Abstract
Xeroderma pigmentosum (XP) is an autosomal recessive genetic disorder. Afflicted patients show extreme sun-sensitivity and skin cancer predisposition. XP is in most cases associated with deficient nucleotide excision repair (NER), which is the process responsible for removing photolesions from DNA. Measuring NER activity by nucleotide incorporation into repair patches, termed ‘unscheduled DNA synthesis (UDS)’, is one of the most commonly used assays for XP-diagnosis and NER research. We have established a rapid and accurate procedure for measuring UDS by replacement of thymidine with 5-ethynyl-2'-deoxyuridine (EdU). EdU incorporated into repair patches can be directly conjugated to fluorescent azide derivatives, thereby obviating the need for either radiolabeled thymidine or denaturation and antibody detection of incorporated bromodeoxyuridine (BrdU). We demonstrate that the EdU incorporation assay is compatible with conventional techniques such as immunofluorescent staining and labeling of cells with micro-latex beads. Importantly, we can complete the entire UDS assay within half a day from preparation of the assay coverslips; this technique may prove useful as a method for XP diagnosis.
INTRODUCTION
Cells are continuously exposed to various types of DNA damage induced by both endogenous and exogenous sources. Consequently, all living organisms have developed DNA repair mechanisms to ensure that the integrity of the genetic information is maintained (1). Nucleotide excision repair (NER) is one of the most versatile DNA repair systems, which deals with the major UV photoproducts, as well as many DNA adducts arising from exposure to a variety of chemical carcinogens (2,3). NER involves removal of damaged nucleotides from the DNA by dual incisions, and filling and sealing of the residual single-strand gaps by DNA polymerases and ligases (1–3).
Xeroderma pigmentosum (XP) is the prototype NER-deficient genetic disorder (2–4), the incidence of which is 1 in 80 000 to 1 000 000 in the general population, depending on the location (5). XP patients suffer from severe photosensitivity, a high incidence of sunlight-induced skin cancer and in some cases neurological abnormalities. In ∼80% of XP cases, the patients have a defect in one of the genes responsible for NER. XP-variant patients, who comprise the residual 20% of XP cases, are proficient in NER but have a defect in post-replication repair (PRR) as they lack the specialized DNA polymerase, polη, which accurately replicates past cyclobutane pyrimidine dimers (CPDs) (6).
Cockayne syndrome (CS) and Trichothiodystrophy (TTD) are also NER-deficient disorders (4,7,8). NER consists of two sub-pathways: global genome repair (GGR) is relatively slow but functions genome wide (3), whereas transcription-coupled repair (TCR) is a faster process that is specialized for the transcribed strand of actively transcribed regions (9,10). With the exception of XPC and XPE, which are used exclusively in GGR (3), all genes defective in patients with XP, the combined features of XP and CS (XP/CS), and TTD are associated with both pathways, whereas in CS only TCR is compromised (7,9–11). NER-deficient patients have been so far assigned to at least 11 complementation groups (XP-A to G, CS-A and B, TTD-A and ERCC1), which correspond to 11 different NER damage recognition and incision proteins (3,4).
The most commonly used assay for assessing NER-deficiency entails measuring nucleotide incorporation levels associated with DNA-repair activity. GGR, which contributes to ∼90% of NER activity, is assessed by damage induced non-S-phase gap-filling DNA replication, termed ‘unscheduled DNA synthesis (UDS)’ (12), while TCR activity can be measured by ‘recovery of RNA synthesis (RRS)’ levels after DNA-damaging treatment (13). Currently, a variety of methods for measuring UDS have been established (8,12,14–17). For inducing DNA damage 254-nm UVC-irradiation is commonly used and cells are then incubated in the presence of either radiolabeled thymidine or a nucleoside analog. The most frequently used methods in XP-diagnostic laboratories are: (i) 3H-thymidine incorporation, followed by evaluation of grain counting after autoradiography of tissue-culture coverslips—this technique is accurate and compatible with most immunostaining-based methods, but the procedures are labor-intensive and time-consuming (15,16). (ii) Bromodeoxyuridine (BrdU) incorporation, followed by alkaline denaturation of DNA, and immunofluorescent detection by anti-BrdU antibody—this is a convenient variant of method (i); replacement of 3H-thymidine by BrdU diminishes the time needed for the assay, however it has reduced sensitivity (∼50% reduction of UDS activity is not detectable) (18). (iii) Liquid scintillation counting of 3H-thymidine incorporation into repair patches—although this method is rapid, the quantitative capacity is lower than the other methods as this is a batch assay. Furthermore, replicative DNA synthesis has to be abolished by using non-dividing cells and the DNA replication inhibitor, hydroxyurea (HU), during the assay (12).
