Abstract
We report herein the synthesis and physical and physiological characterization of fully modified 2′-modified-4′-thioRNAs, i.e. 2′-fluoro-4′-thioRNA (F-SRNA) and 2′-O-Me-4′-thioRNA (Me-SRNA), which can be considered as a hybrid chemical modification based on 2′-modified oligonucleotides (ONs) and 4′-thioRNA (SRNA). In its hybridization with a complementary RNA, F-SRNA (15mer) showed the highest Tm value (+16°C relative to the natural RNA duplex). In addition, both F-SRNA and Me-SRNA preferred RNA as a complementary partner rather than DNA in duplex formation. The results of a comprehensive comparison of nuclease stability of single-stranded F-SRNA and Me-SRNA along with 2′-fluoroRNA (FRNA), 2′-O-MeRNA (MeRNA), SRNA, and natural RNA and DNA, revealed that Me-SRNA had the highest stability with t1/2 values of > 24 h against S1 nuclease (an endonuclease) and 79.2 min against SVPD (a 3′-exonuclease). Moreover, the stability of Me-SRNA was significantly improved in 50% human plasma (t1/2 = 1631 min) compared with FRNA (t1/2 = 53.2 min) and MeRNA (t1/2 = 187 min), whose modifications are currently used as components of therapeutic aptamers. The results presented in this article will, it is hoped, contribute to the development of 2′-modified-4′-thioRNAs, especially Me-SRNA, as a new RNA molecule for therapeutic applications.
INTRODUCTION
A large number of chemically modified oligonucleotides (ONs) have been synthesized for use in nucleic-acid-based therapeutics (1). The discovery of RNA interference (RNAi) has reinvigorated interest in this trend (2). Chemically modified ONs are required especially to have resistance toward nuclease degradation and thermal stability of duplex formation for their in vivo applications. Among the modified ONs developed so far, a series of 2′-modified ONs appears to be the most promising due to their higher nuclease resistance and favorable hybridization with complementary RNA (3). For example, 2′-fluoroRNA (FRNA) and 2′-O-MeRNA (MeRNA) are the most typical modifications (4–6). When 2′-hydroxy groups in RNA are substituted, these modified RNAs are highly resistant to nuclease degradation, especially toward ribonuclease (RNase) digestion. In addition, such modifications force the ONs to adopt a higher proportion of the northern conformation in a helical structure, which is one of the contributing factors of stronger hybridization property of these modified ONs. Because of such favorable properties, these ONs are currently used as components of therapeutic aptamers, such as Macugen® (7).
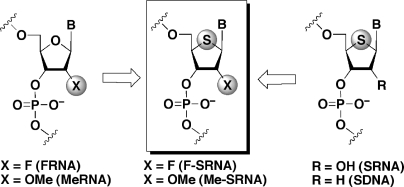
Whereas chemical modification is carried out at the 2′-position of ONs, the 4′-position, namely the furanose ring oxygen, has currently been recognized as an alternative modification site. Our group has been intensely studying the synthesis and application of a series of 4′-thionucleic acids (4′-thioRNA and 4′-thioDNA), which consist of 4′-thioribonucleosides (8) and 2′-deoxy-4′-thioribonucleosides (9). Since 4′-thioRNA (SRNA) and 4′-thioDNA (SDNA) exhibited nuclease resistance, high hybridization and structural similarity to the A-form RNA duplexes (10–12), applications of these ONs for isolating the SRNA aptamers by SELEX (13,14) are now under investigation as well as the development of tools for gene silencing via RNAi machinery (15–17). As part of our research project, we envisioned synthesizing 2′-modified-4′-thioRNAs, i.e. 2′-fluoro-4′-thioRNA (F-SRNA) and 2′-O-Me-4′-thioRNA (Me-SRNA) (Figure 1), which can be considered as a hybrid chemical modification based on the 2′-modified ONs and SRNA. These new ONs are expected to show synergic effects on their potential such as hybridization and nuclease resistance.
Figure 1.
Design of 2′-modified-4′-thioRNAs.
Herein we describe the synthesis of 2′-modified-4′-thioRNAs and their characterization including their hybridization and nuclease resistance. Thus far, numerous chemically modified ONs have been developed and their properties were investigated. However, these studies were generally carried out individually, furnishing no comprehensive comparison with other known modified ONs developed so far. Accordingly, we compared the properties of the newly prepared 2′-modified-4′-thioRNAs with other ONs including FRNA, MeRNA and SRNA, the details of which are discussed below.
MATERIALS AND METHODS
General methods
Physical data were measured as follows: 1H, 13C and 31P NMR spectra were recorded at 270 and 400, and 500 MHz instruments, respectively, in CDCl3, DMSO-d6, or D2O as the solvent with tetramethylsilane or H3PO4 (for 31P NMR) as an internal standard. Chemical shifts are reported in parts per million (δ), and signals are expressed as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet) or br (broad). Mass spectra were measured on JEOL JMS-HX110 spectrometer. TLC was done on Merck Kieselgel F254 precoated plates. Silica gel used for column chromatography was Merck silica gel 5715. S1 nuclease and SVPD were purchased from TaKaRa and MB Biomedicals, respectively. [γ-32P]ATP was purchased from Perkin Elmer.
Synthesis of phosphoroamidite units
Synthetic protocols shown in Schemes 1 and 2 were highlighted in this article. Protocols for synthesis of the other units along with CPG support for F-SRNA and Me-SRNA synthesis were given in the Supplementary Data (Schemes S1, S2 and S3).
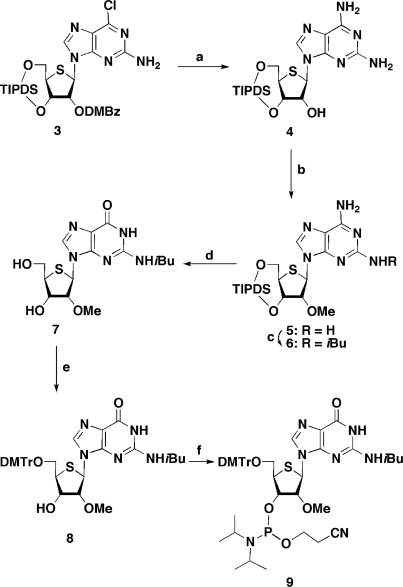
Scheme 1.
Scheme 2.
(a) NH3/EtOH, 80°C then MeNH2, MeOH; (b) CH3I, NaH, DMF, –40°C; (c) isobutyryl chloride, pyridine, –10°C; (d) NaNO2, aqueous AcOH then Et3N•3HF, Et3N, CH2Cl2; (e) DMTrCl, pyridine; (f) 2-cyanoethyl N,N-diisopropylchlorophosphoramidite chloride, iPr2NEt, DMAP, CH2Cl2.
