Abstract
Nuclear and cytoplasmic forms of the yeast exosome share 10 components, of which only Rrp44/Dis3 is believed to possess 3′ exonuclease activity. We report that expression only of Rrp44 lacking 3′-exonuclease activity (Rrp44-exo) supports growth in S288c-related strains (BY4741). In BY4741, rrp44-exo was synthetic-lethal with loss of the cytoplasmic 5′-exonuclease Xrn1, indicating block of mRNA turnover, but not with loss of the nuclear 3′-exonuclease Rrp6. The RNA processing phenotype of rrp44-exo was milder than that seen on Rrp44 depletion, indicating that Rrp44-exo retains important functions. Recombinant Rrp44 was shown to possess manganese-dependent endonuclease activity in vitro that was abolished by four point mutations in the putative metal binding residues of its N-terminal PIN domain. Rrp44 lacking both exonuclease and endonuclease activity failed to support growth in strains depleted of endogenous Rrp44. Strains expressing Rrp44-exo and Rrp44-endo–exo exhibited different RNA processing patterns in vivo suggesting Rrp44-dependent endonucleolytic cleavages in the 5′-ETS and ITS2 regions of the pre-rRNA. Finally, the N-terminal PIN domain was shown to be necessary and sufficient for association with the core exosome, indicating its dual function as a nuclease and structural element.
INTRODUCTION
The exosome complex is implicated in many RNA processing and degradation activities. Ten ‘core’ exosome components are shared between the nuclear and cytoplasmic forms of the complex, and all of these are essential for cell viability (1). The nuclear exosome participates in many RNA degradation and surveillance pathways, as well as processing the precursors to the 5.8S rRNA and other stable RNA species (2). The cytoplasmic exosome functions in mRNA degradation, participating in general mRNA turnover and several activated decay and surveillance pathways (3).
Structural and functional analyses indicate that Rrp44 (Dis3) is the only catalytically active 3′–5′ exonuclease in the yeast exosome core (4–6), whereas the nuclear exosome is associated with a second active nuclease (Rrp6) (7). Rrp44 is related to Escherichia coli RNase R, a member of the RNase II (RNase B) family of hydrolytic exonucleases. As shown in Figure 1A, Rrp44 has an N-terminal PIN-domain (PilT N-terminus—derived from the name of an E. coli protein implicated in pilus formation), which is not shared with RNase R or RNase II. Recent studies reported endonuclease activity associated with the PIN-domains of human Smg6 and yeast Swt1, both of which are implicated in mRNA surveillance (8,9), and the PIN domain protein Nob1 was predicted to be a pre-rRNA endonuclease (10,11). The PIN domain in Rrp44 is followed by a putative RNA-binding, cold-shock domain (CSD), for which functional analyses have not yet been reported. The exonuclease activity resides in the RNB domain and this is abolished by the point mutation D551N (4,6). A C-terminal S1 RNA-binding domain is also important for substrate binding and for activity in vitro and in vivo, and this is impaired by the G916E mutation (6,12).
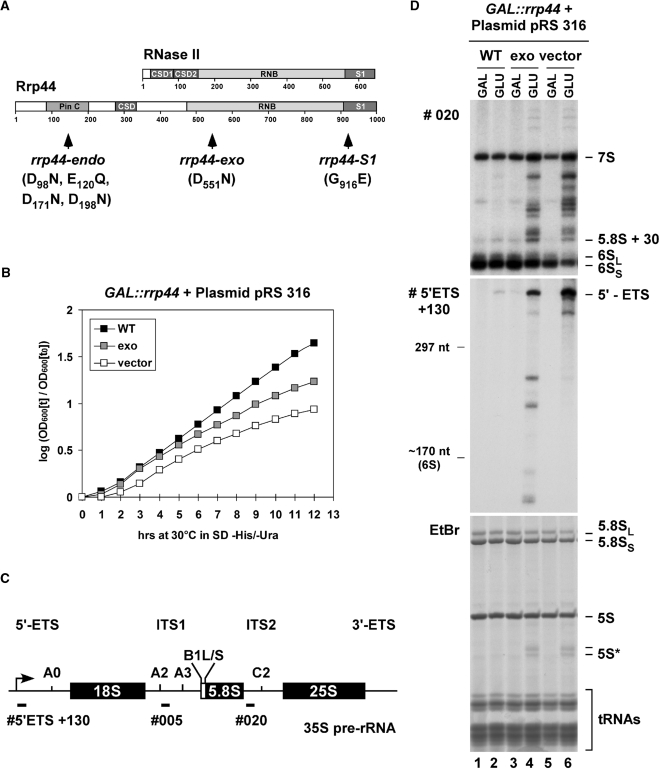
Figure 1.
In vivo analysis of yeast strains expressing Rrp44-exo. (A) Domain structures of E. coli RNase II and S. cerevisiae Rrp44. Positions of Rrp44 point mutations are indicated. (B) Growth analysis of yeast strains expressing Rrp44-exo. A GAL::rrp44 yeast strain transformed with plasmids encoding Rrp44 [WT or D551N (exo)] or the empty vector pRS316 were grown in liquid selective glucose medium at 30°C to analyze growth. (C) Structure of the 35S pre-rRNA with the location of oligonucleotide probes used for Northern hybridization. (D) Northern analysis of pre-rRNA processing in the GAL::rrp44 strain transformed with a plasmid expressing either WT Rrp44 or the mutant (D551N) Rrp44-exo protein, or an empty vector. RNA was isolated from GAL::rrp44 strains grown at 30°C under permissive conditions (GAL) and 8 h after transcriptional repression (GLU). RNA was separated on an 8% polyacrylamide/8 M urea gel and either detected by Northern hybridization with the oligonucleotide probes depicted in Figure 1C or by staining with EtBr.
Strains lacking the exonuclease activity of Rrp44 are viable, whereas the integrity of the exosome complex is essential since depletion of any single core component is lethal (1,4–6). These observations raised several questions, including whether other nucleases in the nucleus or cytoplasm can functionally substitute for the exonuclease activity of Rrp44 and whether other nuclease activities are associated with the core exosome?