In this report, we have employed 5-ethynyl-2′-deoxyuridine (EdU), an alkyne-conjugated nucleoside analogue of thymidine, as an alternative to radioactive thymidine or BrdU (19). In contrast to the BrdU-based UDS assay, EdU can be directly conjugated to fluorescent azide (20), and thus, the assay does not require denaturing of DNA nor the use of antibodies. We have successfully improved the sensitivity, which is one of the weak points of BrdU-based methods. Furthermore, the total time for the assay was dramatically reduced even compared to the BrdU-based assay. Using this technique, a standard UDS assay can be completed within half a day from preparation of the assay coverslips.
MATERIALS AND METHODS
Cell strains and cultures
All the cells used in this report are human primary fibroblasts. Normal 1BR (21), 48BR (21), 142BR (22) and 251BR (22), XP-patient-derived XP12BR (XP-D) (this study), XP15BR (XP-A) (22), XP13BR (XP-C) (this study) and XP20BE (XP-G) (23) and CS-patient-derived CS10LO (CS-B) (24) cells were used. Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS (Gibco-BRL) and penicillin–streptomycin (Wako), and maintained at semi-confluent density during the experiments unless otherwise stated. For labeling with latex-beads, 48BR cells were cultured with a few drops of 0.5-μm diameter polybead Hydroxylate (Polysciences) in DMEM for 3 days.
UV-induced UDS measurement in normal primary human fibroblasts by EdU incorporation
5-ethynyl-2′-deoxyuridine (EdU) is available from Invitrogen (19). To optimize UV-induced UDS by EdU incorporation, effects of UV dose and EdU-incubation period were examined. 48BR cells were cultured on coverslips and maintained at confluent density. Cells were washed with PBS, followed by irradiation with different doses (5–20 J/m2) of UVC (254 nm). After UV irradiation, cells were immediately incubated with serum-free DMEM supplemented with 10 μM EdU for different periods (0.5, 1, 2 and 4 h). Serum-free medium was used since serum often contains thymidine, which competes with EdU for incorporation into DNA. Cells were then washed with PBS, followed by fixation and permeabilization with PBS containing 2% formaldehyde, 0.5% triton X-100 and 300 mM sucrose for 20 min. After extensive washing with PBS, cells were blocked with 10% FBS in PBS for 30 min. Incorporated EdU was detected by fluorescent-azide coupling reaction (Click-iT, Invitrogen) (19). Briefly, cells were incubated for 30 min with azide-conjugated Alexa Fluor 488 dye in TBS supplemented with 4 mM CuSO4. Cells were then washed three times with PBS containing 0.05% Tween-20 (PBST). Coverslips were finally soaked with PBS, fixed with 3.7% formaldehyde in PBS for 20 min, and mounted on glass slides with Aquapolymount (Polysciences). Photographs of the cells were captured with a fluorescent microscope equipped with a CCD camera (BIOREVO9000-KEYENCE), and captured images were processed and analysed with ImageJ software (NIH). At least 50 non-S-phase cells were randomly selected from a single captured field, and the average nuclear fluorescent intensity was calculated. Data points presented in the text are the averages calculated from five different fields.