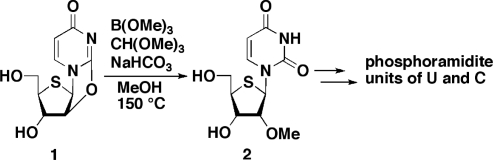
1-(2-O-Methyl-4-thio-β-d-ribofuranosyl)uracil (2)
To a solution of 1 (9) (3.0 g, 12 mmol) in MeOH (120 ml) were added trimethyl borate (64% in MeOH, 4.0 ml, 24 mmol), trimethyl orthoformate (1.4 ml, 12 mmol) and sodium bicarbonate (83 mg). The reaction mixture was heated for 48 h at 150°C in a steel container. The reaction mixture was cooled to room temperature and concentrated in vacuo. The residue was purified by a silica gel column, eluted with MeOH in CHCl3 (0–5%), to give 2 (2.9 g, 86%) as a white foam. An analytical sample was crystallized from EtOH: mp, 177°C; FAB-LRMS m/z 275 (MH+); 1H NMR (D2O) δ: 8.12 (d, 1 H, J = 8.1 Hz), 5.89 (d, 1 H, J = 4.8 Hz), 5.77 (d, 1 H, J = 8.1 Hz), 4.17 (dd, 1 H, J = 4.3 and 5.0 Hz), 3.92 (dd, 1 H, J = 4.3 and 5.0 Hz), 3.78 (dd, 1 H, J = 4.8 and 12 Hz), 3.71 (dd, 1 H, J = 5.4 and 12 Hz), 3.38 (s, 3 H), 3.36–3.33 (m, 1 H); 13C NMR (DMSO-d6) δ: 162.8, 150.8, 141.2, 102.1, 85.3, 70.0, 62.6, 60.6, 57.6, 54.3. Anal. Calcd for C10H14N2O5S: C, 43.79; H, 5.14; N, 10.21. Found: C, 43.78; H, 5.03; N, 10.24.
2,6-Diamino-9-[3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-4-thio-β-d-furanosyl]-9H-purine (4)
Compound 3 (1.4 g, 2.0 mmol) (8) was dissolved in ethanolic ammonia (saturated at 0°C, 80 ml), and the mixture was heated at 100°C for 7.5 h in a steel container. The solvent was removed in vacuo. The residue was dissolved in MeNH2 in MeOH (40%, 60 ml), and the mixture was kept at room temperature for 1.5 h. The solvent was removed, and the residue was coevaporated with MeOH. The residue was purified by a silica gel column, eluted with MeOH in CHCl3 (0–3%), to give 4 (812 mg, 75%) as a white solid: FAB-LRMS m/z 541 (MH+); FAB-HRMS calcd for C22H41N6O4SSi2 (MH+) 541.2448, found 541.2446; 1H NMR (DMSO-d6) δ: 7.89 (s, 1 H), 6.78 (br s, 2 H), 5.90 (d, 1 H, J = 4.5 Hz), 5.86 (br s, 2 H), 5.47 (s, 1 H), 4.35 (dd, 1 H, J = 3.0 and 9.4 Hz), 4.09 (m, 1 H), 4.03–4.00 (m, 2 H), 3.62–3.60 (m, 1 H), 1.06–0.84 (m, 28 H); 13C NMR (DMSO-d6) δ: 160.3, 156.1, 151.4, 134.7, 113.3, 77.5, 72.3, 60.6, 58.2, 48.5, 17.4, 17.2, 17.2, 16.8, 16.7, 12.6, 12.6, 12.5, 11.8.
2,6-Diamino-9-[2-O-methyl-3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-4-thio-β-d-ribofuranosyl]purine (5)
To a mixture of 4 (1.6 g, 3.0 mmol) and methyl iodide (1.3 ml, 21 mmol) in dry DMF (30 ml) was added NaH (60% in oil, 600 mg, 15 mmol) at –40°C. After being stirred for 2.5 h at the same temperature, the reaction was quenched by addition of saturated aqueous NH4Cl. The solvent was removed in vacuo. The residue was diluted with CHCl3, which was washed with H2O and saturated aqueous NH4Cl, followed by brine. The organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by a silica gel column, eluted with MeOH in CHCl3 (0–2%), to give 5 (1.4 g, 85%) as a white foam: FAB-LRMS m/z 555 (MH+); FAB-HRMS calcd for C23H43N6O4SSi2 (MH+) 555.2596, found 555.2595; 1H NMR (DMSO-d6) δ: 8.13 (s, 1 H), 5.71 (s, 1 H), 5.50 and 4.79 (each br s, each 2 H), 4.42 (dd, 1 H, J = 3.4 and 9.7 Hz), 4.15 (dd, 1 H, J = 2.8 and 12.6 Hz), 3.83 (d, 1 H, J = 3.4 Hz), 3.71 (s, 3 H), 3.68 (dd, 1 H, J = 2.8 and 9.7 Hz), 1.15–0.84 (m, 28 H); 13C NMR (CDCl3) δ: 159.7, 155.8, 151.8, 137.1, 114.9, 87.2, 72.3, 59.4, 58.8, 57.9, 49.3, 17.4, 17.3, 17.1, 17.0, 16.9, 13.2, 13.1, 13.0, 12.3.
6-Amino-2-isobutyrylamino-9-[2-O-methyl-3,5-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-4-thio-β-d-ribofuranosyl]purine (6)
A solution of 5 (250 mg, 0.44 mmol) in dry pyridine (5 ml) was cooled to –10°C in an ice–ethanol bath, and isobutyryl chloride (55 µl, 0.53 mmol) was added dropwise to the stirred solution over a period of 10 min at the same temperature. The reaction mixture was stirred for 2 h at the same temperature followed by 2 h at room temperature. The reaction was quenched by addition of ethanol. The solvent was removed in vacuo, and the residue was diluted with CHCl3, which was washed with H2O and saturated aqueous NaHCO3, followed by brine. The organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by a silica gel column, eluted with MeOH in CHCl3 (0–2%), to give 6 (230 mg, 83%) as a white foam: FAB-LRMS m/z 625 (MH+); FAB-HRMS calcd for C27H49N6O5SSi2 (MH+) 625.3024, found 625.3032; 1H NMR (DMSO-d6) δ: 9.81 (br s, 1 H), 8.14 (s, 1 H), 7.26 (br s, 2 H), 5.72 (s, 1 H), 4.33 (dd, 1 H, J = 3.2 and 9.7 Hz), 4.03–4.02 (m, 3 H), 3.63 (s, 3 H), 4.60–3.58 (m, 1 H), 2.93 (m, 1 H), 1.10–0.78 (m, 34 H); 13C NMR (CDCl3) δ: 180.1, 156.9, 153.1, 149.9, 138.9, 117.1, 87.2, 72.5, 59.5, 59.4, 57.8, 49.1, 33.6, 19.3, 19.2, 17.4, 17.3, 17.0, 17.0, 16.8, 13.2, 13.2, 13.1, 13.0, 12.9, 12.3.