MATERIALS AND METHODS
In vivo analyses
Growth and handling of Saccharomyces cerevisiae were by standard techniques. Strains were grown at 25°C or 30°C in YPD or synthetic dropout medium containing 0.67% nitrogen base (Difco) and either 2% glucose or 2% galactose. Yeast RNA extraction and northern hybridization were performed as described (13). Northern signals were generally visualized by autoradiography, with the exception of the lighter exposure in Figure 6B, which was generated by a Fuji FLA-5100 PhosphorImager. Oligonucleotide probes are listed in Table S1.
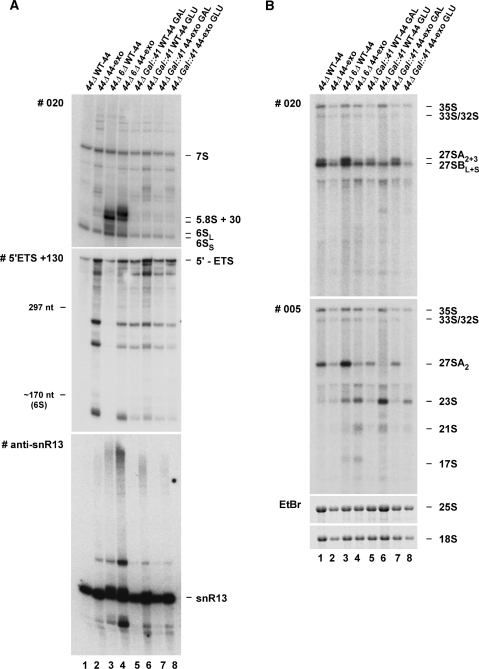
Figure 6.
The N-terminal PIN domain in Rrp44 harbors endonuclease activity in vivo. (A) Northern analysis of pre-rRNA processing in the GAL::rrp44 strain transformed with plasmids expressing either WT or mutant Rrp44 protein, or an empty vector pRS315. The mutants analyzed are rrp44-exo, rrp44-endo and rrp44-endo–exo (see Figure 5). RNA was isolated from GAL::rrp44 strains grown at 30°C under permissive conditions (GAL) and 10 h after transcriptional repression (GLU). RNA was separated on an 8% polyacrylamide/8 M urea gel and analyzed as described in Figure 1D. (B) Northern analysis of pre-rRNA processing in rrp44Δ (lanes 1–3) or rrp44Δrrp6Δ (lanes 4–6) strains transformed with a plasmid expressing either WT or mutant Rrp44 protein. Strains were grown at 30°C (rrp44Δ) or 25°C (rrp44Δ rrp6Δ) and RNA was analyzed as in (A). Two different exposures are shown for the probe recognizing precursors of the 5.8S rRNA (#020).
Rrp44 expression plasmids
The construction of yeast expression plasmids for Rrp44 is described in detail in (6). Briefly, the RRP44 ORF is fused to a C-terminal tag containing a streptavidin-binding peptide (Strep-tag II), TEV cleavage site and two copies of the z-domain of protein A (szz-tag) and cloned into the XhoI restriction sites of either pRS316 (URA3) or pRS315 (LEU2) (14). For both yeast and E. coli expression plasmids (see below), point mutations were created using the QuikChange kit (Stratagene) and deletion of the PIN-domain was achieved by PCR using the oligos listed in Table S1.
Deletion/modification of RRP44 and plasmid shuffling
The BY4741 wild-type (WT) strain was first supplemented with a plasmid carrying RRP44-szz and a URA3 selective marker. The genomic RRP44 ORF was then precisely deleted by homologous recombination with a PCR product generated from the pFA6A-kanMX6 deletion cassette (15). Following construction of rrp44Δ, a second plasmid (pRS315) (14) was introduced that carried LEU2 and expressed either WT or mutant Rrp44. The URA3 plasmid was then counter-selected on plates containing 5-fluoroorotic acid (5-FOA), which is converted to the toxic uracil analogue 5-fluoro-uracil by the action of Ura3. RRP44 was modified in W303 to provide control by the GAL promoter by integration of a PCR product generated from pYM-N27 (16). Oligonucleotides used in generation of PCR cassettes are described in Table S1.
Overexpression and purification of recombinant proteins
E. coli strain BL21(DE3)pLysS (Stratagene) was transformed with plasmids encoding WT or mutant GST-Rrp44 [derived from GST-Dis3sc (17)] or GST alone (pGEX-4T1). Expression, cell lysis and single-step purification on glutathione sepharose was performed as previously described (6).
In vitro assays
Ribonuclease assays were performed with 200 fmol recombinant GST-Rrp44 and 10 fmol 5′- or 3′-end-labeled A30 RNA or 150 fmol 5′-end-labeled 52-nt stem loop RNA substrate (5′-GGCCCCGGGC CCCGUAGAAA AUCUUAGUAA UCCUUCUUAC AUUGCCCGGG GC-3′) in 10 mM Tris–HCl pH 7.6, 75 mM NaCl, 2 mM DTT, 100 μg/ml BSA, 0.8 U/μl RNasin, 4.5% glycerol, 0.05% Nonidet P40, 0.5 mM MgCl2 and 0, 0.5 or 5 mM MnCl2. Prior to addition of the labeled RNA, 10 µl reactions were pre-incubated for 10 min at 30°C. After an additional 1–2 h at 30°C, reactions were mixed with one volume of RNA formamide buffer, heated for 10 min at 65°C and separated on a denaturing 12% polyacrylamide/8 M urea sequencing gel. Reaction products were visualized by autoradiography.
Affinity purification of yeast exosomes
One step purifications of exosomes on IgG sepharose columns were performed as described (18). To isolate exosome complexes, Csl4-TAP strains were grown in YPD medium to OD600 0.7 at 25°C. Each preparation used 1 l of culture, producing 150 μl of the final TEV fraction. Exosomes were purified in buffer TMN150 (20 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.1% NP-40, 5 mM MgCl2) or treated with 800 mM MgCl2 before TEV elution to dissociate endogenous Rrp44. Enzyme concentrations were normalized by immunoblotting using an anti-Rrp6 antibody (19) or anti-peptide antibodies raised against Rrp44 and Rrp43 (this study).