We found that 20 J/m2 UV irradiation followed by 2-h EdU incubation was the optimal condition for the UDS assay. This condition was used for all experiments except for Figures 1A and 5.
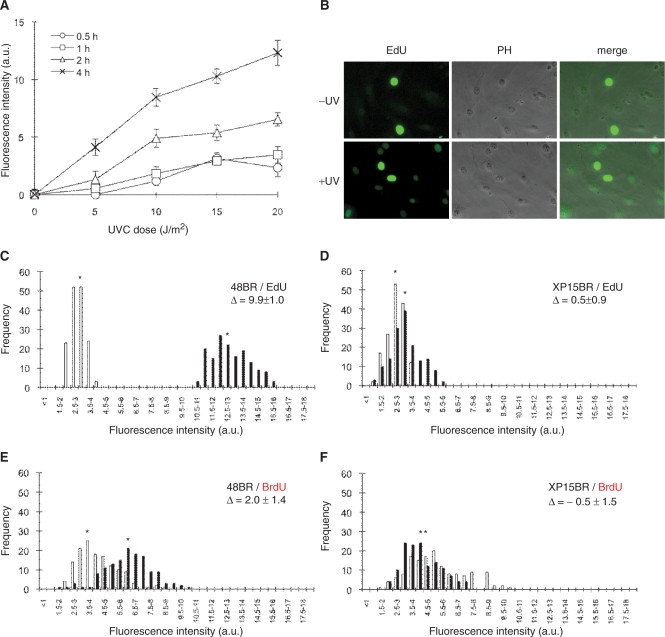
Figure 1.
UV-induced UDS assay in normal human primary fibroblasts. (A) Normal 48BR cells were cultured on coverslips, UVC-irradiated at the indicated doses, followed by incubation with 10 μM EdU for different time periods, fixation and conjugation of fluorescent dye to the incorporated EdU as described in ‘Materials and methods’ section. The intensity of nuclear fluorescence, which is associated with UDS activity, was analysed using a fluorescence microscope and image-processing software. For each data point, at least 250 nuclei were analysed. Points and error bars indicate means of discrete nuclei fluorescent intensity and standard errors, respectively. (B) Typical photos of the EdU assay are shown. 48BR cells were UVC irradiated (20 J/m2, +UV) or mock-treated (−UV), followed by 2-h incubation with EdU. EdU, coupled to Alexa fluor 488-azide; PH, phase contrast. (C–F) UDS performed by incorporation of EdU (C and D) or BrdU (E and F) was compared. XP15BR cells are primary fibroblasts from an XP-A patient. Cells were UVC irradiated (20 J/m2), followed by incubation with 10 μM EdU or 5 μM BrdU for 2 h. Bars represent frequencies of the fluorescence levels in the indicated classes, with (black) or without (white) UVC irradiation. Asterisks indicate the mean values of nuclear fluorescent intensities, which correspond to the UDS levels. Δ represents UDS difference between irradiated and unirradiated samples.
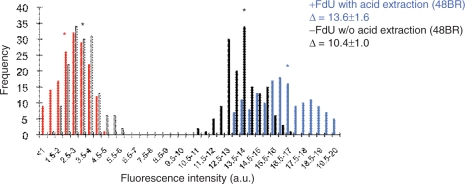
Figure 5.
Improvement of the sensitivity by FdU supplement, longer incubation and acid extraction. Normal 48BR cells were cultured on coverslips, UVC irradiated (20 J/m2), followed by incubation with 10 μM EdU and 1 μM FdU (Sigma) for 4 h. Cells were then fixed as indicated in Figure 2. Acid extraction was performed using Bouin's fixative (Sigma) for 30 min, followed by extensive washing with PBS. Conjugation of fluorescent dye and detection of EdU signals were identical to the experiments in Figure 2. Bars represent frequencies of the fluorescence levels in indicated classes, with (black, identical to Figure 2A; blue, + FdU + 4-h incubation + acid extraction) or without (white, identical to Figure 2A; red, + FdU + 4-h incubation + acid extraction) UVC irradiation. Asterisks indicate the mean values of nuclear fluorescent intensities. Δ represents UDS differences with or without UVC irradiation.
