other Caenorhabditis species, and another 2298 (60%) transcripts had orthologues in three out of four of these species (Figure 3). Only 202 (5%) transcripts had orthologues in P. pacificus as well as in some Caenorhabditis species, 188 (5%) transcripts had orthologues from at least one of the two parasitic nematodes but not P. pacificus, and 116 (3%) transcripts had orthologues from both P. pacificus and a parasitic nematode. Only three transcripts had orthologues from P. pacificus and both parasitic nematodes but not from any species in Caenorhabditis. Finally, 13 transcripts had orthologues in all seven nematode species: rpl-2 (B0250.1), cyn-10 (B0252.4b), rps-13 (C16A3.9), phi-18 (C37C3.2 transcripts b&c), D1054.14, rps-9 (F40F8.10), rpn-6 (F57B9.10b), T10C6.5, cdc-37 (W08F4.8a), W09G12.5 (now known as F38A1.8), rab-30 (Y45F3A.2) and aps-3 (Y48G8AL.14).
Figure 3.
Species composition of orthologous upstream sequence region sets. The upstream regions of C. elegans transcripts were pooled with the upstream regions of their orthologues from the other seven genomes to form orthologous upstream sequence region sets. Only C. elegans transcripts with at least three high-quality orthologues were used, resulting in a total of 3847 sets. Of these, 1027 contained sequence from all four species in genus Caenorhabditis (dark blue), while a total of 2298 of the sets contained sequence from all but one of the four Caenorhabditis species (various shades of light blue). Only 522 of the sets contained sequence from Pristionchus pacificus, Brugia malayi, or Trichinella spiralis; 13 sets contained sequence from all seven species (purple).
Chromosomes III and X were overrepresented among the transcripts in the set, while Chromosomes IV and V were underrepresented (Pearson χ2 P-value <10–15). In contrast, the proportion of transcripts on Chromosomes I and II was not significantly different (Supplementary Figure S3).
Motif discovery
A multi-species high-order Markov background model improved MotifSampler's specificity. MotifSampler can use a high-order Markov background model to reduce the probability that it will return unmasked repeats and other low-complexity sequences as a motif (28). This was important for nematode genomes because they are 57–70% AT and contain much low-complexity sequence.
Extensive testing was done to determine settings for MotifSampler parameters that maximized the sensitivity while minimizing the total number of motifs. We found that the sensitivity was >80% when we retained motifs with MotifSampler scores above the 70th percentile but decreased rapidly for score thresholds above the 80th percentile. The coverage (proportion of bases covered by at least one motif) decreased linearly as we increased the motif score threshold from the 50th to the 90th percentile. Therefore, we retained only the top 30% of motifs found by MotifSampler.
A substantial number of motifs were very wide. Of the total of 158 017 motifs found, 14 bp motifs were the most common of the five widths (Supplementary Figure S4). After overlapping motifs were merged, the distribution of motif widths developed a long tail: many of the motifs were much wider than 14 bp, nearly 4000 motifs were ≥30 bp wide, and the widest motif was 212 bp.
Most motifs were found in all sequences of the orthologous upstream sequence region set. The majority of the upstream sequence region sets consisted of C. elegans and three or four sequences from other Caenorhabditis species (Figure 3). The motif discovery algorithm found 84% of motifs in all species of the sequence set, with the result that most motifs had a species depth (i.e. the number of species in which the motif was found) of four or five, including C. elegans. Four percent of motifs had a depth less than four, 59% of motifs had a depth of four, 33% had a depth of five and 4% had a depth greater than five. All but 20 of the motifs had a depth of at least three. Motifs that were not found in all sequences came from upstream sequence sets in which one or more of the sequences were very different from the others. For example, the motifs were not found on a sequence from one of the more distant species or on a sequence that was highly repetitive.
The conserved proportion of upstream regions varied widely. Of all unmasked bases of C. elegans upstream regions, 45% were covered by at least one motif. The interquartile range of coverage of upstream regions was 36–58%, while a few upstream regions were nearly completely covered with motifs and other upstream regions were only 8% covered. There was a weak negative correlation (r = −0.43) between coverage and upstream length: shorter upstream sequences tended to have higher coverage (i.e. be more highly conserved). The spatial distribution of motifs across the upstream regions was uniform. No significant difference was seen between the distribution of motifs with respect to the ATG and the distribution of motifs with respect to the opposite end of the sequence (KS test, P > 0.2).