GST pull down assays
Equal amounts of GST-bait proteins (∼1 pmol) were immobilized on glutathione sepharose beads and incubated with purified yeast exosomes (15 µl TEV eluate) for 1 h at 4°C in Buffer NB (20 mM Tris–HCl pH 7.6, 150 mM NaCl, 8.7% glycerol, 0.1% Nonidet P40 and 5 mM MgCl2). The beads were then washed five times with buffer NB. The retained proteins were separated on an 8% polyacrylamide/SDS gel and the presence of the core exosome was analyzed by immunoblotting with an antibody specific for Rrp43 (Figure 7D) and Rrp4 (data not shown). Inputs represent 20% of the starting material.
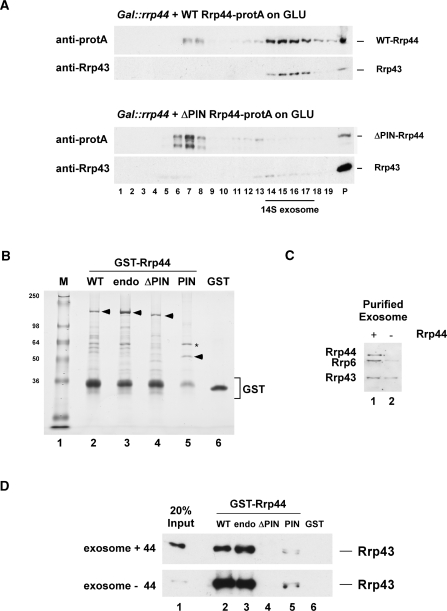
Figure 7.
The N-terminal PIN domain in Rrp44 mediates protein–protein interactions. (A) Sedimentation behavior of WT and Rrp44-ΔPIN on glycerol gradients. Gal::rrp44 strains transformed with plasmids encoding protA-tagged Rrp44 [full-length (WT) or ΔPIN] were grown on glucose for 8.5 h to deplete endogenous Rrp44. Protein extracts from 27.5 ODs of cells were then separated on linear 10–30% glycerol gradients and analyzed by western blotting. (B) Recombinant, GST-tagged WT Rrp44 (WT), Rrp44-endo (endo), Rrp44-ΔPIN (ΔPIN), the PIN domain alone (PIN) and free GST were expressed in E. coli and purified on a glutathione sepharose column. The proteins (indicated by arrowheads) were separated on an 8% polyacrylamide/SDS gel and stained with Coomassie blue. Asterisk: E. coli GroEL. (C) Western blot analysis of TAP-purified exosomes from Csl4-TAP strains grown at 25°C. Exosomes were purified on IgG sepharose at 150 mM NaCl (lane 1) or treated with 800 mM MgCl2 to dissociate endogenous Rrp44 (lane 2). TEV eluates (2.5 μl) were separated on an 8% polyacrylamide/SDS gel and analyzed by immunoblotting with anti-Rrp44, anti-Rrp6 and anti-Rrp43 antibodies. (D) GST-pull down experiment. The GST-tagged Rrp44 constructs or free GST (B) were coupled to glutathione sepharose beads and incubated with purified yeast exosomes (C). Bound material was eluted under denaturing conditions, separated on an 8% polyacrylamide/SDS gel and analyzed by immunoblotting with an anti-Rrp43 antibody.
Glycerol gradient analyses
Yeast lysate from 27.5 ODs of cells was prepared in buffer TMN150 containing 1 mM dithioerythritol and complete EDTA-free protease inhibitors (Roche) and loaded on a linear 4-ml 10 to 30% (w/v) glycerol gradient. After centrifugation for 17 h at 45 000 rpm in a Beckman SW60 rotor, the gradient was harvested manually from the top. Proteins were isolated from the gradient fractions by acetone precipitation, separated on an 8% polyacrylamide/SDS gel and analyzed by immunoblotting.
RESULTS
Yeast strains lacking the exonuclease activity of Rrp44 are viable and retain partial exosome function
To assess the effects of the loss of the enzymatic activity of Rrp44 in vivo, we modified strain BY4741 to express HA-tagged Rrp44 under control of the repressible GAL10 promoter (Table S1), allowing depletion of endogenous Rrp44. This strain was transformed with plasmids expressing either WT-Rrp44 or Rrp44 lacking exonuclease activity (Rrp44-exo, Figure 1A) under the control of the endogenous RRP44 promoter, or with the empty cloning vector (pRS316). To allow purification of exosome complexes to test for in vitro exonuclease activity, the plasmid-expressed proteins also carried C-terminal fusions with a tag containing the streptavidin-binding peptide, TEV cleavage site and two copies of the z-domain of protein A (szz-tag; see Materials and methods section and Table S1).
The growth of individual transformants was then analyzed under repressive conditions on SD -His/-Ura medium at 30°C (Figure 1B). Growth of the strain carrying the empty vector was progressively reduced after transfer to glucose. In contrast, growth was fully restored by expression of WT Rrp44. A reduced rate of growth was maintained by expression of the catalytically inactive Rrp44-exo (Figure 1B). After prolonged growth in glucose medium (24 h), the doubling time of strains expressing only Rrp44-exo was ∼3 h when compared to the WT doubling time of ∼2 h (data not shown). This indicates that the essential function of Rrp44 in vivo does not require its exonuclease activity.
Genetic depletion of Rrp44 or any other core exosome component results in characteristic defects in 3′-maturation of the 5.8S rRNA from the 7S pre-rRNA and in degradation of the excised 5′-external transcribed spacer region (5′-ETS) of the pre-rRNA (20–23). Exosome mutants were also reported to accumulate a 3′-truncated and polyadenylated fragment of the 5S rRNA (24).