N2-Isobutyryl-9-(2-O-methyl-4-thio-β-d-ribofuranosyl)guanine (7)
To a solution of 6 (340 mg, 0.55 mmol) in a mixture of AcOH-H2O (2:1, 9 ml) was added NaNO2 (600 mg, 8.8 mmol), and the mixture was stirred at room temperature. After 30 min, additional NaNO2 (1.2 g, 18 mmol) was added, and the reaction mixture was stirred for additional 38 h at the same temperature. The reaction was neutralized with NaHCO3. The mixture was filtered through a Celite pad and washed with CHCl3. The filtrate was diluted with CHCl3, which was washed with H2O and saturated aqueous NaHCO3, followed by brine. The organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was dissolved in dry CH2Cl2 (6 ml), and Et3N•3HF (270 µl, 1.6 mmol), Et3N (460 µl, 3.3 mmol) were added to the solution. The mixture was stirred for 8 h at room temperature. The solvent was removed, and the residue was coevaporated with MeOH. The resulting precipitate was filtered, washed with MeOH and dried to give 7 (153 mg, 73% in two steps) as a white solid: FAB-LRMS m/z 384 (MH+); FAB-HRMS calcd for C15H22N5O5S (MH+) 384.1341, found 384.1349; 1H NMR (DMSO-d6) δ: 12.6 (br s, 1H), 11.6 (br s, 1H), 8.39 (s, 1H), 5.87 (d, 1H, J = 7.3 Hz), 7.38 (d, 1H, J = 4.5 Hz) 5.21 (t, 1H, J = 5.7 Hz), 4.40 (m, 1H), 4.30 (dd, 1H, J = 3.2 and 7.3 Hz), 3.74–3.73 (m, 1H), 3.60–3.57 (m, 1H), 3.28 (m, 4H), 2.76–2.73 (m, 1H), 1.10, 1.12 (each s, each 3H); 13C NMR (DMSO-d6) δ: 180.1, 154.7, 149.0, 148.2, 137.9, 119.8, 85.9, 69.8, 63.1, 58.4, 57.7, 53.6, 34.7, 18.8, 18.8.
N2-Isobutyryl-9-[-5-O-(4,4′-dimethoxytrityl)-2-O-methyl-4-thio-β-d-furanosyl]guanine (8)
To a solution of 7 (150 mg, 0.40 mmol) in dry pyridine (4 ml) was added DMTrCl (202 mg, 0.60 mmol), and the reaction mixture was stirred for 2 h at room temperature. The reaction was quenched by addition of ice. The mixture was concentrated in vacuo. The residue was diluted with AcOEt, which was washed with H2O and saturated aqueous NaHCO3, followed by brine. The organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by a silica gel column, eluted with MeOH in CHCl3 (0–5%), to give 8 (282 mg, quant.) as a white foam: FAB-LRMS m/z 686 (MH+); FAB-HRMS calcd for C36H40N5O7S (MH+) 686.2648, found 686.2650; 1H NMR (CDCl3) δ: 12.1 (br s, 1H), 8.94 (br s, 1H), 7.91 (s, 1H), 7.50–7.20 (m, 9H), 6.83–6.81 (m, 4H), 5.87 (d, 1H, J = 5.1 Hz), 4.34 (m, 1H), 4.22 (t, 1H, J = 5.1 Hz), 3.77 (s, 6H), 3.66–3.63 (m, 1H), 3.48 (dd, 1H, J = 4.5 and 9.7 Hz), 3.41 (s, 3H), 3.38 (dd, 1H, J = 6.3 and 9.7 Hz), 2.93 (br s, 1H), 2.37–2.31 (m, 1H), 1.10–1.07 (m, 6 H); 13C NMR (CDCl3) δ: 178.9, 158.6, 155.5, 148.4, 147.5, 144.5, 138.2, 135.6, 135.5, 130.0, 128.0, 127.9, 127.0, 121.6, 113.2, 87.5, 86.6, 73.1, 64.9, 60.0, 58.8, 55.2, 50.5, 36.1, 18.8, 18.7.
N2-Isobutyryl-9-[3-O-{2-cyanoethoxy(diisopropylamino)phosphino}-5-O-(4,4′-dimethoxytrityl)-2-O-methyl-4-thio-β-d-furanosyl]guanine (9)
To a solution of 8 (1.5 g, 2.1 mmol) in dry CH2Cl2 (21 ml) were added N,N-diisopropylethylamine (0.95 ml, 5.4 mmol), DMAP (33 mg, 0.27 mmol) and 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (0.91 ml, 4.1 mmol), and the reaction mixture was stirred for 4 h at 0°C. The reaction was quenched by addition of ice. The reaction mixture was diluted with AcOEt, which was washed with H2O and saturated brine. The organic layer was dried (Na2SO4) and concentrated in vacuo. The residue was purified by a silica gel column, eluted with hexane/AcOEt (1:1), to give 9 (1.4 g, 73%) as a white foam: FAB-LRMS m/z 886 (MH+); FAB-HRMS calcd for C45H57N7O8PS (MH+) 886.3727, found 886.3716; 31P NMR (CDCl3) δ: 151.3 and 149.1.
Synthesis of ONs
Support bound chemically modified ONs were synthesized on an Applied Biosystem 3400 DNA synthesizer using the corresponding phosphoramidite units at a 1.0 µmol scale following the standard procedure described for oligoribonucleotides. Each of phosphoramidite units was used at concentration of 0.1 M in dry acetonitrile, and the coupling time was extended to 10 min for each step. After completion of the synthesis, the CPG support was treated with concentrated NH4OH or NH4OH/EtOH (3:1) at 55°C for 16 h. In the case of CPG supports loaded FRNA1, FRNA2 and F-SRNA2, these were treated with methanolic ammonia (saturated at 0°C) at room temperature for 24 h. Then, the support was filtered off. The filtrate was concentrated and the ON protected by a DMTr group at the 5′-end was chromatographed on a C-18 silica gel column with a linear gradient of acetonitrile (from 5% to 40%) in 0.1 N TEAA buffer (pH 7.0). The fractions containing the full-length ON were combined and concentrated. The residue was treated with aqueous acetic acid (70%) for 20 min at room temperature. The solution was concentrated and the residue was purified on reversed-phase HPLC, using a J′sphere ODS-M80 column (4.6 × 150 mm, YMC) with a linear gradient of acetonitrile (from 10% to 40%) in 0.1 N TEAA buffer (pH 7.0). The structure of each RNA was confirmed by measurement of MALDI-TOF/MASS spectrometry on a Voyager-DE pro. MeRNA1: calculated mass, C157H206N55O103P14 4942.89 (M–H); observed mass, 4946.24. MeRNA2: calculated mass, C155H205N50O103P14 4847.86 (M–H); observed mass, 4847.36. FRNA1: calculated mass, C142H161F15N55O88P14 4762.59 (M–H); observed mass, 4753.7. FRNA2: calculated mass, C140H160F15N50O88P14 4667.56 (M–H); observed mass, 4669.50. SRNA2: calculated mass, C140H175N50O88P14S15 4877.28 (M–H); observed mass, 4880.65. Me-SRNA1: calculated mass, C157H206N55O88P14S15 5182.54 (M–H); observed mass, 5180.99. F-SRNA2: calculated mass, C140H160F15N50O73P14S15 4907.22 (M–H); observed mass, 4907.02.
Determination of hybridization ability
Thermally induced transitions were monitored at 260 nm on a Beckman DU 650 spectrophotometer. Samples were prepared as follows. Duplex formation: a solution containing an appropriate ON and a complementary sequence (3 µM each) in a buffer of 10 mM Na cacodylate (pH 7.0) containing 100 mM NaCl were heated at 90°C for 5 min, then cooled gradually to room temperature and used for the thermal denaturation study. The sample temperature was increased 0.5°C/min.
Circular dichroism (CD) measurements
CD spectra were obtained at 25°C on a Jasco J720. The solution containing samples (3 µM each) in a buffer of 10 mM Na cacodylate (pH 7.0) containing 100 mM NaCl was prepared, and the sample spectra were subtracted from the buffer spectrum. The molar ellipticity was calculated from the equation [θ] = θ/cl, where θ is the relative intensity, c the sample concentration and l the cell path length in centimeters.