Validation
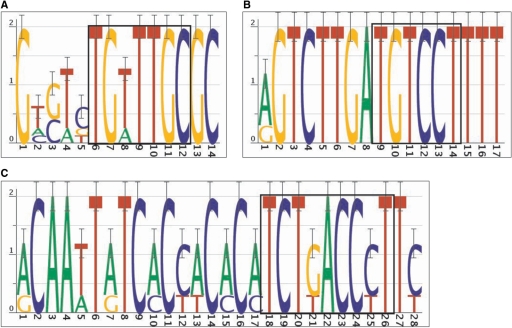
Discovered motifs were compared to experimentally proven TFBSs from the literature to gauge the success of the motif discovery process. For the 44 experimentally proven sites in the upstream regions under examination, 36 (82%) overlapped with motifs by at least 50% of the TFBS width, and 29 (66%) overlapped a motif completely. A complete list of experimentally proven sites and all cisRED motifs that overlapped them is shown in Supplementary Table S5. For example, the following sites were found: the PHA-4 site near tph-1 (ZK1290.2b) (29) (Figure 4A), a DAF-12 site near lit-1 (W06F12.1c) (30) (Figure 4B) and an ‘Early-2’ motif near K07C11.4 described by Gaudet et al. (4) (Figure 4C). Of the eight known sites that were not found, seven were poorly conserved and one was a low-complexity PHA-4 site.
Figure 4.
Examples of experimentally proven sites. (A) A motif that overlaps a PHA-4 site upstream of tph-1 (ZK1290.2b). (B) A motif that overlaps a DAF-12 site upstream of lit-1 (W06F12.1c). (C) A motif that overlaps an ‘Early-2’ site upstream of K07C11.4. Locations of experimentally proven sites are indicated by black boxes. cisRED URLs are indicated in Table 1.
Motif P-values and information content were uncorrelated with motif function. We assigned a preliminary score to each motif using a simplified version of the scoring function described by Robertson et al. (20) in an attempt to evaluate its significance with respect to gene regulation. This score measured two parameters: depth of the motif (relative to the depth of the input set, which was from four to eight), and the average conservation of the bases (weighted by evolutionary distance, with more distant species weighted more heavily). The width of the motif was not included in the scoring function because experimentally proven TFBSs are as narrow as 6 bp and as wide as 16 bp. Each motif was then assigned a P-value indicating its rank in the distribution of scores of all 158 017 motifs. However, we found no relationship between the P-values and the functionality of the motifs; motifs overlapping experimentally proven sites were as likely to have a high P-value as a low P-value.
Motif information content [IC; a measure of the degree of conservation (25)] ranged from 0.7 bits to a perfectly conserved 2 bits with an interquartile range of 1.45–1.75 (Supplementary Figure S6). As was the case for the scoring function, IC was not useful in discriminating motifs that overlapped TFBSs; we observed no difference in the distribution of average IC between motifs overlapping experimentally proven sites and all motifs.
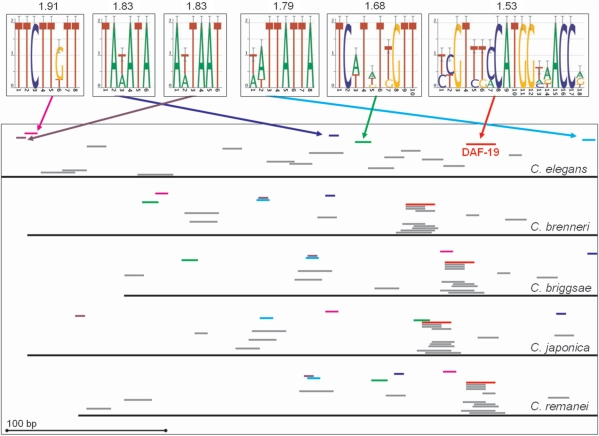
Functional regulatory elements were not the most highly conserved portions of the upstream regions. For example, we found 20 motifs in the 371 bp upstream region of xbx-1 (F02D8.3) and its orthologues in C. briggsae, C. remanei, C. brenneri and C. japonica, resulting in a coverage of 62% (Figure 5). This upstream region also contained an experimentally proven DAF-19 site (31), which was found by our method. However, five of the other motifs were more strongly conserved than the DAF-19 site (indicated by consensus sequence logos (32); average IC also shown for each).