Northern analysis of the GAL::rrp44 strain also expressing WT Rrp44 (Figure 1D, lanes 1 and 2) revealed similar levels of 3′ extended pre-5.8S rRNA and the 5′-ETS in galactose (GAL, lane 1) and glucose (GLU, lane 2) media. Expression of only Rrp44-exo resulted in the accumulation of truncated 5S rRNA (labeled as 5S*) and the 7S pre-rRNA plus truncated fragments (Figure 1D, lane 4). Accumulation of longer 3′-extended 5.8S species was greater in the Rrp44-depleted strain, whereas the shortened 6S pre-rRNA was reduced, indicating a greater inhibition of processing following depletion of Rrp44 than in the rrp44-exo strain. Accumulation of the full length 5′-ETS was greater in the Rrp44-depleted strain (Figure 1D, lane 6) than in the rrp44-exo strain (Figure 1D, lane 4), which also showed prominent 5′-ETS fragments that were absent from the Rrp44-depleted strain. These observations are consistent with residual degradation activity associated with Rrp44-exo.
Rrp44-depleted strains also exhibit defects at earlier steps in pre-rRNA maturation on the pathway of 18S rRNA synthesis (25). These effects are likely to be indirect, since 18S rRNA synthesis involves only endonuclease activities, and similar defects are seen in many strains with late-acting defects in the 25S and 5.8S rRNA synthesis pathway (26). Northern analyses of high molecular weight pre-rRNA precursors (35S, 27SA2 pre-rRNA and the aberrant 23S RNA) showed only very modest defects in the strain expressing Rrp44-exo (Figure S1, and see below).
The exonuclease activity of Rrp44 appears to play an important role in maturation of the 7S pre-rRNA, and in degradation of the excised 5′-ETS pre-rRNA region and truncated 5S rRNA. In contrast, the exonuclease activity is less required for the, presumably indirect, role of Rrp44 in early pre-rRNA processing steps. However, strains lacking the exonuclease activity of Rrp44 are viable and showed less RNA processing defects than the Rrp44-depleted strain on the pre-rRNA substrates.
Loss of the exonuclease activity of Rrp44 has additive effects with loss of Rrp6
Constructs expressed under the GAL promoter are never fully repressed, and so a low level of Rrp44 will always be expressed in a GAL::rrp44 strain. To avoid this problem, the RRP44 ORF was precisely deleted in strain BY4741 supplemented with a plasmid carrying RRP44 and a URA3 selective marker. Following construction of rrp44Δ, a second plasmid was introduced that carried LEU2 and expressed either Rrp44 or Rrp44-exo. Mitotic segregants that had lost the URA3 plasmid containing wild type RRP44 were then isolated on plates containing 5-FOA, which selectively kills cells expressing Ura3. The rrp44Δ strains showed an increased doubling time when complemented by expression of Rrp44-exo (3 h when compared to 2 h for WT-Rrp44, Figure 2), similar to the results obtained with GAL::rrp44 strains (Figure 1B).
Figure 2.
Growth analysis of yeast strains expressing Rrp44-exo. Yeast strains carrying either rrp44Δ or rrp44Δ rrp6Δ were transformed with plasmids expressing WT Rrp44 (WT) or Rrp44 with the D551N mutation (exo). To analyze growth, strains were grown in selective liquid glucose medium at 30°C (rrp44Δ) or 25°C (rrp44Δ rrp6Δ), which are their respective optima.
‘Since Rrp44 is essential for viability, whereas its exonuclease activity is largely dispensable, it seemed likely that it was partially redundant with the activities of one or more other nucleases. An obvious possibility was the nuclear, exosome-associated exonuclease Rrp6. Indeed, the rrp44-exo mutation was reported to result in synthetic lethality with an rrp6Δ (4). However, this was not observed in our strains (Figures 2 and 3). To test for synthetic lethality between rrp44-exo and rrp6Δ, the RRP6 ORF was first deleted in the rrp44Δ strain complemented with a plasmid carrying RRP44 and a URA3 selective marker (see above). A second plasmid (pRS315) carrying LEU2 and expressing either Rrp44 or Rrp44-exo was subsequently introduced and the URA3 plasmid was then counter-selected on 5-FOA containing SD -His/-Leu plates. The growth of individual transformants was then analyzed on SD -His/-Leu medium at 25°C (Figure 2). The loss of Rrp44-exonuclease activity reduced the growth of RRP6 and rrp6Δ by a similar proportion, relative to the same strains expressing Rrp44 (Figure 2).
Figure 3.
The rrp44-exo mutation is not synthetically lethal with deletion of Rrp6. (A) and (B). In vivo analyses of yeast strains expressing Rrp44-exo in the presence or absence of the exosome components Rrp6 or Rrp41. RNA was separated on an 8% polyacrylamide/8 M urea gel (A) or an 1% agarose/glyoxal gel (B) and analyzed as described in Figure 1D.
Northern analysis was further used to compare the RNA processing phenotypes of rrp44Δ rrp6Δ double mutant strains complemented by plasmids expressing either WT Rrp44 or Rrp44-exo (Figure 3). Yeast strains carrying rrp6Δ exhibit a distinctive accumulation of the 3′-extended precursor 5.8S + 30 (27) (Figure 3A, lane 3), which is found as a short (S) and a 5′-extended long (L) form. Expression of only Rrp44-exo in the rrp6Δ strain leads to accumulation of a longer form of this precursor (Figure 3A, lane 4). To confirm the identity of the extended 5.8S species, primer extension was performed using a probe located across the 5.8S-ITS2 boundary (data not shown). This revealed identical 5′ ends, showing that the species detected in the rrp6Δ rrp44-exo double mutant is 3′ extended by ∼10 nt relative to the 5.8S + 30 RNA seen in rrp6Δ single mutant strains.
Loss of Rrp6 has little effect on degradation of the excised 5′-ETS (Figure 3A, lane 3) and the phenotype of the rrp6Δ rrp44-exo strain (Figure 3A, lane 4) resembled that of rrp44-exo alone (Figure 3A, lane 2). Both 3′ extended and truncated forms of the box C + D snoRNA snR13 were previously observed in strains with defects in nuclear RNA surveillance (28,29). These forms of snR13 were also detected in both the rrp6Δ and rrp44-exo single mutants (Figure 3A, lanes 3 and 2), with stronger accumulation in the double mutant strain (Figure 3A, lane 4).
