Abstract
We present a simple technique for visualizing replication of individual DNA molecules in real time. By attaching a rolling-circle substrate to a TIRF microscope-mounted flow chamber, we are able to monitor the progression of single-DNA synthesis events and accurately measure rates and processivities of single T7 and Escherichia coli replisomes as they replicate DNA. This method allows for rapid and precise characterization of the kinetics of DNA synthesis and the effects of replication inhibitors.
INTRODUCTION
DNA replication is a fundamental biological process that requires the coordinated activities of a large number of enzymes organized in a multiprotein assembly termed the replisome. Using a variety of systems and approaches, researchers aim to understand how the various proteins work together and how their interactions affect replication activities (1–4). Traditionally, biochemical DNA replication assays rely on agarose gel electrophoresis and/or the incorporation of radioactive nucleotides to measure rates of DNA synthesis and processivity of replication events. From a practical standpoint, these techniques involve multiple time-consuming steps as well as the use of hazardous materials. Furthermore, these techniques average over a large ensemble of molecules, making it inherently difficult to obtain distributions of enzymatic rates and processivities, essential data for complete characterization of replisomal activities. Accurate determination of processivity typically involves either trapping or dilution techniques, both of which can inadvertently affect reaction concentrations and dissociation kinetics. Also, rare events and short-lived intermediates are not easily observed in bulk-phase techniques, obscuring some of the dynamic interactions and states that occur during DNA replication.
Information on rate, processivity and short-lived intermediate states can be obtained by observing individual replisomes at the single-molecule level. In recent years, numerous single-molecule techniques have been developed to characterize the activity of nucleic-acid enzymes such as DNA polymerases and helicases (5–9). Most studies have relied on the mechanical manipulation of individual proteins or on the imaging of fluorescent tags to report on the catalytic activity of replication proteins. Although quite powerful, these techniques are generally limited to observing a single-DNA substrate at a time, making it difficult to accumulate statistics for larger multiprotein complexes. Furthermore, the experimental setups can be prohibitively complex and expensive for the general user. Recently, we have demonstrated a multiplexed single-molecule replication assay based on flow-stretching of tethered bacteriophage λ DNA and used the technique to characterize helicase–primase and helicase–polymerase interactions in leading-strand synthesis (10–12). In these experiments, the difference in extension of single-stranded (ss) and double-stranded (ds) DNA at low pN forces is exploited to observe an enzyme acting on DNA. The requirement for a net conversion between ss and dsDNA makes it challenging to apply this method to coordinated replication where both DNA strands at the fork are copied. Using replication loops as an observable, we have recently used this flow-stretching technique to study the processes controlling the dynamics of loop formation at the T7 replication fork (13). Nevertheless, these experiments present only an indirect readout of replication-fork dynamics.
Here, we present a simple single-molecule assay for measuring coordinated DNA replication by individual replisomes in real time. We employ the rolling-circle DNA amplification scheme, as it facilitates highly processive DNA synthesis. Rolling-circle replication has been proven a useful method for monitoring real-time synthesis in bulk-phase assays (14–16) and at the single-molecule level to characterize telomere mimic sequence ssDNA (17). Briefly, we couple the 5′-end of the lagging strand of a rolling-circle substrate to the surface of a flow chamber and introduce replication components into the flow cell to initiate DNA synthesis. By applying a constant, laminar flow through the chamber we hydrodynamically stretch the growing DNA. Application of a low concentration of intercalating stain during the reaction allows us to directly image the time-dependent length of dozens of growing DNA molecules in real time. As a proof of principle, we characterize fully reconstituted replisomes from two systems, bacteriophage T7 and Escherichia coli, some of the largest protein complexes studied to date at the single-molecule level. Bacterial and viral DNA replication are obvious targets for therapeutic inhibition, and this assay readily lends itself to studying replication inhibitors, a principle demonstrated by examining the effects of a dideoxynucleotide triphosphate on the rate and processivity of T7 DNA replication.
MATERIALS AND METHODS
Rolling-circle substrate preparation
Substrate was prepared as described previously (18). A 66-mer tail oligonucleotide 5′-Biotin-T36AATTCGTAATCATGGTCATAGCTGTTTCCT-3′ (Integrated DNA Technologies) was annealed to M13mp18 ssDNA (New England Biolabs) and extended with 64 nM T7 polymerase in reaction buffer (see below) at 37°C for 12 min. The reaction was quenched with 100 mM EDTA, cooled, and the filled M13 extracted via phenol–chloroform extraction.