Figure 5.
Example of high-coverage upstream sequence region with an experimentally proven site. The upstream regions of xbx-1 (F02D8.3) and its orthologues in C. briggsae, C. remanei, C. brenneri and C. japonica are indicated by black lines. The ATG of each transcript or putative orthologue is at the right edge of the figure. The logos of the top six most-conserved motifs and their IC are shown; the locations of these motifs in each upstream sequence are indicated by coloured bars. The locations of the remaining motifs are indicated by grey bars. Motifs are sorted by IC with the most conserved motif at the top. The experimentally proven DAF-19 site is indicated. The cisRED URL is indicated in Table 1.
Annotation to reveal similarity to known TFBSs
Five percent of the motifs were similar to TFBSs previously characterized in C. elegans. Motifs for which the C. elegans sequence displayed some similarity to one of 13 sets of TFBSs in C. elegans were identified and assigned a P-value indicating the significance of the similarity. We found that 36 of the motifs that overlapped experimentally proven sites by at least 5 bp could be annotated using this procedure. These could be separated into two groups: 20 motifs had very significant annotation P-values of <0.00015, and the other 16 had less significant annotation P-values (P > 0.0009). Given this, the stringent threshold of 0.00015 was used for the ORegAnno binding sequence annotations. Four of the TFs had no annotated motifs below this threshold; sequences that were the same as or similar to these TFBSs appeared frequently enough among the non-conserved parts of the upstream regions that they could not be applied to the motifs with confidence. The TFs that were not annotated successfully were: PHA-4, DAF-12, the ‘Heat Shock Element’ described by GuhaThakurta et al. (33), and the ‘Late-2’ element described by Gaudet et al. (4). A total of 7650 TF-motif combinations were annotated, representing 7449 different motifs; several motifs were annotated as similar to more than one TFBS. The most commonly annotated TFBS was DAF-19: 1305 motifs were annotated as similar to a DAF-19 site (Supplementary Table S7).
Eleven percent of the motifs were similar to TFBSs from TRANSFAC; 15% were similar to TFBSs from JASPAR. In order to determine whether any of the motifs were similar to binding sequences identified in species other than C. elegans, the same procedure was used to annotate the motifs using binding sequences from TRANSFAC and JASPAR. TRANSFAC contained binding sequences for 319 different TFs, which were mainly characterized in mammalian species. We chose a stringent threshold (P < 10−5) and annotated 17 740 (11%) motifs as similar to 221 TRANSFAC TFBSs. The most commonly annotated TFBS was PAX5/BSAP: 969 motifs were similar to this site (Supplementary Table S7).
The annotation results using TFBSs from JASPAR overlapped substantially with the TRANSFAC results because the two databases use some of the same sources (34). However, because the binding sequences in JASPAR were non-redundant, we chose a higher P-value threshold (P < 10−4) for the JASPAR annotations, and annotated 23 331 (15%) motifs as similar to binding sites of 39 TFs. As with the TRANSFAC results, the most commonly annotated TFBS was BSAP/PAX5: 2041 motifs were similar to this site based on JASPAR binding sequence examples (Supplementary Table S7). In total, 40 396 (26%) motifs were annotated with at least one TFBS from one of the three databases.
cisRED web interface
All data and results discussed here, including orthologous upstream sequence region sets for each transcript, motifs found, and annotations, are available via the web interface at www.cisred.org (20) (Supplementary Document S8). URLs for motifs in figures are shown in Table 1. Additionally, all WABA and MotifSampler data are available on request.
Table 1.
Figure URLs
All results are available via the cisRED web interface. URLs of motifs in figures are indicated.
Applications
Several examples of applications of the information in the cisRED C. elegans database to current questions in nematode genomics, gene annotation, evolution, and gene regulation are illustrated below.