High molecular weight RNA was also analyzed (Figure 3B). The rrp6Δ mutation alone resulted in increased levels of the 27SA2 pre-rRNA, the first committed intermediate on the pathway of 5.8S and 25S rRNA synthesis. The aberrant 23S, 21S and 17S RNAs were also accumulated (Figure 3B, lane 3); these are known targets for degradation by Rrp6 and the TRAMP5 polyadenylation complex (24,25,30). In contrast, rrp44-exo alone reduced the level of 27SA2 but had little effect on 23S (Figure 3B, lane 2). The double mutant showed an intermediate phenotype.
Since the exonuclease activity of Rrp44 is dispensable for growth, even in the absence of Rrp6, we considered the possibility that the essential role of the other, apparently non-catalytic components of the core exosome might lie in restraining and controlling an otherwise over-promiscuous exonuclease activity of Rrp44. To test this model, the core exosome component Rrp41 was placed under the control of a GAL promoter in the rrp44Δ strain expressing either intact Rrp44 or Rrp44-exo. However, following transfer to glucose medium, the GAL::rrp41 strains showed similar growth inhibition upon expression of Rrp44 or Rrp44-exo (data not shown). Northern analyses (Figure 3A and B, lanes 5–8) also indicated that the loss of Rrp44 exonuclease activity did not suppress or give synergistic defects when combined with depletion of Rrp41. We conclude that the phenotype of Rrp41 depletion is neither dependent on, nor attenuated by, the exonuclease activity of Rrp44.
Rrp44 exonuclease activity is essential in the absence of the cytoplasmic 5′–3′ exonuclease Xrn1
Since the exonuclease activity of Rrp44 was apparently not strongly redundant with Rrp6, we tested for synthetic lethal interactions with other exonucleases.
The non-essential, 5′–3′-exonuclease Xrn1 (31) plays a major role in cytoplasmic mRNA turnover and is synthetically lethal with the cytoplasmic cofactors for the exosome, due to synergistic inhibition of mRNA degradation (32,33). Deletion of XRN1 was synthetically lethal with rrp44-exo (Figure 4A). We conclude that loss of the 3′-exonuclease activity of Rrp44 is synthetic-lethal with loss of the cytoplasmic 5′–3′ exonuclease Xrn1, probably due to synergistic inhibition of mRNA degradation. In contrast, loss of the PIN-domain associated endonuclease activity of Rrp44 (see below) was not synthetic-lethal with xrn1Δ (Figure 4B).
Figure 4.
The Rrp44-exo mutation, but not the Rrp44-endo mutation, is synthetically lethal with loss of Xrn1. (A) In vivo analysis of Gal::rrp44 strains expressing WT-Rrp44 or Rrp44-exo in the absence of the cytoplasmic 5′–3′ exonuclease Xrn1. Strains were streaked for single colonies on selective plates containing either 2% galactose or 2% glucose and incubated at 30°C. (B) In vivo analysis of rrp44Δ strains expressing WT or mutant Rrp44 in the absence of Xrn1. Strains were streaked for single colonies on selective plates with or without 5-FOA, which selects for loss of the plasmid carrying URA3 and WT-RRP44, and incubated at 25°C.
In addition to Rrp44, yeast contains two other proteins with homology to the RNase II family. Of these, Dss1 is mitochondrial and seemed unlikely to function redundantly with Rrp44. In contrast, Ssd1 is localized to the cytoplasm (34,35) and shows genetic interactions consistent with functions in RNA turnover and/or surveillance (36,37). Laboratory strains of yeast are polymorphic for Ssd1 synthesis (38); strains derived from S288c that were used for the systematic sequencing project and construction of the gene deletion collection, including BY4741 that we used for our initial functional analyses of Rrp44, harbor the full-length protein (Ssd1-v), whereas the widely used W303 strain expresses a truncated version of the protein (Ssd1-d). In comparison to GAL::rrp44 strains derived from BY4741, expression of Rrp44-exo in the W303 background consistently supported slightly less efficient growth when intact Rrp44 was depleted by incubation on glucose medium (data not shown). To determine whether this was due to the lack of intact Ssd1, the full-length form of Ssd1 was expressed from a plasmid. However, this failed to clearly improve growth of W303 strains expressing only Rrp44-exo.
Strain differences between BY4741 and W303 have a modest but reproducible impact on sensitivity to loss of Rrp44 exonuclease activity. Similar genetic background effects may be responsible for the fact that we found strains lacking the exonuclease activity of both Rrp44 and Rrp6 to be viable in BY4741, whereas they were previously reported to be synthetic lethal in strain BMA 64, which is related to W303 (4).
The PIN domain of Rrp44 shows endonuclease activity
Comparison of the RNA processing phenotype of Rrp44-depletion to that shown by strains expressing Rrp44-exo (Figure 1D) suggested that some residual nuclease activity remained. Recent analyses of the PIN domain proteins, human Smg6 and yeast Swt1, had revealed that robust in vitro endonuclease activity required the presence of manganese ions (8,9). Four conserved acidic residues (Aspartate/D or Glutamate/E) coordinate the metal ion in the active site. All four residues are present in the N-terminal PIN domain of Rrp44 (Figure 1A and see below) and we therefore assessed whether Rrp44 also exhibited endonuclease activity in the presence of Mn2+ (Figure 5).
Figure 5.
