Abstract
Although some experiments suggest that the ribosome displays specificity for the identity of the esterified amino acid of its aminoacyl-tRNA substrate, a study measuring dissociation rates of several misacylated tRNAs containing the GAC anticodon from the A site showed little indication for such specificity. In this article, an expanded set of misacylated tRNAs and two 2′-deoxynucleotide-substituted mRNAs are used to demonstrate the presence of a lower threshold in koff values for aa-tRNA binding to the A site. When a tRNA binds sufficiently well to reach this threshold, additional stabilizing effects due to the esterified amino acid or changes in tRNA sequence are not observed. However, specificity for different amino acid side chains and the tRNA body is observed when tRNA binding is sufficiently weaker than this threshold. We propose that uniform aa-tRNA binding to the A site may be a consequence of a conformational change in the ribosome, induced by the presence of the appropriate combination of contributions from the anticodon, amino acid and tRNA body.
INTRODUCTION
There are 45 different Escherichia coli elongator aa-tRNAs, and in order for protein synthesis to proceed in an efficient manner, the ribosome must treat these aa-tRNAs as roughly equivalent substrates. Indeed, both binding (1) and kinetic (2) experiments show very similar properties. Uniform binding requires the presence of post-transcriptional modifications (1) as well as the presence of sequence elements within the body of the tRNA (3). What is less clear is the role of the esterified amino acid; although the presence of an esterified amino acid is necessary for some tRNAs to achieve uniformity (1), it is less certain whether the identity of the amino acid side chain is important. Several studies suggest this may be the case. Various 2′(3′)-O-aminoacyl derivatives of CpA with different amino acid side chains display a large range of apparent Km values in a peptidyltransferase assay (4), suggesting that the binding of the corresponding aa-tRNAs to the ribosome depends on the identity of esterified amino acid. The same conclusion can be reached from the fact that the IC50 values of a series of puromycin derivatives which differed in the identity of the amino acid side chain moiety varied substantially in an eukaryotic in vitro translation system (5). Furthermore, X-ray crystal structures of aa-tRNA substrate analogs bound to the A site of the 50S subunit of Haloarcula marismortui ribosomes indicate that the side chain of the amino acid fits into an asymmetric, hydrophobic cleft in the 23S ribosomal RNA (rRNA) that would be expected to show specificity for different amino acid substrates (6,7). While these data argue that the ribosome does show specificity for the side chain of the esterified amino acid, we had previously reported that four different tRNAs each esterified with three different non-cognate amino acids bound to the ribosomal A site just as well as the corresponding aa-tRNAs esterified with their cognate amino acid (8), indicating a lack of specificity by the ribosome.
In light of this contradictory data, it seemed possible that in our previous experiments using misacylated tRNAs, any effects of the amino acid side chain and tRNA body on binding to the A site were somehow masked (8). This may have occurred because only a few tRNA bodies were used, and all contained the GAC anticodon, which may have dominated the binding of the misacylated tRNAs to the ribosome (9). To see if this was true, ribosomal binding of a more diverse set of tRNA bodies was examined and the strength of the codon•anticodon complex with ribosomes was reduced by either substituting the weaker GAA anticodon into each tRNA or by using mRNAs that contained a destabilizing 2′-deoxynucleotide in the codon. With this expanded set of substrates, an important property of aa-tRNAs binding to the ribosomal A site was discovered, and clear evidence was obtained that the ribosomal A site shows specificity for both the esterified amino acid and the tRNA body.
MATERIALS AND METHODS
tRNA preparation
tRNA templates for E. coli tRNA3Gly(GAC), tRNA1Ile(GAA), tRNALys(GAC), tRNAMet(GAA), tRNATrp(GAC), tRNATrp(GAA), tRNA2Tyr(GAC) and tRNA2Tyr(GAA) (anticodon substitution shown in parentheses) were prepared by PCR amplification of plasmid DNA as described in (10). tRNA templates for the remaining tRNAs were prepared by primer extension of overlapping DNA oligomers. In vitro transcription (11) was performed as described in (12), except 16 mM GMP was used instead of 20 mM GMP. tRNAs were purified on 10% denaturing polyacrylamide gels.
3′ labeling and aminoacylation reactions
tRNAs were 3′-[32P]-labeled using [α-32P]-ATP (3000 Ci/mmole, Amersham) and terminal tRNA nucleotidyl transferase, according to (13). After labeling, tRNAs were extracted with phenol:chloroform, precipitated with ethanol, and stored in T.E. Buffer (10 mM Tris–HCl, pH 7 and 100 µM EDTA) at −20°C. tRNAs were aminoacylated, purified and stored as described in (12), except 150 µM of unlabeled amino acid was used instead of [3H] amino acid.
mRNA preparation
mRNA fragments were purchased from Dharmacon (Lafayette, CO), deprotected and purified by denaturing PAGE. The mRNAs were derivatives of the initiation region of the T4 gp32 mRNA (14), and had the sequence: 5′-GGC AAG GAG GUA AAA AUG XXX GCA CGU, where XXX was GUC or GdUC in experiments using GAC anticodon-substituted tRNAs and UUC or UdUC in experiments using GAA anticodon-substituted tRNAs.