BrdU incorporation assay
Cells were cultured and UVC irradiated (20 J/m2), and incubated in conditions identical to the EdU-incorporation assay described above (5 μM BrdU, instead of EdU, was supplemented). Cells were then washed with PBS, followed by fixation and permeabilization in PBS containing 2% formaldehyde, 0.5% tritonX-100 and 300 mM sucrose for 20 min. After extensive washing with PBS, cells were treated with 4 M HCl in PBS for 15 min for denaturing DNA. Cells were extensively washed with PBS for neutralization, followed by blocking with 10% FBS in PBS for 30 min. Incorporated BrdU was detected by 1-h incubation with mouse anti-BrdU antibody (BD, diluted 1:150 in PBST). Cells were then washed three times with PBST, followed by 1-h incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen, diluted 1:500 in PBST). Coverslips were finally soaked in PBS, fixed with 3.7% formaldehyde in PBS for 20 min, and mounted on glass slides with Aquapolymount (Polysciences). Image capture and analyses were performed as described above.
3H-thymidine incorporation assay
Details of the assay have been previously described (12). Cells were cultured in 1% DMEM for 3 days (3 × 105 cells per 5-cm-diameter dish). They were then incubated for 1 h with 10 mM hydroxyurea (HU), UV irradiated and incubated for a further 3 h with 1% DMEM containing 10 μCi/ml 3H-thymidine and 10 mM HU. 3H-thymidine incorporated into acid-insoluble materials was measured by liquid scintillation counting.
Immunostaining
48BR and XP20BE cells were mix-cultured at 1:1 ratio (48BR alone for ki67 staining). UV irradiation, EdU incubation and fixing and permeabilization steps were as described above. For antibody detection, cells were blocked with 10% FBS in PBS for 30 min, followed by 1-h incubation with mouse monoclonal anti-XPG (8H7, Santa Cruz Biotechnology), or rabbit monoclonal anti-ki67 (SP6, Thermo scientific) antibodies (diluted 1:100 in PBST). Cells were then washed three times with PBST, followed by 1-h incubation with secondary antibodies: Alexa Fluor 594-conjugated goat-anti-mouse-, and goat-anti-rabbit-IgG, for XPG and ki67 detection, respectively (Invitrogen, diluted 1:1000 in PBST). To avoid unnecessary nuclear fluorescence, DAPI staining was omitted. After extensive washing with PBST, cells were fixed for 20 min with 3.7% formaldehyde in PBS. Subsequently, EdU detection was carried out as described above.
RESULTS
Effects of UV dose and EdU-incubation period on measurement of UV-induced UDS in normal human primary fibroblasts
48BR normal primary human fibroblasts were cultured on coverslips and subsequently exposed to different doses of UVC, followed by incorporation of EdU for different time periods. UVC treatment elicited UDS activity as shown by an increase in nuclear fluorescence, after conjugation of fluorescent azide to incorporated EdU (Figure 1A and B). As reported in previous publications employing 3H-thymidine labeling and autoradiography (15,16), strong signals from scheduled DNA synthesis in S-phase cells can be distinguished from much weaker cell-cycle independent unscheduled EdU incorporation, resulting from repair synthesis (Figure 1B). Additionally, the fluorescent-azide coupling reaction seems specific to incorporated EdU, as we barely detected any non-specific cytoplasmic staining or DNA-replication-independent nuclear signals (Figure 1B). We observed that the nuclear fluorescent intensities were proportional to both UV dose and EdU-incubation time, indicating that this end-point satisfactorily represents the amount of incorporated EdU, which is directly convertible to semi-quantitative UDS measurement (Figure 1A). We were able to detect nuclear fluorescent signals at relatively low UV exposures, and with short time-periods of post-UV EdU incubation, demonstrating that this technique is sensitive enough to detect low levels of UDS activity.