Recombinant Rrp44 exhibits two distinct ribonuclease activities in vitro. (A) The ribonuclease activity of recombinant GST-Rrp44 is altered by the presence of manganese ions. Two hundred fmol recombinant protein (WT or Rrp44-exo) and 10 fmol 5′-end-labeled A30 RNA substrate were incubated for 60 min at 30°C in the presence of 0.5 mM MgCl2 and in the absence or presence of 5 mM MnCl2. Reaction products were separated on a 12% polyacrylamide/8 M urea gel and visualized by autoradiography. Control: mock-digested substrate lacking recombinant protein. (B) Ribonuclease activity of recombinant GST-Rrp44-exo on 5′-end-labeled (lanes 1–4) or 3′-end-labeled (lanes 5–8) A30 RNA substrate. Assay conditions were as described in (A). (C) Endonucleolytic activity of recombinant GST-Rrp44 on a 5′-end-labeled RNA substrate with a stable terminal stem structure. Two hundred femtomoles recombinant protein (WT or mutants, see below) and 150 fmol RNA were incubated for 2 h at 30°C in the presence of 0.5 mM MgCl2 and 0.5 mM MnCl2. Reaction products were analyzed as described above. The regions corresponding to the stem structure are indicated with black lines on the gel (C) and cartoon (D). The region corresponding to the loop is indicated in gray. The asterisks mark two degradation products of unclear origin that occur when the substrate is incubated with any form of recombinant GST-Rrp44 or other GST-tagged proteins (data not shown). Rrp44-exo: Rrp44 with D551N mutation. Rrp44-endo: Rrp44 with four individual amino-acid exchanges (D91N, E120Q, D171N and D198N) in the metal-coordinating centre of the N-terminal PIN domain. Rrp44-endo–exo: Rrp44 with 4 PIN mutations plus D551N. Rrp44-exo-S1: Rrp44 with D551N plus a G916E mutation in the S1 RNA-binding domain.
In vitro nuclease assays were first performed in the absence or presence of 5 mM Mn2+ using recombinant, GST-tagged proteins and a 5′-end-labeled A30 RNA substrate (Figure 5A). Protein preparations used are shown in Figure S2. Consistent with previous results (6), WT Rrp44 (lane 2) exhibited strong 3′-exonucleolytic activity in the absence of Mn2+, completely degrading the substrate to a 4 nt product. In contrast, the Rrp44-exo mutant showed little degradation activity (lane 3). However, in the presence of Mn2+, both WT and mutant proteins cleaved the RNA, generating very similar degradation products (lanes 5 and 6). From these results, we concluded that Rrp44 harbors two distinct ribonuclease activities, which respond differently to Mn2+ ions under in vitro conditions. The presence of Mn2+ inhibited 3′-exonuclease activity, whereas a second activity was stimulated.
To further characterize the novel activity, we performed in vitro nuclease assays using the exonucleolytically inactive Rrp44-exo mutant and 5′- and 3′-labeled A30 RNA substrates (Figure 5B). Rrp44-exo generated a ladder of degradation intermediates from both substrates in the absence of added Mn2+ ions (lanes 2 and 6), likely due to co-purification of a low level of Mn2+ bound to the recombinant protein, but the degradation activity was strongly stimulated in the presence of added Mn2+ (lanes 4 and 8). The ability of the Rrp44-exo mutant to generate degradation intermediates from both 3′-labeled and 5′-labeled substrates shows that it either carries both 5′-exonuclease and 3′-exonuclease activities or, more likely, possesses an endonuclease activity.
To assess if this potential endonuclease activity was associated with the PIN domain of Rrp44, point mutations were introduced at each of the four conserved active-site amino acids (D91N, E120Q, D171N, D198N), to create the Rrp44-endo mutant. In addition, the Rrp44-exo mutation was combined with the four PIN-domain mutations (Rrp44-endo–exo) and with a point mutation in the S1 RNA-binding domain (Rrp44-exo-S1) (6).
In vitro nuclease assays were performed using recombinant, GST-tagged proteins and a 5′-labeled substrate RNA derived from the 3′ region of the mouse 5.8S rRNA, which has a well defined, stable terminal stem structure (Figure 5C and D). WT Rrp44 (Figure 5C, lane 2) partially degraded the substrate to a 10 nt product. In contrast, Rrp44-exo generated a set of fragments with end-points in the loop-region of the substrate RNA (Figure 5C, lane 3). These fragments were not generated by either the Rrp44-endo (Figure 5C, lane 4) or Rrp44-endo–exo proteins (lane 5), indicating that their formation required a functional PIN domain. Rrp44-exo-S1 did generate the loop fragments, but lacked the major 10 nt product, suggesting that S1 RNA-binding activity might be more important for the exonuclease activity of Rrp44 than for endonuclease activity. A prominent product that is truncated by ∼10 nt was detected in some samples. This appears to be due to a contaminating nuclease that is associated with purification of GST-fusion constructs from E. coli, as the same band was seen in analyses of other fusion proteins (data not shown). The truncated fragment appears to be very susceptible to degradation by the exonuclease activity of Rrp44, giving rise to the observed variations in signal. The PIN domain alone did not show in vitro activity. However, when purified from E. coli the PIN domain co-purified, apparently stoichiometrically, with the bacterial chaperone GroEL (indicated with an asterisk in Figure 7B), strongly indicating that it is largely misfolded. We conclude that recombinant Rrp44 shows an endonuclease activity that requires Mn2+ ions and the metal-binding amino acids of the PIN domain.
The effects of the rrp44-endo mutation were also tested on viability. In the GAL::rrp44 strain, expression of Rrp44-endo clearly supported growth when WT Rrp44 was depleted by growth on glucose medium. In contrast, expression of Rrp44-endo–exo failed to support growth (data not shown), indicating that the endonuclease and exonuclease activities of Rrp44 share some redundant essential function. In plasmid shuffle experiments, expression of Rrp44-endo, but not Rrp44-endo–exo, supported viability in an rrp44Δ rrp6Δ double mutant strain (data not shown, but see Figure 6B, lane 6). A low number of rrp44Δ strains complemented by the plasmid expressing Rrp44-endo–exo could be isolated by plasmid shuffle (data not shown). Rare cells are therefore able to adapt to loss of both the endonuclease and exonuclease activities of Rrp44 by epigenetic or physiological mechanisms.
Northern analyses were performed on the GAL::rrp44 strain expressing Rrp44-endo and Rrp44-endo–exo during Rrp44 depletion (Figure 6A). These showed that the 5′-extended forms of the 5.8S rRNA and truncated forms of the 5′-ETS observed in strains expressing Rrp44-exo were suppressed in the rrp44-endo strain and altered in the rrp44-endo–exo strain. In particular, 5′-ETS fragments accumulating in the rrp44-exo strain (Figure 6A, lane 4) were clearly reduced in strains expressing Rrp44-endo–exo (Figure 6A, lane 8). This suggested that the endonuclease activity of Rrp44 is involved in formation of intermediate species that are stabilized by loss of Rrp44 3′-exonuclease activity. The longer form of the 5.8S + 30 RNA seen in the rrp6Δ strain expressing Rrp44-exo (Figure 6B, lane 5) was not observed in the rrp6Δ strain expressing Rrp44-endo (Figure 6B, lane 6). Accumulation of the truncated form of the 5S rRNA, seen in rrp44-exo strains, was not suppressed in the rrp44-endo–exo strain. However, the truncated 5S was accumulated in rrp6Δ strains, even in the presence of the rrp44-endo mutation.