Protein purification
His6-tagged E. coli AlaRS was purified as described previously (13,15). His6-tagged yeast PheRS was purified from a plasmid provided by D. Tirrell (California Institute of Technology, CA) using the protocol for ‘Preparation of cleared E. coli lysates under native conditions’ (Qiagen). His6-tagged E. coli EF-Tu was purified from a plasmid kindly provided by R. Green (Johns Hopkins University, MD), as described (16). The histidine tag was removed using TEV protease. The final protein sample was dialyzed into 50 mM Na2HPO4 (pH 7.5), 10 mM MgCl2, 100 µM GDP, 5 mM DTT, and 10% glycerol (Dialysis Buffer). EF-Tu was stored in 10 µl aliquots at −80°C. The concentration of protein was determined by the Bradford assay.
Ribosome purification
Tight-coupled 70S ribosomes were isolated and purified as described in (17). The purified ribosomes were suspended in 50 mM Hepes (pH 7.0), 30 mM KCl, 70 mM NH4Cl, 1 mM DTT and 10 mM MgCl2 (RB Buffer-10) and stored at −80°C. The fraction of active ribosomes was determined by binding site titrations of the P and A sites using transcribed tRNA2AVal. Reported ribosome concentrations are for active ribosomes.
A site binding of tRNAs
Dissociation rate constants of tRNA release from the ribosomal A site were measured using a filter binding assay similar to that described in (17). Ribosomes were activated by incubation at 42°C for 10 min. Then, 2 μM ribosomes were pre-incubated with 10 μM mRNA and 10 μM deacyl-tRNAfMet in 50 mM Hepes (pH 7.0), 30 mM KCl, 70 mM NH4Cl, 1 mM DTT and 15 mM MgCl2 (RB Buffer-15) at room temperature. 3′-[32P]-labeled aa-tRNAs were enzymatically loaded into the A site of the ribosome–mRNA–tRNAfMet complex using E. coli EF-Tu in the following way: EF-Tu stored in its GDP-bound form was converted to its GTP-bound form immediately before use by incubating 5.2 μM EF-Tu, 20 μM GTP, 40 μg/ml pyruvate kinase and 3 mM phosphoenolpyruvate in RB Buffer-15 for 30 min at 37°C. Ternary complex was formed by incubating 2.3 μM activated EF-Tu•GTP with 200 nM aa-[32P]-tRNA in RB Buffer-15 for 15 min on ice. A total of 20 nM EF-Tu•GTP•aa-[32P]-tRNA was mixed with 100 nM ribosome•mRNA•tRNAfMet for 2 min in RB Buffer-15 at room temperature. EF-Tu not bound to the ribosome was removed by passing the reaction through an S-300 HR column (Amersham) equilibrated in RB Buffer-10. Dissociation from the A site was then initiated by mixing 5 µl of the column flow-through (ribosome•tRNA complexes) with 250 µl chase solution containing RB Buffer-10 and 30 nM of the appropriate unlabeled chase tRNA. Aliquots (18 µl) were removed at desired times and filtered through a dual membrane system as described in (17). koff values were then determined from a semilogarithmic plot of fraction bound versus time. koff values for deacylated 3′-[32P]-tRNAs were determined similarly, except tRNAs were loaded without the aid of EF-Tu (1,17). Control experiments verified that ribosomes remained active for aa-tRNA binding throughout the course of the experiments. Also, previous control experiments have shown that any observed increase in A site•tRNA stabilization upon tRNA aminoacylation is not due to a movement from the A/A state (deacylated tRNA) to the A/P state (aa-tRNA), because the same increase in stabilization was observed when the P site was fully blocked using a non-hydrolyzable Met-(2′-NH2)tRNAfMet (17). Lastly, experiments using the single deoxynucleotide-substituted mRNAs, GdUC and UdUC, used 20 µM tRNAfMet.
RESULTS
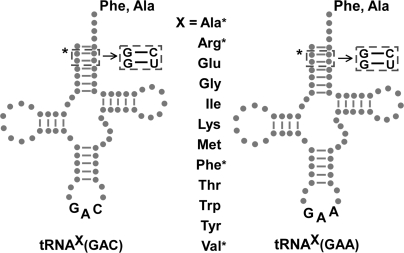
The GAC and GAA anticodons were introduced into 12 different unmodified E. coli tRNA transcripts to create a set of two cognate [tRNAVal(GAC) and tRNAPhe(GAA)] and 22 anticodon-substituted tRNAs (Figure 1). Two different anticodons were used to compare the effect of the tRNA bodies in the background of both ‘strong’ GAC•GUC and ‘intermediate’ GAA•UUC anticodon•codon interactions (3). The use of different anticodons also tested whether effects on ribosome binding due to the identity of the tRNA body were dependent on the anticodon substitution. Nine of the tRNA bodies were chosen because they had been used in previous studies (1,3,8); the remaining three (tRNAMet, tRNA1Ile and tRNATrp) were added to increase the diversity of tRNA sequences.
Figure 1.
tRNAs used in this study. The GAC (valine) and GAA (phenylalanine) anticodons were transplanted into 12 different E. coli tRNA bodies (gray), where the tRNA body ‘X’ is defined as the tRNA sequence excluding the anticodon. The resulting 24 tRNA mutants could all be aminoacylated with phenylalanine by PheRS. The recognition elements for alanyl-tRNA synthetase (G2-C71, G3-U70) were introduced into the eight tRNAs (four different tRNA bodies, two anticodons) indicated by the asterisk to also permit aminoacylation with alanine.