Assays for S1 nuclease stability
Each RNA sample labeled with 32P at the 5′-end (5 pmol) was mixed with the corresponding unlabeled RNA (100 pmol). The RNA sample was incubated in S1 nuclease buffer (30 mM sodium acetate, 0.28 M NaCl, 1 mM ZnSO4, pH 4.6) supplemented with S1 nuclease (0.17 U/µl or 51 U/µl) at 37°C. At appropriate periods, 2 µl aliquots of the reaction mixture were added to 10 µl of loading buffer (1× TBE, 7 M urea, 0.05% bromophenol blue, 0.05% xylene cyanol). The mixtures were then analyzed by electrophoresis on 20% polyacrylamide gel containing 7 M urea. Radioactive densities of the gel were visualized by a Bio-imaging analyzer (Bas 2500, Fuji Co., Ltd).
Assays for SVPD stability
Each RNA sample labeled with 32P at the 5′-end (5 pmol) was mixed with the corresponding unlabeled RNA (100 pmol). The RNA sample was incubated in a buffer (40 mM Tris–HCl, 8 mM MgCl2, 5 mM DTT, pH 7.5) supplemented with SVPD (6.8 × 10−5 U/µl) at 37°C. At appropriate periods, 2 µl aliquots of the reaction mixture were added to 10 µl of loading buffer (1× TBE, 7 M urea, 0.05% bromophenol blue, 0.05% xylene cyanol). The mixtures were then analyzed by electrophoresis on 20% polyacrylamide gel containing 7 M urea. Radioactive densities of the gel were visualized by a Bio-imaging analyzer (Bas 2500, Fuji Co., Ltd).
Assays for human plasma stability
Each RNA sample labeled with 32P at the 5′-end (5 pmol) was mixed with the corresponding unlabeled RNA (100 pmol). The RNA sample was incubated in PBS (40 µl) containing 50% human plasma at 37°C. At appropriate periods, 3 µl aliquots of the reaction mixture were added to 16 µl of loading buffer (1 × TBE, 7 M urea, 0.05% bromophenol blue, 0.05% xylene cyanol). The mixtures were then analyzed by electrophoresis on 20% polyacrylamide gel containing 7 M urea. Radioactive densities of the gel were visualized by a Bio-imaging analyzer (Bas 2500, Fuji Co., Ltd).
RESULTS AND DISCUSSION
Chemistry
As reported in our previous article (18), we have synthesized 2′-deoxy-2′-fluoro-4′-thiouridine, -thiocytidine and -thioadenosine, the components of F-SRNA. However, despite further attempts to prepare 2′-deoxy-2′-fluoro-4′-thioguanosine, we did not obtain the desired compound in sufficient quantity to carry out ON synthesis. For the synthesis of a series of 2′-O-Me-4′-thioribonucleosides, the components of Me-SRNA, uridine, cytidine, adenosine and guanosine derivatives were all prepared according to methods similar to those used for their 4′-O-congener (19,20). Thus, treatment of 2,2′-O-anhydro-4′-thiouridine (1) (9) with a mixture of trimethyl borate and trimethyl orthoformate in the presence of a catalytic amount of sodium bicarbonate afforded 2′-O-Me-4′-thiouridine (2) in 86% yield (Scheme 1). The resulting 2 was converted into its cytosine derivative, and then the corresponding phosphoramidite units, respectively, using the standard procedure (details of the synthetic scheme and experimental procedures are given in the Supplementary Data; Scheme S2). In the case of the purine derivatives, a direct methylation of the 2′-hydroxy group was employed. In Scheme 2, the synthesis of the 2′-O-Me-4′-thioguanosine units is shown as an example, since the chemical conversion of the nucleobase differs slightly from that of our previous method (10). Thus, immediately after the Pummerer reaction (8), compound 3 was heated in NH3/EtOH, followed by MeNH2/MeOH at room temperature to give the diamino derivative 4. When the resulting 4 was treated with MeI in DMF in the presence of NaH at –40°C, the 2′-O-Me derivative 5 was obtained in 85% yield. Selective protection of the 2-amino group of 5 by isobutyryl chloride afforded 6. The remaining 6-amino group of 6 was then hydrolyzed via diazotization to give the guanine derivative. Since a partial deprotection of the TIPDS group on the sugar portion took place during the reaction, the resulting mixture was subsequently treated with Et3N•3HF to give 7 in 73% yield in two steps. Protection of the 5′-hydroxy group with a dimethoxytrityl (DMTr) group, followed by reaction with N,N-diisopropylchlorophosphoramidite in the presence of Hünig′s base afforded the 2′-O-Me-4′-thioguanosine phosphoramidite unit 9. The corresponding adenosine unit was also prepared in a similar manner (details of the synthetic scheme and experimental procedures are given in the Supplementary Data; Scheme S3). Using these phosphoramidite units, we synthesized 2′-modified-4′-thioRNAs along with other ONs in a DNA/RNA synthesizer following the standard procedure. The sequences used in this study are shown in Figure 2. The series of ON1s includes Me-SRNA (Me-SRNA1), which is the same as that used in our previous report (10), while the sequence of ON2s includes F-SRNA (F-SRNA2), since the 4′-thioguanosine phosphoramidite unit of this modified ON was not available. For a comprehensive comparison of their properties, MeRNAs (MeRNA1 and MeRNA2), FRNAs (FRNA1 and FRNA2), and SRNAs [SRNA1 (10) and SRNA2] along with natural RNAs (RNA1 and RNA2) and DNAs (DNA1 and DNA2) were also prepared.
Figure 2.
Sequences of modified ONs.
Duplex stability and structural aspects
The thermal stabilities of the synthetic ONs with complementary RNA (cRNA1 or cRNA2) and DNA (cDNA1 or cDNA2) were measured by ultraviolet melting experiments in a buffer of 10 mM sodium cacodylate (pH 7.0) containing 100 mM NaCl (Table 1). A duplex of RNA1:cRNA1 had a Tm value of 66.2°C and that of DNA1:cRNA1 showed a lower Tm value (56.2°C and ΔTm = –10°C). When RNA1 was changed to the modified RNAs, the duplexes were all stabilized compared to RNA1:cRNA1. Among the duplexes examined (a series of ON1:cRNA1), FRNA1:cRNA1 showed the highest Tm value (77.4°C, ΔTm = +11.2°C). The rank order of the Tm's with the complementary RNA (cRNA1) was FRNA > Me-SRNA = SRNA > MeRNA > RNA > DNA, where the order of FRNA > MeRNA > RNA > DNA was identical with that of the previous report (6). Although there was no synergistic effect in Me-SRNA1 in terms of hybridization with RNA, the advantage of this modified RNA was obvious from the comparison of the Tm values of ON1:cRNA1 with those of ON1:cDNA1, to wit, the modified ONs, which hybridize with RNA, generally form a stable duplex with DNA. In fact, both MeRNA1 and FRNA1 formed a thermally stable duplex with cDNA1 just as they did with cRNA1. On the other hand, Me-SRNA1 formed a less stable duplex with cDNA1 (42.4°C; ΔTm = −9.2°C). Thus, the rank order of the Tm values with the complementary DNA (cDNA1) was changed to the following: FRNA > DNA > MeRNA > RNA > SRNA > Me-SRNA. As we previously reported (10), this property is characteristic of SRNA (SRNA1:cRNA1 = 73.3°C versus SRNA1:cDNA1 = 43.6°C; ΔTm = 29.7°C), and was further enhanced by methylation of the 2′-OH group, namely Me-SRNA (Me-SRNA1:cRNA1 = 73.3°C versus Me-SRNA1:cDNA1 = 42.4°C; ΔTm = 30.9°C).