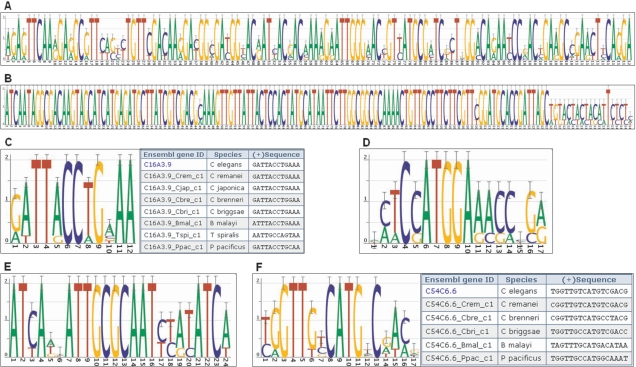
Some wide motifs were unannotated protein-coding exons. There were 3918 motifs ≥30 bp wide. While many of these were in coding exons belonging to other transcripts of the same gene, others represented novel findings. Some of the wide motifs resembled protein-coding exons even though no coding exon was annotated by WormBase in that location. For example, a 120 bp motif was found immediately upstream of the ATG of Y73B3A.12, a member of the Calmodulin family (Figure 6A). It had a depth of six species, occurring in all species except C. briggsae and P. pacificus. A BLASTX (35) search for the C. elegans motif sequence returned many matches to Calmodulin genes of various species, which indicated that this region of the C. elegans genome is likely to be a coding exon that was not annotated by WormBase.
Figure 6.
Examples of applications. (A) A 120 bp motif upstream of Y73B3A.12, a member of the Calmodulin family. (B) A 143-bp motif upstream of grd-7 (F46H5.6). (C) A deeply conserved element upstream of rps-13 (C16A3.9) with a table showing motif sequences in all eight species. (D) A DAF-19-like site upstream of kin-2 (R07E4.6b). (E) An ATF4-like site upstream of Y34B4A.10. (F) A DAF-19-like site upstream of C54C6.6 with a table showing motif sequences in four species from genus Caenorhabditis, plus B. malayi and P. pacificus. cisRED URLs are indicated in Table 1.
Some highly conserved wide motifs may be noncoding RNA genes. A 143 bp motif was found upstream of grd-7 (F46H5.6) (Figure 6B), and all but five of the bases were perfectly conserved among four species of Caenorhabditis (this transcript had no acceptable C. japonica orthologue). The C. briggsae sequence included a 1 bp insertion, causing a shift in the consensus sequence logo (32) at the 125th base of the motif. A BLAST search for this sequence returned no matches. However, WormBase indicated that the motif overlapped a predicted noncoding RNA gene near the 3′ UTR of unc-10 (T10A3.1b). This finding provides support for the predicted RNA gene in that location and its strong conservation in three other species suggests that it is functional. It also provides a hypothetical function for other very wide motifs that do not appear to be protein-coding.
Several very highly conserved motifs occurred in all eight nematode species. Thirteen transcripts had high-quality orthologues in all seven non-annotated species, and were associated with 115 motifs that occurred in all eight species. For example, a highly conserved motif was found in the 5′ UTR of rps-13 (C16A3.9) (Figure 6C). Of the 12 bases that make up the motif, seven bases were perfectly conserved in all eight species.
Annotated motifs provided new information regarding TFBS locations and evolution of TF binding and function. The motif annotation process, which used TF binding sequences for both mammalian and C. elegans TFs, returned many novel binding site candidates. For example, a motif similar to a DAF-19 binding site was found near kin-2 (R07E4.6b; Figure 6D). The annotation results can also be used to suggest novel binding site candidates for uncharacterized TFs that are orthologues of characterized mammalian TFs. For example, a human ATF4-like motif was found near Y34B4A.10 (Figure 6E). Finally, the annotation process revealed information concerning the conservation of TFBSs in the more distant nematode species. For example, a DAF-19-like site near the uncharacterized gene C54C6.6 (Figure 6F) showed that the site was strongly conserved in P. pacificus and weakly conserved in B. malayi.