To assess the relative stability of each of the 24 deacylated tRNAs in the ribosomal A site, the dissociation rate constant of each tRNA from the A site was measured using a filter binding assay (17). In our experiments, aminoacylated tRNAs were loaded into the ribosomal A site enzymatically by EF-Tu, while deacylated tRNAs were loaded non-enzymatically. There is abundant evidence that non-enzymatically loaded tRNAs occupy the A site in the same way they do in normal translation. Not only are non-enzymatically loaded aa-tRNAs fully active in translation, but their koff values are the same as when they are loaded using EF-Tu (17,18). In addition, rRNA footprinting experiments show that both non-enzymatically loaded aa-tRNA and deacyl-tRNA bind ribosomes similarly to aa-tRNA introduced with EF-Tu (19). Since kon values for different tRNAs binding to the A site do not vary significantly (1), the koff values used here correlate well with equilibrium dissociation constants that are more commonly used to study ‘non enzymatic’ tRNA binding to ribosomes (3). koff and KD experiments have been widely used to define many of the basic properties of aa-tRNA binding to the ribosomal A site, including establishing the genetic code (20), demonstrating the role of post transcriptional modifications (21,22), and investigating the properties of various tRNA mutations (23). The assay, however, does not measure a mechanistically relevant step of translation. In normal translation, aminoacyl-tRNAs in the A site do not dissociate from their cognate codons into solution at an appreciable rate, but instead exit the A site into the P site as peptidyl tRNAs at a rate that is about 105-fold faster than koff. Nevertheless, these non-physiological assays appear to accurately reflect the interactions which aa-tRNA makes with the A site and, in some cases, correlate remarkably well with the translational performance of aa-tRNAs (24,25).
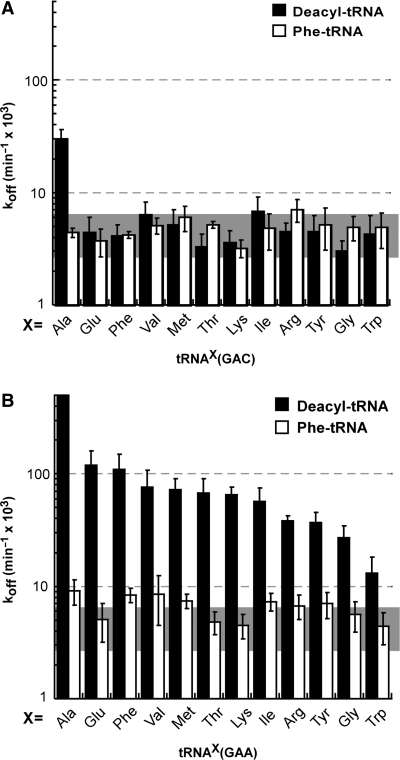
Two major conclusions may be drawn from the comparison of the stabilities of the deacylated tRNAs in the A site (Table 1, Figure 2). First, each tRNA displays a slower koff when it contains the GAC anticodon than when it contains the GAA anticodon, clearly demonstrating the non-equivalent binding contributions of different codon•anticodon pairs (3). Interestingly, however, the difference in stability between each GAC/GAA tRNA pair varies in a manner that depends on the tRNA body. For example, the difference in koff values between tRNATrp(GAC) and tRNATrp(GAA) is only 3-fold, but the difference between tRNAPhe(GAC) and tRNAPhe(GAA) is 27-fold. This suggests that the relative contribution of the anticodon is different for different tRNA bodies. Second, the variation in koff among the different tRNA bodies is much broader in the GAA anticodon background than in the GAC anticodon background. The deacyl-tRNA(GAA)s show about a 40-fold range of koff values, with some dissociating faster than the cognate tRNAPhe(GAA) and others actually dissociating more slowly. For example, the very weak tRNAAla(GAA) has a koff at least 4-fold faster than tRNAPhe(GAA), but the koff for tight tRNATrp(GAA) is more than 8-fold slower than tRNAPhe(GAA). In contrast, except for tRNAAla(GAC), all of the deacyl-tRNA(GAC)s display koff values within 2-fold of the cognate deacyl-tRNAVal(GAC). Taken together, these results indicate that different tRNA bodies do interact differently with the ribosome; however, these effects are only observed in the weaker GAA anticodon background. Presumably, the tRNA body effects still exist in the GAC anticodon-substituted tRNAs but are somehow masked by the very stable GAC anticodon•codon interaction.
Table 1.
Effects of aminoacylation with phenylalanine and anticodon substitution on koff (min−1 × 103) from the ribosomal A site
| tRNA | GAA anticodon |
GAC anticodon |
||
|---|---|---|---|---|
| Deacyl- | Phe- | Deacyl- | Phe- | |
| tRNA2Ala | >500a | 9.1 (2.3) | 30 (±6)a | 4.4 (0.4)b |
| tRNA2Arg | 38 (5) | 6.7 (1.6) | 4.5 (0.8) | 7.0 (1.6) |
| tRNA2Glu | 120 (39) | 5.1 (1.9) | 4.4 (1.6) | 3.7 (1.0) |
| tRNA3Gly | 27 (8) | 5.6 (1.7) | 3.0 (0.7) | 4.9 (1.2) |
| tRNA1Ile | 57 (18) | 7.3 (1.3) | 6.8 (2.3) | 4.8 (1.7) |
| tRNALys | 65 (11) | 4.5 (1.1) | 3.6 (1.0) | 3.2 (0.6) |
| tRNAMet | 73 (17) | 7.4 (1.1) | 5.2 (1.8) | 6.0 (1.5) |
| tRNAPhe | 110 (39) | 8.4 (1.2) | 4.1 (1.1) | 4.2 (0.3)b |
| tRNA3Thr | 68 (23) | 4.8 (1.1) | 3.3 (1.0) | 5.2 (0.4)b |
| tRNATrp | 13 (5) | 4.4 (1.4) | 4.3 (1.9) | 4.9 (1.7) |
| tRNA2Tyr | 37 (8) | 7.0 (1.8) | 4.5 (1.7) | 5.2 (2.1) |
| tRNA2AVal | 76 (31) | 8.5 (4.0) | 6.3 (1.9) | 5.1 (0.8)b |
Figure 2.