Sensitivity and resolution of the EdU-based UDS assay
We evaluated the resolution as well as the sensitivity of the EdU-based UDS assay, by comparing the results obtained from the EdU technique with that of conventional autoradiography and BrdU-based methods. Many publications that have utilized autoradiography for the UDS assay report UDS levels for the most severe XP cases (i.e. XP-A or XP-G/CS) as <5–10% of normal levels (25). These can be distinguished from cell strains from XP patients, typically from XP-C or XP-D groups, which often have UDS levels of 20–40% of residual UDS activity (intermediate UDS level). The intensity-distribution plots for the EdU-based UDS assay for both normal 48BR (UDS positive, Figure 1C) and XP-deficient XP15BR (UDS negative, Figure 1D) fibroblasts resemble those of published autoradiography-based experiments, though we acknowledge that the background UDS levels were relatively higher than that from autoradiography (this point is discussed later). The EdU assay consistently produced a satisfactory SD of 10–15%.
We compared the relative sensitivities of the EdU assay and the BrdU-based method. Though BrdU has been commonly used for S-phase labeling, publications using it for UDS assays are quite limited because of its restricted sensitivity and resolution (18) (Figure 1E and F). Although the histograms of normal 48BR and XP-deficient XP15BR fibroblasts could be discriminated by the BrdU-based assay (Figure 1E and F), the resolution as well as SD were both markedly improved in the EdU method, suggesting that EdU is a more sensitive substitute for BrdU in the UDS assay.
UV-induced EdU incorporation in NER-deficient primary fibroblasts
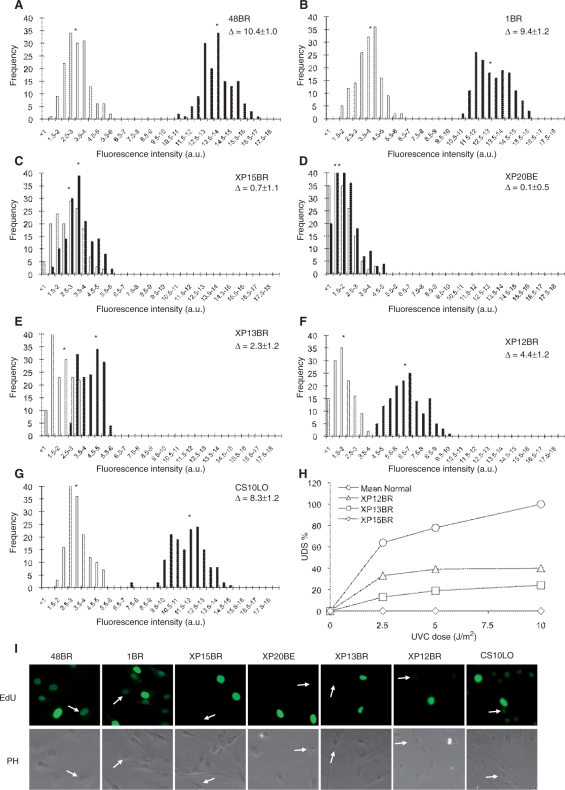
To apply the EdU technique to XP diagnosis, we estimated the levels of UV-induced EdU incorporation in several NER-deficient primary fibroblasts as well as normal controls (Figure 2A–G for histograms, and Figure 2I for typical photos). Since XP is a genetically heterogenous disorder, both the NER genes affected and the types of mutations determine the magnitude of the defect in UDS. We measured UDS in XP15BR (XP-A), XP20BE (XP-G/CS), XP13BR (XP-C), XP12BR (XP-D) and CS-patient-derived CS10LO (CS-B) cells, and compared the results with those obtained using 3H-thymidine incorporation. With 20 J/m2 UVC irradiation, we observed substantial UDS in normal fibroblasts, 48BR (Figure 2A) and 1BR (Figure 2B), as measured by EdU incorporation. We barely detected any UDS in severe XP-patient-derived XP15BR (XP-A, Figure 2C) and XP20BE (XP-G, Figure 2D) fibroblasts, while we detected marginal (∼20% of normal) UDS activity in XP-C patient-derived XP13BR (Figure 2E) fibroblasts.