Coverslip surface preparation
To minimize non-specific interactions between the glass surface and proteins, we covalently couple high-molecular-weight polyethylene glycol (PEG) to the surface, including a small fraction of biotin moieties for tethering of our DNA substrates. First, the glass is coupled to the alkoxy group of an aminosilane, creating a surface with reactive amine groups that can subsequently be coated with a polymer of choice (19). By coupling a mixed population of biotinylated and non-biotinylated succinimidyl propionate-PEG to the amine-functionalized glass, we coat the coverslip in a layer of PEG displaying a mixture of biotin and non-reactive methyl groups. The biotinylated coverslip is incubated with a 1 mg/ml solution of streptavidin (Sigma) in PBS (pH 7.5) immediately before use in the experiment.
Flow-cell construction
Chamber construction was done as described previously (12). Briefly, a 2-mm channel was cut out of double-sided adhesive tape (Grace BioLabs) and attached to a quartz slide with pre-drilled holes and a functionalized coverslip (12,19). Polyethylene tubes (0.76 mm inner diameter, 1.22 mm outer diameter; Becton Dickinson) were inserted into the holes, and the entire chamber was sealed with quick-dry epoxy. The flow cell was incubated with 20 mM Tris pH 7.5, 2 mM EDTA, 50 mM NaCl, 200 μg/ml bovine serum albumin (BSA) and 0.005% Tween to block the surface and further prevent non-specific interactions.
TIRF microscope setup
The flow cell was placed on an Olympus IX-71 inverted microscope and connected to a syringe pump (Harvard Apparatus) used to control the flow of buffer (20). An airspring was inserted between the flow cell and pump to dampen flow fluctuations. SYTOX Orange (Invitrogen), present in the replication buffer was excited through a 60× oil-immersion TIRF objective (Olympus, NA = 1.45) with a 532 nm solid-state laser (Coherent Compass 215M-75). The resulting fluorescence was collected back through the objective, passed through an emission filter (Chroma HQ600/75 m) to eliminate residual laser light and recorded on an EM–CCD (Hamamatsu).
Replication reactions
Substrate DNA was flowed into the chamber at 25 pM for 30 min at 0.025 ml/min and excess DNA was removed via washing with buffer. For reactions performed at 37°C, a home-built aluminum block was affixed to the top quartz surface with high thermal conductivity paste and heated with a cartridge heater (Omega) controlled by variable power supply (Elenco). By measuring the buffer temperature in the flow chamber we were able to control the cartridge heater to achieve 37°C at the observed region.
T7 reaction
Replication reactions were performed and proteins purified as described previously (13,21,22). Reaction buffer contained 40 mM Tris pH 7.5, 50 mM potassium glutamate, 10 mM magnesium chloride (MgCl2), 100 μg/ml (BSA) with 5 mM dithiothreitol (DTT), 600 μM deoxyribonucleotides (dNTPs, New England Biolabs), 300 μM ATP (Sigma), 300 μM CTP and 15 nM SYTOX Orange added immediately before beginning the reaction. Proteins were added as: 5 nM gp4 (hexameric), 40 nM polymerase (1:1 gp5: thioredoxin) and 360 nM gp2.5. DideoxyGTP (ddGTP, Roche) was added at indicated concentrations immediately before starting the reactions.
E. coli reaction
DnaB helicase, DnaC helicase loader, DnaG primase, αεθ polymerase, β processivity clamp and the τ2γ1δδ′χψ and τ3δδ′χψ clamp loader assemblies were purified as described (12). PriA, PriB, DnaT and SSB were prepared from overproducing strains by methods similar to those described by Marians (23). Replication reactions were performed as described previously (12,24). Reaction buffer contained 50 mM HEPES pH 7.9, 12 mM magnesium acetate (MgOAc2), 80 mM potassium chloride (KCl) and 100 μg/ml BSA with 10 mM DTT, 40 μM dNTPs, 200 μM UTP, GTP and CTP, 1 mM ATP and 15 nM SYTOX Orange added before the reaction. Proteins were added as: 30 nM DnaB (hexameric), 180 nM DnaC (monomeric), 30 nM αεθ, 15 nM τ2γ1δδ′χψ or τ3δδ′χψ, 30 nM β (dimeric), 300 nM DnaG, 250 nM SSB (tetrameric), 20 nM PriA, 40 nM PriB and 480 nM DnaT.