Effects of anticodon changes, different tRNA bodies and phenylalanylation on koff from the ribosomal A site (data from Table 1). (A) Deacylated and phenylalanylated tRNAs containing the GAC (valine) anticodon. (B) Deacylated and phenylalanylated tRNAs containing the GAA (phenylalanine) anticodon. The gray horizontal band represents the range of stabilities of cognate, purified aa-tRNAs in the A site: (2.7 × 10−3 to 6.4 × 10−3 min−1 (1)) and defines the threshold. 0.5 min−1 is the upper limit of the assay.
Next, the dissociation rates were determined when all 24 tRNAs were aminoacylated with phenylalanine. By using a common amino acid and a common anticodon, any tRNA body effects could be examined using substrates expected to fully occupy the ribosomal A site. As shown in Table 1 and Figure 2, the effect of the addition of the phenylalanine is much greater with the 12 tRNA(GAA)s than the 12 tRNA(GAC)s. In agreement with previous experiments (1), Phe-tRNAPhe(GAA) displays an ∼14-fold slower koff than deacyl-tRNAPhe(GAA). For the remaining 11 non-cognate tRNA(GAA)s, phenylalanylation slows the koff over a 3- to 50-fold range, such that all 12 aa-tRNAs dissociate from the A site with very similar koff values (6.1 ± 1.8 × 10−3 min−1). Thus, it appears that, in contrast to their deacylated counterparts, differences in stability because of the tRNA body are not observed in Phe-tRNA(GAA)s. The data for the 12 Phe-tRNA(GAC)s is quite different: with the exception of tRNAAla(GAC), the koff values of the tRNA(GAC)s were unaffected by phenylalanylation. These aa-tRNAs display an average koff of 4.5 ± 1.2 × 10−3 min−1, which is within error of the average for the deacyl-tRNA(GAC)s (4.9 ± 1.0 × 10−3 min−1).
Taken together, the data in Table 1 appear to suggest that tRNA binding to the ribosomal A site cannot be understood in terms of a model that assigns consistent thermodynamic values to the amino acid, the tRNA body, or the anticodon. For example, if one evaluates the contribution of the esterified phenylalanine by the difference between the aminoacyl and deacyl tRNAs, then Phe seems to contribute at least 50-fold to the binding of Phe-tRNAAla(GAA), only 7-fold to the binding of Phe-tRNAAla(GAC), and nearly nothing to the remaining tRNA(GAC)s. Similarly, tRNA body effects are large among the deacyl tRNA(GAA)s, but disappear with the Phe-tRNA(GAA)s or either form of the tRNA(GAC)s. In other words, the thermodynamics of tRNA binding to the ribosomal A site appears complex and non-additive.
A clue for understanding this apparent non-additivity is that there seems to be a maximal stability for aa-tRNA binding to the A site. The koff values for all 24 of the Phe-tRNAs in Table 1 are nearly the same (3.2 × 10−3 to 9.5 × 10−3 min−1) and are similar to the narrow range of koff values previously determined for eight fully modified tRNAs aminoacylated with their cognate amino acid (2.7 × 10−3 to 6.4 × 10−3 min−1, Ref. 1), as well as to 12 unmodified misacylated tRNAs esterified with different amino acids (4.2 × 10−3 to 19 × 10−3 min−1, Ref. 8). Although removing the amino acid or the posttranscriptional modifications can significantly increase the koff value (1), an aa-tRNA with a slower koff has never been observed. This lower threshold in the value of koff is not some limitation of the assay itself. Similar filter binding experiments measuring dissociation of a 5′-[32P]-labeled mRNA fragment give koff values ∼10-fold slower than the threshold (17; data not shown). In addition, the koff values of several aa-tRNAs can be reduced below the threshold by adding paromomycin (data not shown), a drug that increases the stability of tRNA in the A site (26). Thus, the lower threshold in koff values is a characteristic of the interaction between the A site and its aa-tRNA substrates and not the assay. The existence of this threshold explains the apparent non-additive effects discussed above. When a tRNA interacts with the A site in a way that is stable enough to reach the threshold, the effects of structural features that further improve the stability are not observed. For example, most of the deacyl tRNAPhe(GAC)s have already reached the maximal stability, so the stabilization due to the phenylalanine is not observed.