Expression of only Rrp44 lacking both the endonuclease and exonuclease activity resulted in a phenotype that closely resembled that seen on depletion of Rrp44 (vector control in Figure 6A, lane 10). Together the data indicate that the PIN domain of Rrp44 exhibits endonuclease activity in vitro and in vivo.
The PIN domain links Rrp44 to the core exosome
The archaeal exosome lacks a homologue of Rrp44, and yeast Rrp44 can be removed from the remaining nine components of the exosome by washing with high salt. This indicates that Rrp44 associates with a highly stable, nine component exosome core structure. A two-hybrid interaction was detected between the PIN domain of human Rrp44 and the RNase PH-homologue OIP2 (the human homologue of yeast Rrp43) (39) suggesting that the PIN domain might be involved in protein–protein interactions that tether Rrp44 to the core structure. This model would be consistent with the absence of PIN domains from E. coli RNase R and II (Figure 1A), which function as monomers.
The association of Rrp44 with the exosome core was initially tested by glycerol gradient centrifugation of yeast cell lysates (Figure 7A). WT, protein A-tagged Rrp44 largely co-sedimented with the core exosome component Rrp43 in ∼14S complexes (fractions 14–17), with a small peak corresponding to the position of free Rrp44 (fractions 7–8). In contrast, protein A-tagged Rrp44 lacking the PIN-domain (Δ aa 86–203), was expressed at levels substantially below that of WT Rrp44, but predominantly sedimented as free protein. In Rrp44-depleted cells, Rrp43 becomes enriched in ∼60S gradient fractions, reflecting stabilized exosome association with pre-ribosomes (L. Milligan and D.T., unpublished results), giving rise to the strong signal in the pellet fractions on the gradient shown.
Binding of Rrp44 to the core exosome was also assessed by in vitro binding assays using recombinant GST-tagged Rrp44 constructs expressed in E. coli (Figure 7B) and core exosome purified from yeast, with or without removal of Rrp44 by washing with 800 mM MgCl2 (Figure 7C). The GST-tagged constructs were bound to glutathione sepharose and incubated with the exosome preparations. The association of the exosome with the GST-fusion proteins was assessed by western blotting using antibodies specific for Rrp43 (Figure 7D) and Rrp4 (data not shown). Exosome binding to the full-length Rrp44 (FL) (Figure 7D, lane 2) was substantially stronger than to Rrp44 lacking the N-terminal PIN domain (ΔPIN) (Figure 7D, lane 4), which was not above the GST background (Figure 7D, lane 6). Notably, exosome binding was not affected by four individual mutations in the PIN domain (Rrp44-endo, see Figure 5) (Figure 7D, lane 3). The PIN domain alone (PIN) (Figure 7D, lane 5) gave a lower signal, but this clearly was above the GST background. As noted above, the low signal may reflect poor folding of the isolated PIN domain, as suggested by its copurification with bacterial GroEL. The signal from the Rrp44-depleted exosome (exosome –44) was stronger than for exosome purified at lower salt concentrations (exosome +44). However, some binding to Rrp44 was seen even without the high-salt wash, suggesting that Rrp44 is substoichiometric in the purified exosome.
We conclude that the PIN domain of Rrp44 plays a dual role in that it harbors endonuclease activity and also functions in tethering Rrp44 to the remaining nine subunit core of the exosome.
DISCUSSION
Genetic depletion of RRP44 is lethal (21), whereas strains expressing the Rrp44-exo mutant are viable with only a modest growth defect, at least in some strain backgrounds, showing that its exonuclease activity is dispensable for growth. It is, of course, difficult to be certain that any mutation fully inhibits enzymatic activity in vivo. However, this seems probable, as the D551N mutation (rrp44-exo) abolished detectable in vitro activity on more than five different substrates (4–6) while the equivalent mutation (D209N) abolished the activity of E. coli RNase II (40).
The exonuclease activity of Rrp44 does play an important role in pre-rRNA processing, participating in both the maturation of the 7S pre-rRNA to 5.8S rRNA and the degradation of the excised 5′-ETS region. However, on both substrates the phenotype of strains expressing only Rrp44-exo was substantially weaker than that seen in strains lacking Rrp44. The only substrate tested that showed apparently identical accumulation in Rrp44-depleted and rrp44-exo strains was a 3′-truncated form of the 5S rRNA, which was previously identified as a TRAMP/exosome substrate (24).
Strain heterogeneity was observed in sensitivity to the rrp44-exo mutation, which conferred a stronger growth phenotype in strain W303 than in a BY4741 strain background, even though both are widely used ‘wild-type’ strains. We tested W303 because it expresses only a truncated form of the RNase II-related protein Ssd1, and we speculated that this might show genetic interactions with loss of the exonuclease activity of Rrp44. In the event, expression of full-length Ssd1 failed to suppress the sensitivity of W303 to the loss of Rrp44 exonuclease activity.
The essential function of Rrp44 might be the maintenance of the structure of the exosome complex or binding to either substrates or cofactors. Recombinant GST-Rrp44-exo protein was able to bind to RNA (6), and the yeast exosome containing Rrp44-exo might thus still be able to recruit RNAs that are normally substrates for Rrp44. Substrates recognized and bound by Rrp44-exo might then be degraded by other nucleases. Loss of Rrp44 exonucleolytic activity is synthetic lethal (sl) with deletion of the nonessential cytoplasmic 5′–3′ exonuclease Xrn1, due to the synergistic inhibition of cytoplasmic mRNA degradation (33). This indicates that the rrp44-exo mutation blocked the 3′ degradation of at least some mRNA species in vivo. In contrast, BY4741 strains expressing only the Rrp44-exo mutant were viable in the absence of the other exosome-associated 3′-exonuclease Rrp6. This finding differs from previous observations made in a W303-related strain (BMA 36) (4), presumably reflecting the differences in strain background. Recent analyses (41) indicate that functional interactions between Rrp6 and Rrp44 are important for only a subset of Rrp6 substrates.