Table 1.
Thermal stability of duplexesa
| With cRNAl or cRNA2 |
With cDNAl or cDNA2 |
|||
|---|---|---|---|---|
| ON | Tm (°C) | ΔTm (°C)b | Tm (°C) | ΔTm (°C)b |
| RNA1 | 66.2 | – | 51.6 | – |
| DNA1 | 56.2 | −10.0 | 59.1 | +7.5 |
| MeRNAl | 70.3 | +4.1 | 52.0 | +0.4 |
| SRNA1 | 73.3 | +7.1 | 43.6 | −8.0 |
| Me-SRNAl | 73.3 | +7.1 | 42.4 | −9.2 |
| FRNA1 | 77.4 | +11.2 | 64.2 | +12.5 |
| RNA2 | 60.1 | – | 44.7 | – |
| DNA2 | 50.1 | −10.0 | 49.5 | +4.8 |
| FRNA2 | 72.1 | +12.0 | 57.8 | +13.1 |
| SRNA2 | 64.2 | +4.1 | 33.4 | −11.3 |
| F-SRNA2 | 76.1 | +16.0 | 51.7 | +7.0 |
| MeRNA2 | 63.8 | +3.7 | 42.4 | −2.3 |
aTm values were given as an average of three independent experiments.
bΔTm values were calculated based on the Tm values of RNA1:cRNA1 (RNA2:cRNA2) or RNA1:cDNA1 (RNA2:cDNA2).
In the case of F-SRNA (see a series of ON2s), this modified ON seems to have a synergistic effect of both FRNA and SRNA for its hybridization. Thus, the duplexes of FRNA2:cRNA2 and SRNA2:cRNA2 showed Tm values of 72.1°C and 64.2°C, respectively. When FRNA2 (or SRNA2) was changed to F-SRNA2, the duplex consisting of F-SRNA2:cRNA2 had the highest Tm value of 76.1°C. Therefore, the rank order of the Tm's with the complementary RNA (cRNA2) was F-SRNA > FRNA > SRNA > MeRNA > RNA > DNA. For the hybridization with cDNA2, F-SRNA showed an intermediate value between FRNA and SRNA. Although F-SRNA increased the Tm values with both RNA and DNA in the same way as FRNA, the difference in Tm values between F-SRNA2:cRNA2 and F-SRNA2:cDNA2 was 24.4°C. This difference was larger than that between FRNA2:cRNA2 and FRNA2:cDNA2 (ΔTm = 14.3°C) and close to the difference between SRNA2:cRNA2 and SRNA2:cDNA2 (ΔTm = 30.8°C). From these results, it can be concluded that either Me-SRNA or F-SRNA prefers RNA as a complementary partner much more than DNA, which is similar to the behavior of SRNA. In addition, F-SRNA, the hybrid of FRNA and SRNA, formed the thermally most stable duplex with RNA.
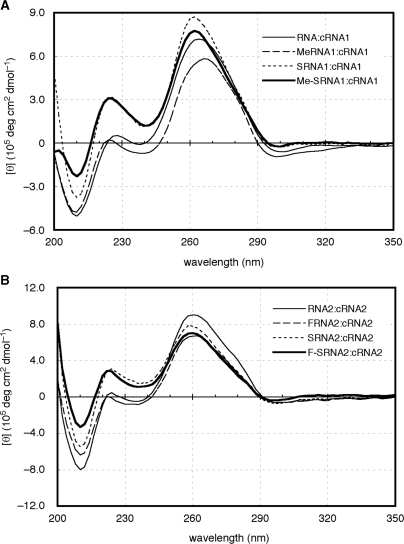
To investigate the structural aspects of the duplexes consisting of the modified ONs, their CD spectra were measured together with those of the natural duplexes. As can be seen in Figure 3A, the CD spectrum of RNA1:cRNA1 had a strong positive band near 260 nm and a negative band near 210 nm, which is characteristic of the A-conformation. Although the ellipticity at 260 nm of MeRNA1:cRNA1 was smaller than those of others, all duplexes had a positive band near 260 nm in each CD spectrum, indicating therefore their A-form structures. In the CD spectra of a series of ON2:cRNA2 (Figure 3B), all duplexes also had strong positive bands near 260 nm, indicating that they adopt the A-form structure. When the sequence of complementary RNA (cRNA1 or cRNA2) was changed to the complementary DNA (cDNA1 or cDNA2), the data suggests that all duplexes adopt the A-like form similar to that of the natural RNA:DNA duplex (Supplementary Data; Figure S1). From these results, there were no obvious structural changes in Me-SRNA and F-SRNA for their duplex formation as was the case for SRNA, MeRNA and FRNA.
Figure 3.
CD spectra of duplexes consist of ON1s:cRNA1 (A) and ON2s:cRNA2 (B).
As mentioned in the Introduction section, a number of sugar-modified ONs have been developed. However, few are known to possess the type of modification of the sugar portion as do Me-SRNA (a combination of 2′-OMe and 4′-thio) and F-SRNA (a combination of 2′-F and 4′-thio). To the best of our knowledge, the ON containing 2′-deoxy-2′-fluoro-4′-thioarabinouridine (4′S-FANA; a combination of 2′-araF and 4′-thio) is the only one known having such a modification pattern (21). Although 4′S-FANA formed a duplex with its complementary RNA in the A-form structure, the resulting duplex was fairly destabilized thermally compared with the RNA:RNA and FANA:RNA duplexes. Thus, no synergistic effect in terms of thermal stability was observed in 4′S-FANA. Contrary to 4′S-FANA, both Me-SRNA and F-SRNA formed a thermally stable duplex with the complementary RNA. Several reasons have been suggested as to why chemical modification of ONs, such as 2′-OMe, 2′-F and 4′-thio, improve their hybridization ability (5,10,22–25): (i) formation of highly organized hydration: (ii) favorable gauche and anomeric effects: (iii) hydrophobic interactions in the minor groove: and (iv) restricted sugar conformation favoring C3′-endo. In their 1H-NMR spectra and crystal structural analysis, all nucleoside units of Me-SRNA and F-SRNA are shown to prefer the C3′-endo conformation just as their 4′-O-congener (18). In this point, however, there is no additional rationale for why 2′-modified-4′-thioRNA, especially F-SRNA, improves and enhances their hybridization relative to the individual chemical modifications. It is hoped that this will be elucidated by crystal structural analysis, which is under investigation, and will be reported elsewhere.