If the threshold in koff values is masking elements stabilizing the interaction between a tRNA and the A site, it should be possible to reveal these elements by destabilizing the complex by some other means. A convenient way to do this is to introduce a 2′-deoxyribose nucleotide into the A site codon. The X-ray crystal structures of the 30S ribosomal subunit indicate that all three 2′-hydroxyl groups in the A site codon participate in hydrogen bonding with the ribosomal RNA bases G530, A1492 and A1493 (9). Indeed, when individual 2′-deoxyribose substitutions were introduced into different positions in the A site codon, the koff of tRNAVal(GAC) increased from 3- to 7-fold, depending upon the position of the deoxyribose substitution (24). Since only the interaction with rRNA is affected, the binding of the tRNA to the A site is destabilized without significantly compromising the integrity of the anticodon•codon helix. Considering this, koff values of four tRNA bodies with either GAA or GAC anticodons were determined using cognate codons which contained a deoxynucleotide in the central position (Table 2). As expected, ribosomal binding of all eight of the deacyl tRNAs was destabilized by the deoxynucleotide-substituted codon. In the case of the deacyl tRNA(GAA)s, binding was destabilized sufficiently to make their koff values difficult to measure. However, the four deacyl tRNA(GAC)s were destabilized from 3- to 13-fold, such that they no longer bound at the threshold of koff values. Quite strikingly, when the eight tRNAs were aminoacylated with phenylalanine, all of their koff values were reduced to the threshold value. Therefore, for the tRNA(GAA)s, the stabilizing effect of the esterified phenylalanine appears much greater when assayed with the UdUC codon than with the UUC codon. In addition, for the tRNA(GAC)s, the stabilizing effect of the phenylalanine that was not seen using the GUC codon is revealed (‘unmasked’) when the destabilizing GdUC codon is used. A similar, but more pronounced, effect was observed when the tRNA stabilities were measured in the presence of the double-2′-deoxynucleotide codon GdUdC (data not shown). While these experiments clearly established that the phenylalanine does indeed contribute to the stability of binding of Phe-tRNA(GAC)s to the A site, the tRNA(GAA) data suggests that the amount that the phenylalanine contributes to binding is not known and may be greater than previously thought. Thus, the existence of the threshold in measured koff values complicates the use of this assay to quantitatively evaluate individual structural features that contribute to the binding of aa-tRNA to the A site.
Table 2.
Effects of aminoacylation and anticodon substitution on koff (min−1 × 103) from A site using codons containing a deoxynucleotide at the central position
| tRNA | GAA anticodon |
GAC anticodon |
||
|---|---|---|---|---|
| Deacyl- | Phe- | Deacyl- | Phe- | |
| tRNA2Ala | >500 | 8.4 (±3.3) | 88 (47) | 7.0 (2.2) |
| tRNA2Arg | 330 (64) | 7.2 (1.4) | 46 (22) | 7.2 (1.4) |
| tRNAPhe | >500 | 13 (2) | 53 (15) | 6.3 (2.3) |
| tRNA2AVal | >500 | 11 (5) | 61 (30) | 6.1 (2.2) |
All koff values were measured with complementary codons. Values (±SD) are for at least three separate experiments. 0.5 min−1 is the upper limit of the assay.
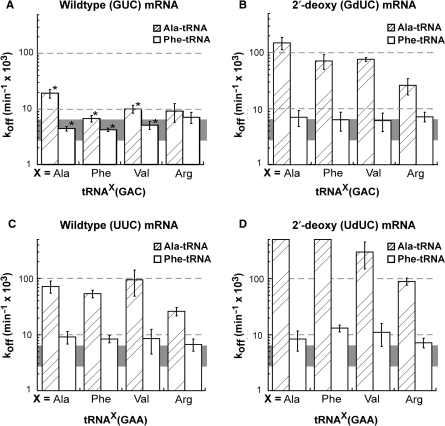
The results of the above experiments prompted a reinterpretation of our previously published experiments which used misacylated aa-tRNAs to demonstrate that the ribosomal A site showed no specificity for the esterified amino acid (8). Since these experiments were performed with tRNA(GAC)s to permit their misacylation and thus showed koff values at the threshold, it now seems likely that any amino acid specificity effects were masked. Therefore, to reassess whether the ribosome shows specificity for the esterified amino acid, koff values for the alanylated forms of the eight tRNAs examined in Table 2 were determined using either all ribose or 2′-deoxynucleotide codons. By lacking the aromatic ring, alanine would not be able to stack on A2486 in the ribosomal A site and thus would be expected to bind less well. As reported in Table 3 and shown graphically in Figure 3, most of the Ala-tRNAs do indeed dissociate faster than their Phe-tRNA counterparts, clearly establishing that the ribosomal A site does show specificity for the esterified amino acid. The only exceptions were three of the tRNA(GAC)s paired with the all-ribose codon, where both the alanylated and the phenylalanylated forms reached the threshold and thus showed no difference. Interestingly, two of these three tRNAs were the very ones which we had used earlier to conclude that the A site showed no amino-acid specificity (8). The narrow dynamic range of the koff assay makes it difficult to accurately determine the relative stabilizing effect of phenylalanine versus alanine. However, if only those Phe-tRNAs that exceed the threshold and those Ala-tRNAs that are not too weak to be measured are considered, an average of 11-fold difference in koff between esterified Phe and Ala is obtained. Thus, as expected, phenylalanine ring stabilizes the binding of aa-tRNAs to the A site.
Table 3.