Data analysis
To obtain length trajectories and minimize the contribution from transverse Brownian fluctuations of the DNA, we calculated intensity projections by summing over a narrow rectangular box of pixels (∼2 μm wide) along the length of the DNA. The end of the DNA was defined by an intensity threshold set to 65% of the maximum intensity. Linear interpolation around the threshold determined the DNA end position to sub-pixel resolution. Rate was determined by linear regression of DNA length plotted versus time. Processivity was determined by deconvolving the length of the M13 DNA substrate from the final length of the extended DNA and converted to basepairs by determining the known length of tethered λ-phage DNA (48.5 kb) with our camera and using the calibrated pixel size in basepairs (here, 1 pixel = 785 bp).
RESULTS AND DISCUSSION
Experimental outline
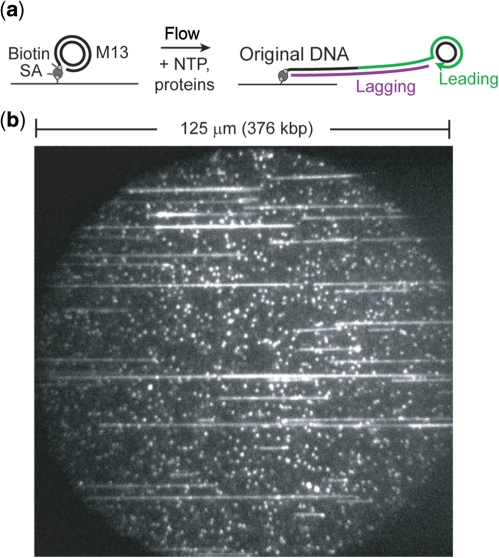
A single-stranded, 7.2-kb circular M13mp18 DNA is primed and its complementary strand synthesized to form dsDNA. Upon completion of synthesis of the entire circular substrate, the polymerase acting at the 3′-end will encounter the 5′-end of the original primer and synthesis will continue by displacing the previously synthesized DNA as ssDNA (Figure 1a). In the presence of protein activities required for priming and other lagging-strand processes, the displaced ssDNA tail will be effectively converted into dsDNA by repetitive priming and synthesis of lagging-strand Okazaki fragments. To allow surface immobilization and single-molecule observation of the replication substrates, we used a 5′-biotinylated ‘tail’ primer in constructing the M13 rolling-circle substrate. This substrate is purified and introduced into a flow chamber constructed with a biotin–streptavidin-functionalized coverslip (12). The filled, surface-attached M13 can serve as a replication template upon addition of nucleotides and proteins. Flowing low picomolar concentrations of M13 DNA into the chamber results in hundreds of DNA molecules in a single field of view (125 μm × 125 μm), each of which can serve as a substrate for the introduced proteins (Figure 1b). As the replication reaction proceeds, the DNA attaching the M13 circle to the surface is extended and stretched fully by hydrodynamic flow of buffer. We are able to visualize individual dsDNA molecules through total-internal reflection fluorescence (TIRF) microscopy by adding SYTOX Orange dsDNA intercalating stain to the reaction buffer, allowing for observation of coupled leading- and lagging-strand synthesis as a processively lengthening, stained dsDNA molecule. Addition of the necessary replication proteins resulted in the lengthening of the M13 template indicating processive DNA synthesis (see Supplementary Data). Trajectories of DNA length as a function of time reported on rates of DNA synthesis (Figure 2), and total product length at the end of synthesis was used to measure processivity.
Figure 1.
(a) Schematic of single-molecule rolling-circle assay. ‘SA’, streptavidin. Upon leading-strand synthesis, DNA is displaced from the circle as the replisome ‘rolls’ around the template. The emerging ‘tail’ is converted to dsDNA via lagging-strand synthesis. As a result, the DNA that couples the M13 circle to the surface increases in length and is extended in the direction of flow. (b) Example field of view. Note both the length and number of products. Each flow cell has thousands of such fields, allowing for large numbers of products to be observed in a single experiment.