DISCUSSION
The application of WABA to the seven non-C. elegans genomes revealed information about the recently sequenced genomes of C. brenneri and C. japonica. All four species in genus Caenorhabditis had similar overall numbers of matches to C. elegans WormPep sequences (Figure 2). However, compared to C. briggsae and C. remanei, there was a disproportionately small number of WormPep sequences that had one match and a large number with two matches in the C. brenneri genome. This anomaly may be because the draft genome sequence of C. brenneri is derived from a strain that was inbred and yet heterozygous over 30% of its genome. As alleles are highly differentiated in this species, the genome assembly contains alternative forms of many genes that were assembled independently (36). C. japonica had 16% fewer matches to C. elegans WormPep protein-coding sequences than the other Caenorhabditis species, and had fewer high-quality orthologues. This may have been due to both the greater evolutionary distance between C. elegans and C. japonica and the poorer genome assembly of C. japonica, which was released very recently and was still in draft stages (Supplementary Table S1). High-quality orthologues among the more distant nematode species were even more rare; only 14% of examined C. elegans transcripts had high-quality orthologues in Pristionchus pacificus, Brugia malayi or Trichinella spiralis. In addition to interference from the low quality of these genome assemblies, the WABA algorithm may be too stringent to find orthologues if the genomes are too distant. In order to minimize the impact of genomic anomalies and maximize the likelihood of finding evolutionarily conserved upstream motifs, we limited this investigation to transcripts with at least three high-quality orthologues. The resulting collection of orthologous upstream sequence region sets was strongly conserved and included only 17% of WormPep transcripts.
Of the 132 bidirectional promoters examined in this study, 86% were conserved among the species of genus Caenorhabditis. The majority of bidirectional promoters in C. elegans have previously been found to be conserved in C. briggsae (37); given the high rate of conservation, bidirectional promoters must be an important mechanism for controlling gene regulation among gene pairs. Some gene pairs on bidirectional promoters are coexpressed while others have a mutually exclusive gene expression pattern (37). Documentation of the conserved elements in these promoters, in combination with the examination of the expression patterns of the transcripts involved, may help to clarify these mechanisms of gene regulation.
While the large majority of orthologous regions in the other species were associated with only one C. elegans transcript, some functionally related groups of C. elegans transcripts had fewer orthologous representatives in the unannotated nematode genome sequences. Most cases of overlapping orthologues in the unannotated genomes belonged to large gene groups such as serpentine receptors. This may be because the four other species of Caenorhabditis are associated with different types of decaying matter (38); C. elegans may have more of these types of receptors to help it find its specific type of food while the other species may have expanded different receptor families. In some cases, two C. elegans genes with overlapping orthologues were side by side (on the same or opposite strand), which suggests that a gene duplication event occurred in C. elegans after C. elegans diverged from the other Caenorhabditis species.
The transcripts that had a sufficient number of orthologues to be used in this analysis had a different chromosomal distribution from the entire set of WormPep transcripts, suggesting that certain regions of the genome are more highly conserved than others (Supplementary Figure S3). Chromosome III is known to be rich in genes with yeast orthologues (9) and essential genes (39) such as those required for cell division (40). A detailed analysis of synteny in the C. elegans and C. briggsae genomes has previously revealed that orthologues are overrepresented on Chromosomes III and X and underrepresented on Chromosome V (41).
Because regulatory elements are not readily distinguishable from other conserved upstream elements, the primary goal of this study was to catalogue all conserved elements of the upstream regions. We did not preface the motif discovery procedure with a multiple sequence alignment so as to avoid the preconditions that conserved elements be in the same order (with respect to the distance from the ATG) and contained within alignable sequence. We tested several motif discovery algorithms and found that while MotifSampler was the most suitable program for this purpose, a high-order background model was essential because nematode intergenic sequence frequently contains low-complexity sequence.
In order to assess the effectiveness of the motif discovery procedure, we compared discovered motifs to experimentally proven TFBSs from ORegAnno. The motif discovery algorithm was highly successful at finding experimentally proven sites, with a sensitivity of 82%. The upstream regions of the positive controls were only characterized with respect to locations of TFBSs (or predicted TFBSs; in some cases, the binding TF is not known). No sections of these upstream regions have been definitively shown not to have regulatory function. Because it is not possible to estimate the false positive rate without true negatives, we only used sensitivity and coverage to choose the threshold for motif inclusion.
We found 20 motifs upstream of xbx-1 (F02D8.3), of which five were more highly conserved than the one corresponding to the DAF-19 site (Figure 5). Because functional analyses of promoter sequences tend to reveal only a few short TFBSs [see for example (4,6,29,33)], it seems unlikely that all of this conserved sequence has regulatory function. However, because the upstream sequence of xbx-1 is uncharacterized other than the DAF-19 site, it is possible that some of the other motifs also have regulatory function.