Comparison of aminoacylation of alanine and phenylalanine on koff (min−1 × 103) from A site
| tRNA | GAA anticodon |
GAC anticodon |
||||||
|---|---|---|---|---|---|---|---|---|
| UUC mRNA |
UdUC mRNA |
GUC mRNA |
GdUC mRNA |
|||||
| Ala- | Relative koff Ala-/Phe- | Ala- | Relative koff Ala-/Phe- | Ala- | Relative koff Ala-/Phe- | Ala- | Relative koff Ala-/Phe- | |
| tRNA2Ala | 73 (18) | 8 | >500 | ≥60 | 19 (3) | 4 | 150 (37) | 21 |
| tRNA2Arg | 26 (4) | 4 | 89 (13) | 12 | 9.1 (3.4) | 1 | 26 (8) | 4 |
| tRNAPhe | 54 (8) | 6 | >500 | ≥38 | 6.7 (0.9) | 2 | 71 (21) | 11 |
| tRNA2AVal | 95 (46) | 11 | 300 (150) | 27 | 9.7 (1.6) | 2 | 76 (6) | 12 |
Figure 3.
Alanylated tRNAs are less stable in the ribosomal A site than phenylalanylated tRNAs. (A) GAC anticodon, all-ribose mRNA (GUC). (B) GAC anticodon, 2′-deoxy mRNA (GdUC). (C) GAA anticodon, all-ribose mRNA (UUC). (D) GAA anticodon, 2′-deoxy mRNA (UdUC). As in Figure 2, the gray horizontal band defines the tight-binding threshold while 0.5 min−1 is the weakest binding that the assay can detect.
Finally, the data in Figure 3 suggest a small but consistent effect of the tRNA body on the stability of aa-tRNA in the A site. In cases where koff is not at the threshold and when the amino acid is held constant, tRNAAla derivatives are consistently weaker and tRNAArg derivatives are consistently tighter than the other two tRNAs. This agrees with the somewhat larger differences seen with the deacylated tRNA(GAA) versions of the same four tRNAs seen in Table 1.
DISCUSSION
In an attempt to dissect the multiple structural features of an aa-tRNA that contribute to the energetics of aa-tRNA•A site binding, the koff assay was used here to evaluate a set of tRNAs with differing anticodons and esterified amino acids. While the data indicate that the ribosome shows specificity for the esterified amino acid and the tRNA body, the data cannot be explained in terms of a set of additive contributions to the overall affinity. Instead, it appears that aa-tRNAs show a minimal attainable value for koff. When this threshold of maximal stability is reached, the effects of additional structural elements known to stabilize the binding of aa-tRNA to the A site are not observed. Thus, no added stability of an esterified amino acid was seen for tRNAs with tight codon•anticodon interactions and no effect of a tighter codon•anticodon interaction could be seen for tRNAs esterified with a tight amino acid. Conversely, when the complex of an aa-tRNA to the A site is sufficiently destabilized, the ‘hidden’ thermodynamic effect of such structural elements is revealed. For example, mRNAs containing 2′-deoxynucleotide substituted codons showed a difference in koff between esterified alanine and phenylalanine.
What, physically, does this threshold effect represent? A likely possibility is that the ribosome must undergo a conformational change in order to release the A site-bound aa-tRNA from the ribosome. If the rate of this isomerization became limiting in our assay, it would result in the narrow range of koff values for any aa-tRNA that can fully induce the conformational change. Alternatively, the threshold of aa-tRNA dissociation could be the result of some other pathway, such as transient dissociation of the ribosomal subunits and release of the aa-tRNA. However, in the high magnesium concentrations used in this study, the 70S ribosome is fully formed, and uncatalysed rates of subunit exchange are very slow (27–29), so we consider this pathway less likely. Indeed, several conformational rearrangements by both the ribosome and the aa-tRNA during the normal mechanism of delivery of aa-tRNA to the A site are well established. X-ray crystal structures show that the 30S ribosomal subunit undergoes a change from an ‘open’ to a ‘closed’ conformation upon binding a cognate anticodon stem loop (26). Also, binding of an aminoacyl-tRNA analog to the A site has been shown to induce a conformational change in the 50S subunit, where an induced-fit mechanism in the peptidyl transferase center was proposed to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA (30). Differences in the 70S ribosome structure with and without bound tRNA further suggest that a conformational change occurs (31). Moreover, cryoelectron microscopy and flourescence data indicate that the 70S ribosome undergoes at least one isomerization upon ternary complex binding (32,33). Lastly, conformational changes have been proposed to occur when the aa-tRNA moves from EF-Tu into the A site (34–37). Thus, it is reasonable to posit that a conformational change is also required for the slow, non-enzymatic release of aa-tRNA from the A site and that this could limit the measured koff.