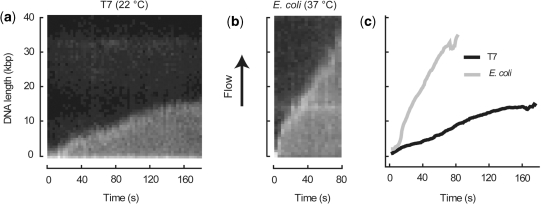
Figure 2.
Kymographs of example DNA molecules from (a) T7 and (b) E. coli replication experiments. Endpoint trajectories are plotted vs. time to obtain rates of synthesis by fitting with linear regression (c). Rates of shown traces are: 99.4 bp s−1 (T7) and 467.1 bp s−1 (E. coli).
T7 DNA replication
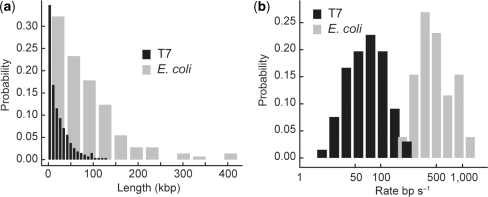
Our single-molecule rolling-circle assay is broadly applicable to general DNA replication. We examined two common model systems of DNA replication, the bacteriophage T7 and E. coli replisomes. After binding our rolling-circle substrate to the surface of the chamber, we introduced the replisomal proteins in replication buffer containing the intercalating dye SYTOX Orange and observed replication as the solution entered the field of view. The T7 replisome can be reconstituted with four proteins: gp4, helicase/primase; gp5 + thioredoxin, polymerase; gp2.5, ssDNA-binding protein (1,13,21) At room temperature (22°C), we observed T7 replication at 75.9 ± 4.8 bp s−1 with an average processivity of 25.3 ± 1.7 kbp (Figure 3). Rates of synthesis were consistent with the previously reported value of 80 bp s−1 at room temperature (13).
Figure 3.
(a) Length distributions of replication products, with means of 25.3 ± 1.7 kbp (T7) and 85.3 ± 6.1 kbp (E. coli). (b) Rate distributions of single molecules, with means of 75.9 ± 4.8 bp s−1 (T7) and 535.5 ± 39 bp s−1 (E. coli). The log of the rates are plotted to allow simultaneous display of both broad distributions.
E. coli DNA replication
The E. coli replisome is significantly more complex (1,2) with 13 core proteins (DnaB, helicase; DnaG, primase; αεθ, polymerase; β, processivity clamp; τ2γ1δδ′χψ or τ3δδ′χψ, clamp loader; SSB, ssDNA-binding protein) and four additional proteins, DnaC, PriA, PriB and DnaT to assist helicase loading on the 5′-blocked DNA substrate (24). Replication by the E. coli replisome at 37°C was observed at 535.5 ± 39 bp s−1, consistent with the previously reported 600 bp s−1 (25–27), with an average processivity of 85.3 ± 6.1 kbp (Figure 3). Replication products with lengths of 350 kbp or greater were observed, demonstrating the high processivity of the E. coli replisome. The dramatic increase in rate and processivity seen with the E. coli as compared to T7 proteins was not merely due to temperature: increasing the temperature of the T7 reaction to 37°C increased the rate to 141.5 ± 8.9 bp s−1 and the processivity to 51.6 ± 9.4 kbp.
Several lines of evidence lead us to conclude that we are observing true coordinated replication. As our assay measures coordinated DNA synthesis by observing replication of the lagging strand, omitting components required for coordination (DnaG primase or χ/ψ using different clamp loader complexes) results in no observable replication. Leading-strand synthesis alone produces no net gain of dsDNA as the circle ‘rolls’, and neither T7 gp5 + thioredoxin or E. coli αεθ is capable of strand invasion synthesis by polymerase activity alone, thus any aberrant or uncoupled replication is not observed in our assay, which visualizes exclusively dsDNA. These facts combined with rate values very similar to those published support our conclusion that each observed replication product is from a single, complete replisome.