While this study has focused on characterizing conserved elements, there is clearly much more to what constitutes a regulatory element than just conservation. Both TFBSs (42) and TFs (43) have been shown to be conserved among C. elegans, C. briggsae and C. remanei. For the highly conserved transcripts studied here, we did not find regulatory elements to be more conserved than other portions of the upstream regions. There was no difference in the distribution of average IC between motifs overlapping experimentally proven sites and all motifs (Supplementary Figure S6). Thus, attempts to assign a score to each motif indicating the probability that it had regulatory function were unsuccessful. In light of these results, we decided to retain all motifs that we identified, regardless of their conservation score.
Experimentally proven sites that were not found were poorly conserved or highly degenerate, and so were not reported by the motif discovery algorithm. Not all TFBSs were conserved; many of the experimentally proven sites had low IC while others were not found at all using our parameters for motif discovery. Additionally, some of the experimentally proven sites that our method did not identify may have been outside of the region we examined on the orthologous sequences, and there may be other ways to regulate transcription of the orthologues, perhaps using different TFs with a parallel function. The AT-rich sites such as PHA-4 (29) are highly degenerate and extremely common in the genome. Nematodes must have a way to distinguish functional from non-functional sites in vivo, perhaps via histone modifications (44).
In a preliminary comparison of conserved regions in C. elegans and C. briggsae, Siepel et al. (45) found that 18–37% of the genomes were conserved, but considered this to be an underestimate because they used phastCons-aligned regions. They anticipated that improved results could be generated by using additional nematode genomes. They suggested that highly conserved elements may contain multiple overlapping binding sites, be under protein-coding or RNA structural constraints, or have ‘as-yet-undiscovered functions’. They also suggested that some conserved regions may have ‘mutational rather than selectional explanations’ and may be ‘shielded from mutations or subjected to hyperefficient repair’. The results described here were generated with eight nematode genomes. Consistent with their suggestion that alignment might underestimate conservation, we found that conserved elements identified using motif discovery resulted in a median coverage of 45% of the upstream regions. This proportion represents the amount of upstream sequence that was conserved to approximately the same degree as TFBSs, some of which are highly degenerate. Again consistent with their discussion, many wide motifs were in annotated or unannotated protein-coding exons belonging to the same gene. Protein-coding motifs can often be recognized by their codon-like conservation pattern in which every third base is poorly conserved because it can be substituted by several different nucleotides without changing the amino acid sequence (Figure 6A); protein-coding regions also tend to have significant results following a BLASTX (35) search. Motifs that appear to be protein-coding but are not annotated could be used to refine C. elegans gene models. Some wide non-protein coding motifs were in 5′ and 3′ UTRs and may be target sites of RNA binding proteins or microRNAs, while others may represent noncoding RNA genes (Figure 6B).
Most motifs were found in all sequences of the input set, with the result that most motifs have a species depth of four or five including C. elegans. The motif discovery algorithm preferred depth over conservation; if the best available version of the motif on one of the sequences was quite different from the others, it was included rather than excluded. This provided us with an opportunity to observe the evolution of conserved upstream elements among the more distant nematode species. Several motifs were found in all eight species and were very highly conserved (Figure 6C), suggesting the presence of ancient genomic elements near essential genes.
Motifs for which the C. elegans sequence displayed a significant similarity to a characterized TFBS were annotated as such. We observed that conserved sequences similar to a wide variety of mammalian TFBSs appeared in C. elegans upstream regions. This annotation is preliminary and the intention was not to exhaustively annotate occurrences of TFBSs from TRANSFAC or JASPAR, but merely to assess which ones seemed to occur frequently among conserved parts of upstream regions as compared to non-conserved parts of upstream regions. There was substantial overlap between the annotation results using TRANSFAC and JASPAR, as JASPAR is a more thoroughly curated subset of TRANSFAC. The results from the two databases were consistent. For example, the most commonly annotated TF was the same for TRANSFAC and JASPAR (PAX5/BSAP) (Supplementary Table S7). Similarly, CREB was the fourth most commonly annotated TF from JASPAR and the third most commonly annotated TF from TRANSFAC.