Figure 2.
UV-induced EdU incorporation in non-S-phase cells is NER repair replication specific. (A–G) Levels of UV-UDS in NER-proficient and NER-deficient cells were examined. Primary fibroblasts were cultured on coverslips, UVC-irradiated (20 J/m2), followed by 2-h incubation with 10 μM EdU. Levels of EdU incorporation were analysed as in Figure 1A and presented as in Figure 1. (H) UDS assay performed by 3H-thymidine incorporation. Quiescent cells were UVC irradiated at the indicated dose and 3H-thymidine incorporation was measured for 3 h after UVC irradiation in the media containing 10 mM hydroxyurea. UDS levels were normalized and expressed as percentages of the UDS in the normal cells at 10 J/m2. Mean Normal represents the average UDS levels of the four different normal cell lines (1BR, 48BR, 142BR and 251BR). Asterisks indicate the mean values of nuclear fluorescent intensities, which correspond to the UDS levels. Δ represents UDS difference between irradiated and un-irradiated samples. (I) Typical photos of the EdU incorporation examined in Figure 2A–G are shown (2 h after 20 J/m2 UVC irradiation). Arrows indicate non-S-phase cells. 48BR, normal; 1BR, normal; XP15BR, XP-A; XP20BE, XP-G; XP13BR, XP-C; XP12BR, XP-D; CS10LO, CS-B.
XP12BR is homozygous for R511Q mutation in the XPD gene (our unpublished data), and, as shown in Figure 2F, we found ∼40% of normal UDS activity. In Figure 2H, we show the data obtained with the same set of cell strains using the 3H-thymidine incorporation. In good agreement with the EdU assay, UDS was barely detectable in XP15BR, but were about ∼20% and ∼40% of normal in XP13BR and XP12BR, respectively. These results show good agreement between the two methods and demonstrate that cells exhibiting intermediate UDS activity can be distinguished from both normal and severe UDS defects by the EdU-based assay.
We then further examined whether CS-deficient fibroblasts were distinguishable from normal cell lines by the UDS assay. CS is only compromised in the TCR pathway of NER, and this is not precisely diagnosed by UDS assays, since the deficiency is usually subtle. Though Figure 2G indicates a modest (∼20%) reduction of the UDS level compared to the normal fibroblasts, we cannot conclude this to be significant and is likely to fall within the normal range.
Levels of nuclear fluorescence after UVC irradiation in severe NER-deficient fibroblasts (Figure 2C and D) were virtually the same as those in unirradiated samples, suggesting that the background fluorescent signals resulting from minor EdU incorporation or non-specific fluorescent conjugation do not affect the analyses. We conclude that the EdU-based UDS assay provides a specific measurement of nucleotide incorporation associated with the repair synthesis step of NER.
Compatibilities of the EdU-based UDS assay with other techniques
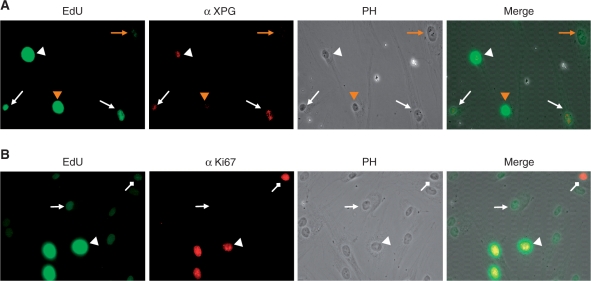
We anticipate applying the EdU-based UDS assay system to clinical diagnoses, in which rigorous intra-assay controls are required. We therefore assessed the compatibility of the UDS assay with other standard techniques that use internal references. We found that fluorescent-azide conjugation to EdU is fully compatible with immunofluorescent staining (Figure 3A and B).