Nuclease stability
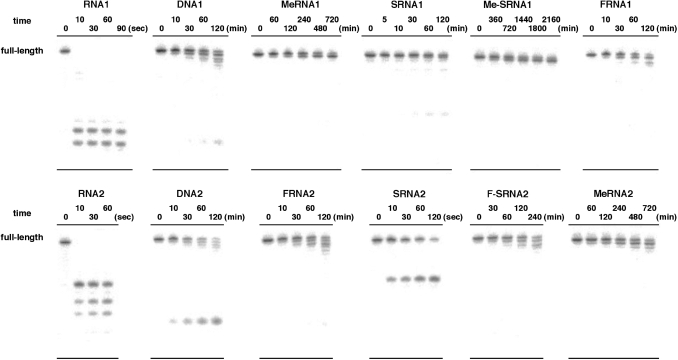
As described above, Me-SRNA and F-SRNA showed a favorable hybridization with RNA arising from the hybrid chemical modification. Thus, we next investigated how these modifications affected their nuclease stability. As is well known, MeRNA is highly resistant against ribonuclease (RNase) digestion due to methylation of the 2′-hydroxy groups. Furthermore, MeRNA also showed higher stability against snake venom phosphodiesterase (SVPD; a 3′-exonuclease) and S1 nuclease (an endonuclease) compared with natural DNA (6). Since FRNA also lacks the 2′-hydroxy groups on its structure, this modified ON was resistant against RNase digestion (26). However, FRNA was degraded as easily as natural DNA in calf serum, probably as a result of 3′-exonuclease activity (5). For SRNA, this modified ON was highly resistant against not only 3′-exo- and endonucleases, but also in human serum, despite possessing a 2′-hydroxy group on its structure (10,27,28). As mentioned above, we and others have already reported the nuclease stability of MeRNA, FRNA and SRNA, compared with natural RNA and/or DNA. However, no comprehensive comparison of their nuclease stability has been reported. Accordingly, we evaluated in detail the nuclease stability of Me-SRNA and F-SRNA along with that of MeRNA, FRNA, SRNA, and natural RNA and DNA.
A series of ON1s and ON2s were labeled at the 5′-end with 32P and incubated in an appropriate buffer in the presence of S1 nuclease (a typical endonuclease), SVPD (a typical 3′-exonuclease), and human plasma. The reactions were then analyzed by PAGE under denaturing conditions, and the half-lives (t1/2) were estimated based on the ratio of full-length RNA at each time interval. The results are summarized in Table 2. In the presence of S1 nuclease (0.17 U/µl), natural RNAs (RNA1 and RNA2) and DNAs (DNA1 and DNA2) were all rapidly cleaved to give t1/2 values of <1 min (the results of PAGE are presented in the Supplementary Data; Figure S2). To our surprise, FRNAs (FRNA1 and FRNA2) were also immediately cleaved, while MeRNAs (MeRNA1 and MeRNA2) were completely stable (>24 h) under the same conditions. The SRNAs, SRNA1 and SRNA2, showed moderate stability, giving t1/2 values of 76.8 and 12.6 min, respectively, which were much higher than the FRNAs. The stability of Me-SRNA and F-SRNA was characteristic. Me-SRNA1 was completely stable as was MeRNA1 (t1/2 > 24 h). To clarify the order of stability, MeRNA1 and Me-SRNA1 were treated in the presence of a higher concentration of S1 nuclease (51 U/µl). As a result, Me-SRNA1 was still stable under the conditions in which MeRNA1 was cleaved to give a t1/2 of 224 min (136 min for MeRNA2). On the other hand, F-SRNA2 was less stable than SRNA2 to give a t1/2 of 6.5 min. Thus, the rank order of S1 nuclease resistance was Me-SRNA >> MeRNA >> SRNA > F-SRNA > FRNA > RNA > DNA. For S1 nuclease stability, it can be concluded that Me-SRNA showed a synergistic effect of MeRNA and SRNA, while F-SRNA did not show any additive effect.
Table 2.
Comprehensive investigation of nuclease stability of chemically modified ONsa
| S1 nuclease |
SVPD |
50% human plasma |
|||
|---|---|---|---|---|---|
| ON | t1/2b | t1/2c | t1/2 | t1/2 | Degradation pattern |
| RNA1 | 25.4 ± 7.2 s | – | 6.0 ± 0.6 min | < 10 s | Endo |
| DNAl | 7.4 ± 1.9 s | – | 3.1 ± 0.1 min | 46.8 ± 7.0 min | Exo |
| MeRNAI | >24 h | 224 ± 47.6 min | 5.3 ± 1.5 min | 187 ± 22.6 min | Exo |
| SRNA1 | 76.8 ± 25.2 min | – | 54.4 ± 2.5 min | 65.7 ± 8.0 min | Endo |
| Me-SRNAl | >24 h | >24 h | 79.2 ± 8.7 min | 1631 ± 60.8 min | Exo |
| FRNA1 | 1.1 ± 0.1 min | – | 5.8 ± l.2 min | 53.2 ± 3.5 min | Exo |
| RNA2 | 13.0 ± 2.4 s | – | 4.3 ± 0.4 min | < 10 s | Endo |
| DNA2 | 6.6 ± 1.3 s | – | 2.7 ± 0.3 min | 31.0 ± 1.7 min | Exo |
| FRNA2 | 20.8+0.4 s | – | 6.0 ± 1.0 min | 51.9 ± 3.1 min | Exo |
| SRNA2 | 12.6 ± 1.7 min | – | 12.8 ± 3.5 min | < 10 s | Endo |
| F-SRNA2 | 6.5 ± 0.5 min | – | 21.0 ± 3.6 min | 120 ± 10.5 min | Exo |
| MeRNA2 | >24 h | 136 ± 11.5 min | 4.5 ± 1.1 min | 435 ± 12.9 min | Exo |
aErrors reflect standard deviation from three independent experiments.
bONs were treated with 0.17 U/µL S1 nuclease.
cONs were treated with 51 U/µL S1 nuclease.
We next compared the stability of each ON against SVPD. Among the ONs examined, either natural ONs (RNA1, RNA2, DNA1 and DNA2) or 2′-modified ONs (MeRNA1, MeRNA2, FRNA1 and FRNA2) were cleaved to give t1/2 values of < 10 min and there were no obvious differences in their stability (the results of PAGE are presented in the Supplementary Data; Figure S3). In our experiment (under the reaction conditions and sequences used), no resistance of MeRNAs against SVPD was observed differing from the previous work in which 2′-O-methyl RNAs showed high stability against SVPD (6). Our results showed that SRNAs (SRNA1 and SRNA2) are more stable than MeRNA and FRNA. In addition, modifications of the 2′-hydroxy group of SRNA, i.e. Me-SRNA and F-SRNA, improved their stability, resulting in the following approximate rank order of SVPD resistance was Me-SRNA > F-SRNA > SRNA > MeRNA, FRNA, RNA and DNA. To develop chemically modified ONs for therapeutic applications, the ONs should be stable in the bodily fluids. Thus, finally, we investigated the stability of ONs in 50% human plasma. Under these conditions, RNA1 was rapidly cleaved (t1/2 < 10 s), while DNA1 was much more stable (t1/2 = 46.8 min). Even in our sequence, MeRNA1 was more stable than DNA1 (6), and the stability of FRNA1 was rather close to DNA1 (5). As we reported, SRNA1 was also relatively stable (10), and it was found that SRNA1 was slightly more stable than FRNA1. As can be seen, the stability of Me-SRNA1 was significantly improved in human plasma (t1/2 = 1631 min), with the rank order of stability in plasma being Me-SRNA >> MeRNA > SRNA > FRNA > DNA >>RNA. In the case of one series of ON2s, SRNA2 was unexpectedly unstable and the stability was the same as that of natural RNA2. When the 2′-hydroxy groups of SRNA2 were fluorinated, the stability of the resulting F-SRNA2 was significantly improved to give a t1/2 of 120 min, which was longer than that of FRNA2. From these results, both Me-SRNA and F-SRNA showed a synergistic improvement with respect to their nuclease stability with one exception (F-SRNA2 against S1 nuclease).