It has been proposed that the conformational rearrangements that occur during aa-tRNA delivery to the A site during normal translation are part of an induced fit mechanism that has evolved to prevent near-cognate aa-tRNAs from participating in peptide bond formation (26,35–39, reviewed in 40). The concept of induced fit asserts that the binding of the correct substrate by an enzyme causes a structural change that promotes the active or productive state of the enzyme, while the binding of incorrect substrates does not (41). When a ternary complex containing a cognate aa-tRNA binds to the ribosome, the 30S and 50S ribosome structures are proposed to rearrange to stimulate GTP hydrolysis in EF-Tu (33,35). We propose here that a similar induced fit mechanism describes the binding of aa-tRNAs to the ribosomal A site. In such a model, an aa-tRNA must contain a minimal set of structural elements to promote and maintain the conformational change in the 70S ribosome needed for A site binding. Such a model can explain the koff data we observe. For example, deacylated tRNAs with weaker anticodons would be unable to fully promote the 70S conformational change. As a result, these tRNAs interact with an ‘open’ conformation of the ribosome and display a range of koff values that is dependent on the thermodynamic contributions available from the tRNA body and the anticodon. However, when a sufficiently stabilizing amino acid or a more stabilizing anticodon is introduced, the conformational change is induced and the ribosome adopts a ‘closed’ conformation and koff reaches the threshold. Such tRNA-induced ribosomal conformational change can explain the non-additivity of tRNA structural components observed in the koff experiments.
The induced fit mechanism for aa-tRNA binding to the A site proposed here is clearly distinct from the induced fit mechanism proposed to occur during the selection of the cognate ternary complex during decoding. However, the two mechanisms share an important structural feature: in both cases, the complex sets of interactions that occur between the anticodon stem loop of the tRNA, the codon and the 16S rRNA are likely to be very similar, if not identical. Extensive structural and biochemical (23,24) experiments on aa-tRNA in the A site support the fact that 16S rRNA makes contacts on the minor groove side of the codon-anticodon helix (9,26). Although no high resolution structure for the ternary complex bound to ribosomes is yet available, biochemical (42) and lower resolution cryoelectron microscopy (33) data indicate that most, if not all, of the same contacts form. While a careful comparison has not yet been made, it is likely that these contacts in the small ribosomal subunit dominate the induced conformational change in both cases. This possibly explains why the results of many off-pathway koff and KD experiments correlate well with the results of on-pathway kinetic experiments performed on the same tRNAs (21,22).
A central conclusion of this work is that the presence and identity of the esterified amino acid on aa-tRNA contribute to the A site binding affinity of aa-tRNA. This observation is consistent with previous biochemical data showing that most deacylated tRNAs bind ribosomes less well than aa-tRNAs (1). The importance of the esterified amino acid identity is also confirmed by assays which show that aa-tRNA analogs bind the ribosome with variable affinities (4). Although we only demonstrated specificity in binding between alanine and phenylalanine, the asymmetric amino acid binding pocket of the ribosome makes it likely that each individual amino acid side chain will contribute differently to A site binding and thus promote ribosome isomerization to different degrees. Our earlier conclusion that no such specificity existed (8) was the result of using misacylated tRNAs with only GAC anticodons. The presence of this strong anticodon was sufficient to induce the ribosomal conformational change, and additional amino acid side chain effects were not observed.
Although the presence and identity of the esterified amino acid is known to be important for the initial binding of aa-tRNA to EF-Tu (12), it seems unlikely that the identity of the esterified amino acid further contributes to the induced fit of the ternary complex during decoding. Throughout the decoding process, the amino acid side chain is believed to remain bound within a pocket in EF-Tu, and thus does not directly interact with the ribosome. However, it seems possible that the identity of the amino acid side chain could influence the rate of accommodation when the 3′ end of the tRNA is released from EF-Tu and passes through the large ribosomal subunit to reach the peptidyl transferase center. If this is true, certain misacylated tRNAs would be expected to show altered rates of decoding. It will be interesting to test this prediction.
The experiments in Tables 1–3 also show varying differences in koff when different tRNA bodies are compared. This effect is most easily seen by comparing deacyl tRNAs, but it can also be seen among aminoacyl tRNAs. However, these comparisons were made using tRNAs containing mutated anticodons, and it is known that anticodon ‘context’ can affect ribosome binding (3). Thus, one cannot conclude from these data whether the observed koff differences are an intrinsic property of the entire tRNA body or simply reflect how well the bodies were able to accommodate the substituted anticodon. Nevertheless, since previous experiments comparing fully modified deacyl tRNAs with cognate codons report significant differences in koff values (1), it seems likely that the A site does bind tRNA bodies with differing affinities.
The A site binding of the transcribed and purified versions of eight of the tRNA bodies used in this study has been measured previously in the presence of their respective parent anticodon (1,8). While the stabilities of seven of the tRNAs are essentially unaffected by removal of their posttranscriptional modifications, transcribed tRNAGlu(UUC) was at least 10-fold less stable in the A site than purified, fully modified tRNAGlu(UUC) (1). In addition, aminoacylation with glutamate has less than a 2-fold effect on either the transcribed or purified form of the tRNA, indicating that the main contributor to A site:tRNAGlu(UUC) binding is the posttranscriptional modifications. Interestingly, we find that this requirement for modification can be alleviated by either introducing a more stable anticodon into the body or by aminoacylating the tRNA with an amino acid belonging to a parent tRNA which requires an amino acid. For example, the stability of tRNAGlu(GAC) in the A site reached the threshold without posttranscriptional modifications or an esterified amino acid. Also, the deacylated form of tRNAGlu(GAA) is as weak as transcribed tRNAGlu(UUC), but the threshold can be reached by aminoacylating with Phe. Thus, the various components of an aa-tRNA (anticodon, tRNA sequence, tRNA modifications, and amino acid) and their respective contributions to aa-tRNA stability in the A site may be interchangeable.