Effects of SYTOX and flow stretching DNA
SYTOX Orange is an intercalating agent and can photodamage DNA (28), making it necessary to determine if its inclusion in our assay affects replication. Based on several control experiments, we conclude that for the systems we examined, SYTOX has no effect on replication. As noted above, rates of DNA synthesis for both T7 and E. coli are consistent with previous measures. Bulk-phase rolling-circle assays we performed showed no dependence on the presence or absence or SYTOX in the replication buffer. In other single-molecule experiments, SYTOX has been shown to have no effect on the size of T7 DNA replication loops or rates of synthesis (13). Performing the single-molecule rolling-circle experiment without SYTOX and staining after the reaction was finished yielded identical processivity distributions (E. coli, 85.6 ± 9.5 kbp). Laser excitation had no effect on DNA synthesis, as products from reactions not illuminated until the end of the experiment displayed identical length distributions as those observed in real time (E. coli, 90.1 ± 13.6 kbp). Also, the force acting on the replication proteins themselves is vanishingly small, as they are located at the circle near the distal end of the DNA and experience effectively no force when stretched (29). Confirming this assumption, we performed the E. coli replication reaction in a tube, stopped the reaction by addition of 100 mM EDTA after 20 min and flowed the mixture into the flow cell. Introduction of SYTOX allowed visualization of the force-free reaction products with consistent length distribution (78.2 ± 11.8 kbp).
Analyzing replication inhibitors
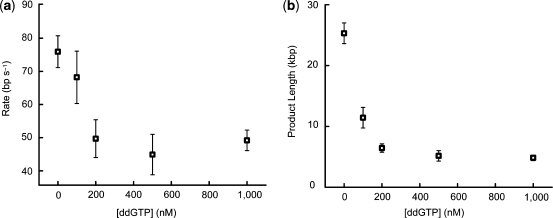
The search for novel antibacterial and antiviral agents remains an important field of research, and DNA replication is an obvious target for pharmacological investigation (30,31). The ease of performing our experiment and the availability of automated fluorescence imaging systems make the assay reported here an ideal method for screening and characterizing replication inhibitors. We demonstrated this concept by measuring rates and processivities of the T7 replication reaction with varying amounts of dideoxyGTP (ddGTP), a chain-terminating nucleotide that can be removed by the exonuclease activity of gp5 (32). Obviously, the incorporation of a single ddGTP depends upon concentration, and each inclusion requires the polymerase to switch between polymerase and exonuclease activities. The relative portion of time needed to excise the dideoxynucleotides increases with the ddGTP concentration and results in a decrease in aggregate rate of DNA synthesis. We observed a rate decrease from 75.9 ± 4.8 bp s−1 in the absence of ddGTP to 49.2 ± 3.1 bp s−1 at 1 μM ddGTP (Figure 4a). In addition, product length decreased from 25.3 ± 1.7 kbp without ddGTP to 4.8 ± 0.5 kbp at 1 μM ddGTP (Figure 4b). It is interesting to note that the decrease in rate and product length scale differently with ddGTP concentration. Product length decreases by ∼5-fold over the concentration range compared to ∼1.5-fold for rate. These results suggest that each ddGTP incorporation increases the probability of replisome dissociation (‘off’ rate), leading to a decrease in total replication processivity. Translating our assay to larger-scale screening or testing would present a simple method for easily analyzed, multiplexed studies of the effects of small molecules on replisomal DNA replication.
Figure 4.
(a) Rates of T7 DNA synthesis at various ddGTP concentrations. (b) Lengths of T7 replication products at various ddGTP concentrations. Values are means and error bars represent standard error of the mean.
CONCLUSIONS
Our assay allows for real-time readout of replication rate and processivity from a single active replisome while increasing dramatically the ease and speed of performing replication experiments with no need for radioactive or hazardous materials. By nature, the rolling-circle method promotes extensive synthesis limited only by the processivity of the replisome rather than the length of the template. This allows us to observe DNA replication up to several hundred kilobasepairs, measuring real values for replisome processivity, numbers difficult to measure precisely with bulk-phase methods due to the exclusion limitations of 50 kbp or fewer in agarose gel electrophoresis. Wide-field imaging allows us to observe hundreds of tethered M13 substrates and study the behaviors of their associated replisomes in a single field of view. Thus, even with modest yields of correct replisomal assembly and activity we are able to construct statistically meaningful (N > 50) rate and processivity histograms from a single 20 min experiment. Components of the assay are made using published methods and can be produced readily in large or reusable quantities. Extension of the assay using multichannel microfluidic devices will allow us to perform a high number of reactions in a short time, a necessary criterion for screening and characterizing the effect of replication inhibitors or other biochemical perturbations. The simplicity of the experiment along with the ability to observe single active replisomes in real time makes our technique a powerful and tractable method for studying the many elements of DNA replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Institutes of Health (GM077248 to A.v.O.); Australian Research Council (to N.E.D.); and Jane Coffin Childs Foundation (to J.J.L.). Funding for open access charge: National Institutes of Health (GM077248).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
T7 replication proteins were a generous gift from Dr Charles Richardson, Harvard Medical School. We are grateful to Charikleia Iaonnou, Allen Lo (University of Wollongong) and Dr Karin Loscha (Australian National University) for the supply of some E. coli proteins.