The exonuclease activities of the exosome in the nucleus (Rrp44 and Rrp6) likely form part of a redundant network of 3′-exonucleases, including the proteins of the Rex family (42), which could, for instance, be needed to fine-tune the balance between pre-ribosomal RNA processing and decay. In the cytoplasm, such a fine balance might not be necessary, since RNA decay is the predominant pathway.
The differences in RNA processing phenotypes between strains depleted of Rrp44 and those expressing Rrp44-exo, suggested that Rrp44 might harbor additional activities. Recent reports of manganese-dependent endonuclease activities associated with other PIN-domain proteins prompted us to determine whether this might also be the case for Rrp44. The endonuclease activity of Rrp44 failed to be observed in previous analyses, probably because in the absence of manganese the exonuclease activity is much more prominent. The concentrations of 0.5 or 5 mM Mn2+ that were used to inhibit exonuclease activity and stimulate endonuclease activity of Rrp44 are much greater than the physiological concentration (43). It seems likely that the in vivo endonuclease activity of Rrp44 is stimulated by one or more of the many exosome cofactors, allowing it to act at lower Mn2+ concentrations. However, we cannot fully exclude the possibility that the PIN domain primarily functions in RNA substrate binding in vivo, with a non-physiological cleavage activity that is induced by high Mn2+ levels in vitro. The range of in vivo substrates for the activity of the Rrp44 PIN domain remains unclear but comparison of the RNA processing phenotypes of strain expressing Rrp44-exo and Rrp44-endo–exo strongly indicates that these are likely to include the 5′-ETS region of the pre-rRNA. However, the putative target sites do not appear to correspond to the known A0 cleavage at position +470. This suggests that the activity of the Rrp44 PIN domain does not act during pre-rRNA maturation, but rather participates in the degradation of the excised spacer region following A0 cleavage. Several early acting ribosome synthesis factors are released from the pre-ribosomes in association with the excised 5′-ETS (44). Very efficient degradation of the 5′-ETS is therefore likely to be important for the release and recycling of ribosome synthesis factors.
Analyses in vivo and in vitro showed that the PIN domain of Rrp44 was both necessary and sufficient for association with the 9 subunit exosome core structure. Previous EM analyses indicated that both the Rrp44 N-terminus (where the PIN domain is located) and the C-terminal region form interactions with the exosome core (45). The data presented here indicate that the N-terminal interactions are more crucial for stable binding. Alterations in the relative positions of the N- and C-terminal regions of Rrp44 seen on association with the core exosome are proposed to be crucial for regulation of its exonuclease activity (45), and interactions between the C-terminal region of Rrp44 and the exosome may play an important role in this regulation.
Within Rrp44, the PIN domain appears to have a dual role, acting to link Rrp44 to the remainder of the exosome in addition to its apparent cleavage activity. This evolution of the region therefore appears to play a crucial part in the functional differences between bacterial RNase R and eukaryotic Rrp44. The structure of the exosome core is related to that of bacterial PNPase (5,46,47). In E. coli, PNPase can associate with RNase E and other proteins in the degradosome complex, that shows both 3′-exonuclease and endonuclease activity (48). The finding that the eukaryotic exosome also possesses both 3′-exonuclease and endonuclease activity underlines the conservation of the RNA processing machinery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Wellcome Trust (to D.T.) and BBSRC (to J.D.B.), a long-term HFSP fellowship (to C.S.) and a PhD studentship from the BBSRC (to E.L.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank J. Houseley for yeast strains and reagents and critical reading of the manuscript.
REFERENCES
- 1.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′–5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 4.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkard KT, Butler JS. A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skruzny M, Schneider C, Racz A, Weng J, Tollervey D, Hurt E. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 2008 doi: 10.1371/journal.pbio.1000008. doi:10..1371/journal.pbio.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica A, Oeffinger M, Dlakic M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell Biol. 2003;23:1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica A, Tollervey D, Dlakic M. The PIN domain of Nob1p is required for 20S pre-rRNA cleavage at site D. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longtine MS, McKenzie A., 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 16.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi E, Hayashi N, Azuma Y, Seki T, Nakamura M, Nakashima N, Yanagida M, He X, Mueller U, Sazer S, et al. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 1996;15:5595–5605. [PMC free article] [PubMed] [Google Scholar]
- 18.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 21.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 27.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen TP, Culbertson MR. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of Yeast nonpolyadenylated snoRNA transcripts. Mol. Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dez C, Dlakic M, Tollervey D. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007;13:1516–1527. doi: 10.1261/rna.609807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larimer FW, Hsu CL, Maupin MK, Stevens A. Characterization of the XRN1 gene encoding a 5′–3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 32.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson J.SJ, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uesono Y, Toh-e A, Kikuchi Y. Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- 35.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 36.Luukkonen BG, Séraphin B. A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicing mutant suppressor. Nucleic Acids Res. 1999;27:3455–3465. doi: 10.1093/nar/27.17.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- 38.Uesono Y, Fujita A, Toh-e A, Kikuchi Y. The MCS1/SSD1/SRK1/SSL1 gene is involved in stable maintenance of the chromosome in yeast. Gene. 1994;143:135–138. doi: 10.1016/0378-1119(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 39.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amblar M, Arraiano CM. A single mutation in Escherichia coli ribonuclease II inactivates the enzyme without affecting RNA binding. FEBS J. 2005;272:363–374. doi: 10.1111/j.1742-4658.2004.04477.x. [DOI] [PubMed] [Google Scholar]
- 41.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 2005;6:R77. doi: 10.1186/gb-2005-6-9-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 45.Wang H.-W, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44-exosome complex suggests routes of RNA recruitment for 3′- end processing. Proc. Natl Acad. Sci. USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- 47.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure Fold Des. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 48.Carpousis AJ, Vanzo NF, Raynal LC. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.