The possibility that different components of an aa-tRNA can be substituted for (and, as such, may compensate for) one another implies that the contribution of each component is independent of the contribution of the others. Although this may be true, the threshold complicates this analysis and disallows a true determination of independence or cooperativeness among aa-tRNA components in A site binding. These results highlight the difficulty of determining actual thermodynamic values for the contributions of the esterified amino acids, as well as the tRNA body and anticodon, to the aa-tRNA•A site interaction.
The experiments reported in this article do not alter the major conclusion of Fahlman et al. (1) that aa-tRNAs show uniform binding to the ribosomal A site and that each aa-tRNA uses a different ‘strategy’ to achieve uniformity. However, the data do alter the interpretation of what uniform binding actually reflects. Structural components of each aa-tRNA, including the anticodon stem loop, the esterified amino acid, and the remainder of the tRNA body, act together to induce a ribosomal conformation change to form the A site complex. All fully modified aa-tRNAs can induce this conformational change if a cognate codon is provided. As a result, they all show a similar ‘threshold’ koff value that reflects the conformational change. However, the way that each individual aa-tRNA combines its structural components to induce the conformational change varies considerably. As a result, altering or deleting parts of a given aminoacyl tRNA can have very different effects upon the value of koff.
FUNDING
This work was supported by National Institutes of Health (grant #R01-GM37552-19 to O.C.U) and the Foundation for Polish Science (to M.O.). Funding for open access charge: The National Institutes of Health (R01-GM37552-19).
Conflict of interest statement. None declared.
REFERENCES
- 1.Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 2.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nature Struct. Mol. Biol. 2005;12:788–793. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 4.Bhuta A, Quiggle K, Ott T, Ringer D, Chladek S. Stereochemical control of ribosomal peptidyltransferase reaction. Role of amino acid side-chain orientation of acceptor substrate. Biochemistry. 1981;20:8–15. doi: 10.1021/bi00504a002. [DOI] [PubMed] [Google Scholar]
- 5.Starck SR, Qi X, Olsen BN, Roberts RW. The puromycin route to assess stereo- and regiochemical constraints on peptide bond formation in eukaryotic ribosomes. J. Am. Chem. Soc. 2003;125:8090–8091. doi: 10.1021/ja034817e. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 7.Schmeing TM, Seila AC, Hansen JL, Freeborn B, Soukup JK, Scaringe SA, Strobel SA, Moore PB, Steitz TA. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nature Struct. Biol. 2002;9:225–230. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 8.Dale T, Uhlenbeck OC. Binding of misacylated tRNAs to the ribosomal A site. RNA. 2005;11:1610–1615. doi: 10.1261/rna.2130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogle JM, Brodersen DE, Clemons W.M., Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 10.Asahara H, Uhlenbeck OC. The tRNA specificity of thermus thermophilus EF-Tu. Proc. Natl Acad. Sci. USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale T, Sanderson LE, Uhlenbeck OC. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004;43:6159–6166. doi: 10.1021/bi036290o. [DOI] [PubMed] [Google Scholar]
- 13.Wolfson AD, Uhlenbeck OC. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc Natl. Acad. Sci. USA. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 15.Wolfson AD, Pleiss JA, Uhlenbeck OC. A new assay for tRNA aminoacylation kinetics. RNA. 1998;4:1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahlman RP, Uhlenbeck OC. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- 18.Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 19.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 20.Nirenberg M, Leder P. RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of sRNA to ribosomes. Science. 1964;145:1399–1407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- 21.Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kothe U, Rodnina MV. Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell. 2007;25:167–174. doi: 10.1016/j.molcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Phelps SS, Jerinic O, Joseph S. Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol. Cell. 2002;10:799–807. doi: 10.1016/s1097-2765(02)00686-x. [DOI] [PubMed] [Google Scholar]
- 24.Fahlman RP, Olejniczak M, Uhlenbeck OC. Quantitative analysis of deoxynucleotide substitutions in the codon-anticodon helix. J. Mol. Biol. 2006;355:887–892. doi: 10.1016/j.jmb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Satoh A, Takai K, Ouchi R, Yokoyama S, Takaku H. Effects of anticodon 2'-O-methylations on tRNA codon recognition in an Escherichia coli cell-free translation. RNA. 2000;6:680–686. doi: 10.1017/s1355838200000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 27.Hirashima A, Kaji A. Factor-dependent release of ribosomes from messenger RNA. Requirement for two heat-stable factors. J. Mol. Biol. 1972;65:43–58. doi: 10.1016/0022-2836(72)90490-1. [DOI] [PubMed] [Google Scholar]
- 28.Pande C, Wishnia A. Pressure dependence of equilibria and kinetics of Escherichia coli ribosomal subunit association. J. Biol. Chem. 1986;261:6272–6278. [PubMed] [Google Scholar]
- 29.Pavlov MY, Antoun A, Lovmar M, Ehrenberg M. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 2008;27:1706–1717. doi: 10.1038/emboj.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmeing T, Huang K, Strobel S, Steitz T. An induced fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 31.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 32.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nature Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 34.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nature Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 35.Gromadski KB, Rodnina MV. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nature Struct. Mol. Biol. 2004;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- 36.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 37.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nature Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 39.Sanbonmatsu KY. Alignment/misalignment hypothesis for tRNA selection by the ribosome. Biochimie. 2006;88:1075–1089. doi: 10.1016/j.biochi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 41.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl Acad. Sci. USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol. Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]