As shown above, we have carried out the comprehensive comparison of nuclease stability of chemically modified ONs. Throughout this investigation, the rank order of stability was proposed. In the presence of human plasma, the high stability of Me-SRNA is worth noting. In addition, it was suggested that the stability of MeRNA is higher than FRNA, and that the stability of SRNA is drastically affected by sequence context. Since human plasma will include multiple nucleases, the degradation pattern of each of the ONs was analyzed based on the results of PAGE. As shown in Figure 4, RNAs were cleaved at a certain position to afford several shorter fragments, which is a consequence of endonuclease activity (probably RNase activity) in human plasma. On the other hand, DNAs afforded fragments corresponding to n − 1 and n − 2 (n = full-length). This mode of degradation can be explained by 3′-exonuclease activity, which is recognized as one of the major factors in ON digestion (29). In addition, one shortest fragment also appeared along with these 3′-exonuclease products. Although we did not determine its structure, we assume it is a labeled phosphate or 5′-monophosphate, which is digested from the 5′-end. Since the 2′-hydroxy groups are substituted in MeRNAs and FRNAs, these modified ONs were resistant against digestion by endonucleases such as RNase and digested gradually by 3′-exonuclease activity, whereas SRNAs were digested by RNase because of the existence of the 2′-hydroxy groups. Both Me-SRNA and F-SRNA were digested by 3′-exonuclease. Therefore, the higher stability of Me-SRNA and F-SRNA is thought to arise from the acquisition of endonuclease resistance relative to SRNA, and improvement of exonuclease resistance, which is suggested by the SVPD stability of SRNA, relative to MeRNA and FRNA. The sequence context of each strand might explain the different stability in human plasma shown by SRNA1 and SRNA2. In serum (probably also in plasma), RNA analogs are predominantly digested by the contribution of RNase A-like activity (30). RNase A is an endonuclease that specifically hydrolyzes RNA after pyrimidine residues. The preference order of cleavage of this enzyme isolated from the pancreas is: UpA > CpA > UpG and CpG > CpC, UpC, CpU and UpU (31). As can be seen in the degradation pattern of SRNA2, this ON is immediately cleaved at the single position between the U(6) and the A(7) positions (UpA) (Supplementary Data; Figure S4). In RNA2, this position was also the major site of cleavage. In contrast, SRNA1 does not have UpA in the sequence, and thus showed higher stability in human plasma. Our results indicate that the UpA sequence is the most cleavable site in human plasma. This information would be helpful in designing RNA therapeutic molecules.
Figure 4.
Hydrolysis pattern of ONs in 50% human plasma. See Materials and methods section for experimental conditions.
Thus far, a large number of chemically modified ONs have been synthesized and their stability against nucleases has been evaluated. Although active site and proposed mechanism of cleavage by nuclease S1 and SVPD used in this study have been suggested (32–34), it is still unclear what kind of chemical modification of ONs appends the nuclease resistance. Our comprehensive comparison indicated useful information on the structural requirement for the nuclease resistance. Under conditions in which natural RNA and DNA rapidly cleaved, FRNA was also cleaved similarly by S1 nuclease. In contrast, MeRNA remained completely intact under the same conditions. Cummins et al. reported that the stability against S1 nuclease imparted by 2′-modifications correlated with the size of the modification (6). Our results indicate that a drastic difference in S1 nuclease resistance may exist between fluoro and methoxy substituents at the 2′-position. In SRNA, this modified ON showed higher stability than RNA, which would arise from structural changes of the sugar ring. These considerations would account for the highest stability of Me-SRNA and lesser stability of F-SRNA versus SRNA. Although F-SRNA was more stable than FRNA because of its 4′-thiosugar construction, this ON was less stable than SRNA due to a smaller substituent at the 2′-position (–OH versus –F). We also examined the stability of 4′-thioDNA (SDNA1; 5′-AGTCCGAATTCACGT-3′) against S1 nuclease, and its t1/2 was estimated to be 19.4 min (the result of PAGE is not shown), which was shorter than that of SRNA1 (–OH versus –H). This data also supported our proposal for S1 nuclease stability. As for the stability against SVPD, there was no obvious difference in RNA, DNA, MeRNA and FRNA. Since SRNA and Me-SRNA were more stable than others, the 4′-thiosugar construction appeared to be a major contributing factor in the resistance. In human plasma (or serum), ONs possessing 2′-OH groups are rapidly cleaved by RNase activity. Therefore, the 2′-OH groups of chemically modified ONs are mostly substituted, and this type of substitution appended complete resistance to RNase digestion. However, chemically modified ONs such as MeRNA and FRNA are generally digested by 3′-exonucleolytic cleavage. Our results indicated that the 2′-O-methylation of SRNA was imparted the highest resistance toward endonucleolytic cleavage, and the 4′-thiolation of MeRNA gave the highest resistance toward exonucleolytic cleavage. From the standpoint of nuclease stability, Me-SRNA showed a synergistic improvement and was extremely stable in human plasma.
CONCLUSION
In this article, we have synthesized 2′-modified-4′-thioRNA, i.e. 2′-fluoro-4′-thioRNA (F-SRNA) and 2′-O-Me-4′-thioRNA (Me-SRNA). These ONs consist of a hybrid chemical modification, and were expected to show synergistic improvement for their hybridization and nuclease stability. Concerning the hybridization, F-SRNA exhibited synergistic improvement to show the highest Tm value in duplex formation with a complementary RNA. Although there was no obvious improvement in the Tm values for Me-SRNA, this modified ON has the advantages of a much more favorable duplex formation with a complementary RNA than with the complementary DNA, which is arising from the character of 4′-thioRNA (SRNA). Nuclease stability is the most important aspect in developing the chemically modified ONs. Thus, we have carried out a comprehensive comparison of nuclease stability of F-SRNA and Me-SRNA, along with FRNA, MeRNA, and natural RNA and DNA. F-SRNA was less stable than MeRNA in human plasma, while Me-SRNA, perhaps one of the most stable ONs, was significantly stable under the same conditions. From these results, Me-SRNA should be a versatile modified ON applicable to antisense molecules against miRNA and siRNA strategies. Further investigations are underway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported in part by Grant-in-Aid for Scientific Research from the Japan Society for Promotion of Science (No. 18109001). Funding for open access charges: Japan Society of Promotion of Science.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Ms Y. Misawa (Hokkaido University) for technical assistance and Ms S. Oka (Center for Instrumental Analysis, Hokkaido University) for technical assistance with MS.