REFERENCES
- 1.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz RT, O’Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 5.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 6.Seidel R, Dekker C. Single-molecule studies of nucleic acid motors. Curr. Opin. Struct. Biol. 2007;17:80–86. doi: 10.1016/j.sbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Strick T, Allemand J, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 8.van Mameren J, Peterman EJ, Wuite GJ. See me, feel me: methods to concurrently visualize and manipulate single DNA molecules and associated proteins. Nucleic Acids Res. 2008;36:4381–4389. doi: 10.1093/nar/gkn412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha T. Structural dynamics and processing of nucleic acids revealed by single-molecule spectroscopy. Biochemistry. 2004;43:4055–4063. doi: 10.1021/bi049973s. [DOI] [PubMed] [Google Scholar]
- 10.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 11.Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol. Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 2008;15:998. doi: 10.1038/nsmb0908-998a. [DOI] [PubMed] [Google Scholar]
- 13.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–340. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smolina IV, Demidov VV, Cantor CR, Broude NE. Real-time monitoring of branched rolling-circle DNA amplification with peptide nucleic acid beacon. Anal. Biochem. 2004;335:326–329. doi: 10.1016/j.ab.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Smolina IV, Cherny DI, Nietupski RM, Beals T, Smith JH, Lane DJ, Broude NE, Demidov VV. High-density fluorescently labeled rolling-circle amplicons for DNA diagnostics. Anal. Biochem. 2005;347:152–155. doi: 10.1016/j.ab.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Demidov VV. Rolling-circle amplification in DNA diagnostics: the power of simplicity. Expert Rev. Mol. Diagn. 2002;2:542–548. doi: 10.1586/14737159.2.6.542. [DOI] [PubMed] [Google Scholar]
- 17.Pomerantz AK, Moerner WE, Kool ET. Visualization of long human telomere mimics by single-molecule fluorescence imaging. J. Phys. Chem. B. 2008;112:13184–13187. doi: 10.1021/jp806696u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 19.Sofia SJ, Premnath VV, Merrill EW. Poly(ethylene oxide) grafted to silicon surfaces: grafting density and protein adsorption. Macromolecules. 1998;31:5059–5070. doi: 10.1021/ma971016l. [DOI] [PubMed] [Google Scholar]
- 20.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Chastain P.D., 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 22.Hamdan SM, Marintcheva B, Cook T, Lee SJ, Tabor S, Richardson CC. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl Acad. Sci. USA. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marians KJ. Phi X174-type primosomal proteins: purification and assay. Methods Enzymol. 1995;262:507–521. doi: 10.1016/0076-6879(95)62042-7. [DOI] [PubMed] [Google Scholar]
- 24.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 25.O'Donnell ME, Kornberg A. Complete replication of templates by Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 1985;260:12884–12889. [PubMed] [Google Scholar]
- 26.Studwell PS, O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem. 1990;265:1171–1178. [PubMed] [Google Scholar]
- 27.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl Acad. Sci. USA. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akerman B, Tuite E. Single- and double-strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res. 1996;24:1080–1090. doi: 10.1093/nar/24.6.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anselmi C, DeSantis P, Scipioni A. Nanoscale mechanical and dynamical properties of DNA single molecules. Biophys. Chem. 2005;113:209–221. doi: 10.1016/j.bpc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Berdis AJ. DNA polymerases as therapeutic targets. Biochemistry. 2008;47:8253–8260. doi: 10.1021/bi801179f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange RP, Locher HH, Wyss PC, Then RL. The targets of currently used antibacterial agents: lessons for drug discovery. Curr. Pharm. Des. 2007;13:3140–3154. doi: 10.2174/138161207782110408. [DOI] [PubMed] [Google Scholar]
- 32.Tabor S, Richardson CC. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.