Because certain characterized TFs in JASPAR, TRANSFAC and ORegAnno had strongly variable or very few binding sequences, we chose to require a C. elegans sequence to be similar to a specific binding sequence rather than generate binding models such as position weight matrices for each TFBS. The limitation of this method was that all mismatches between the C. elegans sequence and a binding sequence were treated equally, which may have generated false positive annotations. Estimating the false positive rate requires a set of true negatives, and such a set is not available. Not all binding sites could be annotated using this method—some TFs, such as PHA-4 and DAF-12, had so many variations in their binding sequences and were so common in the upstream regions that none of the motifs could be annotated with that TFBS at a P-value below the threshold. Motifs were much more likely than non-conserved upstream sequence to be similar to a TFBS. The distribution in scores between the motifs (by definition evolutionarily conserved) and non-conserved upstream sequence was different for most TFs.
A DAF-19-like site was found upstream of kin-2 (R07E4.6b) (Figure 6D). In addition to the high conservation of this site and its strong similarity to a DAF-19 binding site, we have further supporting evidence of its functionality. First, DAF-19 is known to regulate gene expression in ciliated neurons, and kin-2 is expressed in ciliated neurons (46). Secondly, KIN-2 is known to interact with RIC-8 (47), and ric-8 (Y69A2AR.2) has been shown to be regulated by DAF-19 as well (42).
A human ATF4-like motif was found near Y34B4A.10 (Figure 6E). According to WormBase, the C. elegans homologue of the human atf4 gene is atf-5 (T04C10.4) (2). The binding site of C. elegans ATF-5 is uncharacterized; perhaps conserved elements that are similar to the human ATF4 site could be tested for binding with, and regulation via, C. elegans ATF-5.
A DAF-19-like site was found upstream of the uncharacterized transcript C54C6.6 (Figure 6F). This site was shown to have substantial similarity in the more distant nematode species P. pacificus and B. malayi. The conservation of the site in these species suggests that they also have the DAF-19 TF and may use it to regulate the expression of some of the same genes. This example illustrates that annotated motifs can increase our understanding of gene regulation in these species.
CONCLUSIONS
We have shown that WABA is an effective tool for finding orthologues for highly conserved transcripts among nematode genomes. We applied WABA to all annotated protein-coding transcripts from C. elegans; however, only transcripts with at least three high-quality orthologues were included in the motif discovery step. We identified conserved elements in the upstream regions of 3847 C. elegans transcripts (17% of all C. elegans transcripts).
We found that identification of putative regulatory elements via motif discovery among orthologous upstream regions resulted in a sensitivity of 82%, which suggests that most regulatory elements are conserved. However, we also found that the upstream regions also contain numerous other conserved elements, and that regulatory elements are not the most highly conserved elements in these upstream regions. Therefore, while conserved motifs are enriched for regulatory elements, conservation alone can not be used to distinguish regulatory elements from other conserved elements.
All of our results are publicly available via the web interface at www.cisred.org. Gene regulation researchers can use the web interface to see all conserved elements and their annotations for any gene of interest. For work involving laboratory methods such as reporter gene assays and gel shift assays to investigate the regulation of these genes, the cisRED data can immediately focus the search onto conserved and possibly annotated elements in upstream regions.
Many of the conserved elements in the cisRED database are in 5′ and 3′ UTRs of different transcripts; some of these may be candidate targets for RNA binding proteins. Additionally, some of the wide, highly conserved motifs may serve as novel noncoding RNA gene candidates. Those motifs that appear to be protein-coding can be used to refine and expand existing gene models.
Twenty-six percent of the conserved elements were found to be similar to known TFBSs and were annotated as such. These annotations are useful in three important ways. First, they provide novel candidate binding sites for TFs that are already characterized in C. elegans. These sites could be tested by researchers who are interested in targets of the TFs. Secondly, the annotations provide novel binding site candidates for uncharacterized TFs that are orthologues of characterized mammalian TFs. This takes advantage of existing information about TF binding in mammals to expand our understanding of gene regulation in C. elegans. Lastly, the annotations make it possible to assess evolution of TFs, their binding sites, and the process of gene regulation in general by comparing both the TF protein sequence and their predicted binding sites across the different nematode species. The conservation of annotated sites in more distantly related nematodes implies that they have the same TFs as C. elegans and use them to regulate some of the same genes.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the Washington University in St Louis Genome Sequence Center for making the genomes of C. remanei, C. brenneri, C. japonica, P. pacificus and T. spiralis freely available. We thank Obi L. Griffith, Heesun Shin, Bernhard H. G. Sleumer and Foyita Sleumer for comments on the manuscript and useful discussions.