REFERENCES
- 1.Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr. Opin. Chem. Biol. 2006;10:607–614. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Corey DR. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manoharan M. 2′-Carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim. Biophys. Acta. 1999;1489:117–130. doi: 10.1016/s0167-4781(99)00138-4. [DOI] [PubMed] [Google Scholar]
- 4.Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Synthesis and hybridization studies on two complementary nona-(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzales C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 6.Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, Charles M, Guinosso CJ, Cook PD. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng EW, Shima DT, Calias P, Cunningham E.T., Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vescular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 8.Naka T, Minakawa N, Abe H, Kaga D, Matsuda A. The stereoselective synthesis of 4′-β-thioribonucleosides via the Pummerer reaction. J. Am. Chem. Soc. 2000;122:7233–7243. [Google Scholar]
- 9.Inoue N, Kaga D, Minakawa N, Matsuda A. Practical synthesis of 2′-deoxy-4′-thioribonucleosides: substrates for the synthesis of 4′-thioDNA. J. Org. Chem. 2005;70:8597–8600. doi: 10.1021/jo051248f. [DOI] [PubMed] [Google Scholar]
- 10.Hoshika S, Minakawa N, Matsuda A. Synthesis and physical and physiological properties of 4′-thioRNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue N, Minakawa N, Matsuda A. Synthesis and properties of 4′-thioDNA: unexpected RNA-like behavior of 4′-thioDNA. Nucleic Acids Res. 2006;34:3476–3483. doi: 10.1093/nar/gkl491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsugami A, Ohyama T, Inada M, Inoue N, Minakawa N, Matsuda A, Katahira M. Unexpected A-form formation of 4′-thioDNA in solution, revealed by NMR, and the implications as to the mechanism of nuclease resistance. Nucleic Acids Res. 2008;36:1805–1812. doi: 10.1093/nar/gkn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa N, Harashima H, Matsuda A. New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res. 2005;33:2942–2951. doi: 10.1093/nar/gki578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minakawa N, Sanji M, Kato Y, Matsuda A. Investigations toward the selection of fully-modified 4′-thioRNA aptamers: optimization of in vitro transcription steps in the presence of 4′-thioNTPs. Bioorg. Med. Chem. 2008;16:9450–9456. doi: 10.1016/j.bmc.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 15.Hoshika S, Minakawa N, Kamiya H, Harashima H, Matsuda A. RNA interference induced by siRNAs modified with 4′-thioribonucleosides in cultured mammalian cells. FEBS Lett. 2005;579:3115–3118. doi: 10.1016/j.febslet.2005.04.073. [DOI] [PubMed] [Google Scholar]
- 16.Hoshika S, Minakawa N, Shionoya A, Imada K, Ogawa N, Matsuda A. Study of modification pattern–RNA activity relationships by using siRNAs modified with 4′-thioribonucleosides. Chem. Bio. Chem. 2007;8:2133–2138. doi: 10.1002/cbic.200700342. [DOI] [PubMed] [Google Scholar]
- 17.Inoue N, Shionoya A, Minakawa N, Kawakami A, Ogawa N, Matsuda A. Amplification of 4′-thioDNA in the presence of 4′-thio-dTTP and 4′-thio-dCTP, and 4′thioDNA-directed transcription in vitro and in mammalian cells. J. Am. Chem. Soc. 2007;129:15424–15425. doi: 10.1021/ja075953c. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi M, Daidouji S, Shiro M, Minakawa N, Matsuda A. Synthesis and crystal structure of 2′-deoxy-2′-fluoro-4′-thioribonucleosides: substrates for the synthesis of novel modified RNAs. Tetrahedron. 2008;64:4313–4324. [Google Scholar]
- 19.Ross BS, Springer RH, Tortorici Z, Dimock S. A novel and economical synthesis of 2′-O-alkyl-uridines. Nucleosides Nucleotides. 1997;16:1641–1643. [Google Scholar]
- 20.Beigelman L, Haeberli P, Sweedler D, Karpeisky A. Improved synthetic approaches toward 2′-O-methyl-adenosine and guanosine and their N-acyl derivatives. Tetrahedron. 2000;56:1047–1056. [Google Scholar]
- 21.Watts JK, Choubdar N, Sadalapure K, Robert F, Wahba AS, Pelletier J, Pinto BM, Damha MJ. 2′-Fluoro-4′-thioarabino-modified oligonucleotides: conformational switches linked to siRNA activity. Nucleic Acids Res. 2007;35:1441–1451. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubini P, Zürcher W, Egli M. Stabilizing effects of the RNA 2′-substituent: crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem. Biol. 1994;1:39–45. doi: 10.1016/1074-5521(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 23.Lesnik EA, Freier SM. What affects the effect of 2′-alkoxy modification? 1. Stabilization effect of 2′-methoxy substitutions in uniformly modified DNA oligonucleotides. Biochemistry. 1998;37:6991–6997. doi: 10.1021/bi972995c. [DOI] [PubMed] [Google Scholar]
- 24.Auffinger P, Westhof E. Hydrophobic groups stabilize the hydration shell of 2′-O-methylated RNA duplexes. Angew. Chem. Int. Ed. 2001;40:4648–4650. doi: 10.1002/1521-3773(20011217)40:24<4648::aid-anie4648>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 25.Haeberli P, Berger I, Pallan PS, Egli M. Synthesis of 4′-thioribonucleosides and thermodynamic stability and crystal structure of RNA oligomers with incorporated 4′-thiocytosine. Nucleic Acids Res. 2005;33:3965–3975. doi: 10.1093/nar/gki704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono T, Scalf M, Smith LM. 2′-Fluoro modified nucleic acids: polymerase-directed synthesis, properties and stability to analysis by matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res. 1997;25:4581–4588. doi: 10.1093/nar/25.22.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellon L, Barascut J.-L, Maury G, Divita G, Goody R, Imbach J.-L. 4′-Thio-oligo-β-D-ribonucleotides: synthesis of β-4′-thio-oligouridylates nuclease resistance base pairing properties and interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 1993;21:1587–1593. doi: 10.1093/nar/21.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leydier C, Bellon L, Barascut J.-L, Morvan F, Rayner B, Imbach J.-L. 4′-Thio-RNA: synthesis of mixed base 4′-thio-oligoribonucleotides, nuclease resistance, and base pairing properties with complementary single and double strand. Antisense Res. Dev. 1995;5:167–174. doi: 10.1089/ard.1995.5.167. [DOI] [PubMed] [Google Scholar]
- 29.Eder PS, DeVine RJ, Dagle JM, Walder JA. Substrate specificity and kinetics of degradation of antisense oligonucleotides by a 3′ exonuclease in plasma. Antisense Res. Dev. 1991;1:141–151. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- 30.Shimayama T, Nishikawa F, Nishikawa S, Taira K. Nuclease-resistant chimeric ribozymes containing deoxyribonucleotides and phosphorothioate linkages. Nucleic Acids Res. 1993;21:2605–2611. doi: 10.1093/nar/21.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLennan BD, Lane BG. The chain termini of polynucleotides formed by limited enzymic fragmentation of wheat embryo ribosomal RNA. Part 2. Studies of a snake venom rebonuclease and pancreas ribonuclease. Can. J. Biochem. 1968;46:93–107. doi: 10.1139/o68-014. [DOI] [PubMed] [Google Scholar]
- 32.Iwamatsu A, Aoyama H, Dibó G, Tsunasawa S, Sakiyama F. Amino acid sequence of Nuclease S1 from Aspergillus oryzae. J. Biochem. 1991;110:151–158. doi: 10.1093/oxfordjournals.jbchem.a123534. [DOI] [PubMed] [Google Scholar]
- 33.García–Díaz M, Avalos M, Cameselle JC. Alcohol esterification reactions and mechanisms of snake venom 5′-nucleotide phosphodiesterase. Eur. J. Biochem. 1993;213:1139–1148. doi: 10.1111/j.1432-1033.1993.tb17864.x. [DOI] [PubMed] [Google Scholar]
- 34.Suck D. DNA recognition by structure-selective nucleases. Biopolymers. 1997;44:405–421. doi: 10.1002/(SICI)1097-0282(1997)44:4<405::AID-BIP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.