FUNDING
This work was supported by the Michael Smith Foundation for Health Research (MSFHR). S.J.M.J. is a scholar of the MSFHR. M.C.S. is a trainee of the MSFHR. Funding for open access charge: BC Cancer Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 2.Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okkema PG, Krause M. Transcriptional regulation, In WormBook, ed. 2005. The C. elegans Research Community. Available at http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 4.Gaudet J, Muttumu S, Horner M, Mango SE. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GuhaThakurta D, Schriefer LA, Waterston RH, Stormo GD. Novel transcription regulatory elements in Caenorhabditis elegans muscle genes. Genome Res. 2004;14:2457–2468. doi: 10.1101/gr.2961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 10.Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The Genome Sequence of Caenorhabditis briggsae: A Platform for Comparative Genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudhaus W, Kiontke K. Comparison of the cryptic nematode species Caenorhabditis brenneri sp. n. and C. remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditis Elegans group. Zootaxa. 2007;1456:45–62. [Google Scholar]
- 12.Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitreva M, Jasmer DP. Biology and genome of Trichinella spiralis. In WormBook, ed. 2006. The C. elegans Research Community. Available at http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 15.Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DHA. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl Acad. Sci. USA. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 17.Mitreva M, Blaxter ML, Bird DM, McCarter JP. Comparative genomics of nematodes. Trends Genet. 2005;21:573–581. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Kent WJ, Zahler AM. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- 19.Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 2002;9:447–464. doi: 10.1089/10665270252935566. [DOI] [PubMed] [Google Scholar]
- 20.Robertson G, Bilenky M, Lin K, He A, Yuen W, Dagpinar M, Varhol R, Teague K, Griffith OL, Zhang X, et al. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;34:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffith OL, Montgomery SB, Bernier B, Chu B, Kasaian K, Aerts S, Mahony S, Sleumer MC, Bilenky M, Haeussler M, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bryne JC, Valen E, Tang ME, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 26.Baillie DL, Rose AM. WABA success: a tool for sequence comparison between large genomes. Genome Res. 2000;10:1071–1073. doi: 10.1101/gr.10.8.1071. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchal K, Thijs G, De Keersmaecker S, Monsieurs P, De Moor B, Vanderleyden J. Genome-specific higher-order background models to improve motif detection. Trends Microbiol. 2003;11:61–6. doi: 10.1016/s0966-842x(02)00030-6. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 30.Shostak Y, Van Gilst MR, Antebi A, Yamamoto KR. Identification of C. elegans DAF-12-binding sites, response elements, and target genes. Genes Dev. 2004;18:2529–2544. doi: 10.1101/gad.1218504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 32.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.GuhaThakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, Walker DW, Lithgow G, Kim S, Link CD. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 2002;12:701–712. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrière A, Yang S, Pekarek E, Thomas C, Haag ES, Ruvinsky I. Detecting heterozygosity in shotgun genome assemblies: Lessons from obligately outcrossing nematodes. Genome Res. 2009 doi: 10.1101/gr.081851.108. doi: 10.1101/gr.081851.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bando T, Ikeda T, Kagawa H. The homeoproteins MAB-18 and CEH-14 insulate the dauer collagen gene col-43 from activation by the adjacent promoter of the Spermatheca gene sth-1 in Caenorhabditis elegans. J. Mol. Biol. 2005;348:101–112. doi: 10.1016/j.jmb.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 38.Baird SE. Natural and experimental associations of Caenorhabditis remanei with Trachelipus rathkii and other terrestrial isopods. Nematology. 1999;1:471–475. [Google Scholar]
- 39.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 40.Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 41.Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH. Comparison of C. elegans and C. briggsae Genome Sequences Reveals Extensive Conservation of Chromosome Organization and Synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, Newbury CR, Khattra J, Chan S, Go A, et al. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 2006;7:R126. doi: 10.1186/gb-2006-7-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haerty W, Artieri C, Khezri N, Singh RS, Gupta BP. Comparative analysis of function and interaction of transcription factors in nematodes: extensive conservation of orthology coupled to rapid sequence evolution. BMC Genomics. 2008;9:399. doi: 10.1186/1471-2164-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 47.Schade MA, Reynolds NK, Dollins CM, Miller KG. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G alpha(s) pathway and define a third major branch of the synaptic signaling network. Genetics. 2005;169:631–649. doi: 10.1534/genetics.104.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.