Figure 3.
EdU assay is compatible with immunostaining. (A) Normal 48BR and XPG-deficient XP20BE fibroblasts were co-cultured on coverslips, UVC irradiated (20 J/m2), followed by 2-h incubation with 10 μM EdU. Fixed cells were then immunostained with rabbit anti-XPG antibody (red), followed by conjugation of Alexa fluor 488-azide to incorporated EdU (green). (B) UV-induced EdU incorporation in quiescent cells. 48BR cells were UVC irradiated (20 J/m2), followed by 2-h incubation with 10 μM EdU. Fixed cells were then immunostained with rabbit anti-ki67 antibody (red), followed by conjugation of Alexa fluor 488-azide to incorporated EdU (green). Triangle, normal arrow and diamond arrow indicate S-phase, G0 and G1 (or G2/M), respectively. Detailed experimental conditions are described in ‘Materials and methods’ section.
Co-culture of an index fibroblast as well as a target under study on the same coverslip provides a rigorous internal control. In Figure 3A, normal and XPG-deficient fibroblasts were co-cultured, followed by UV-UDS assayed in combination with α-XPG immunostaining. EdU-positive cells (excluding S-phase) perfectly correspond to XPG-positive cells, indicating that two different cell populations can be distinguished in the UDS assay.
Similarly, we examined if there was a cell-cycle preference for the EdU assay: co-immunostaining with a proliferation marker, ki-67, demonstrated that UDS can be detected in quiescent fibroblasts (no Ki67 stain) as well as proliferating populations (Figure 3B).
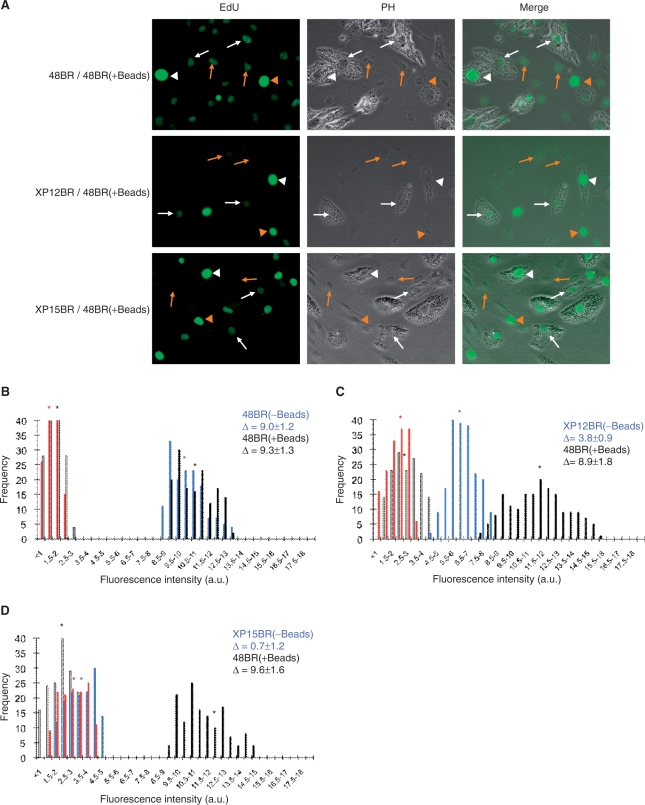
As discussed above, the level of the defect in UDS activity varies between different XP patients. Ideally, ascertaining whether a patient is NER compromised is evaluated by comparing the residual UDS activity of the patient-derived fibroblast with index cells. For this purpose, in a practical UDS-based XP diagnosis, normal fibroblasts have been preloaded with latex-beads in the cytoplasm and co-cultured with patient-derived fibroblasts. The ‘bead-labeled-cells’ provide an internal standard that can be detected under phase contrast microscopy which eliminates sample-to-sample staining variability (15). We tested if this technique is also compatible with the EdU assay. First we checked whether the latex-beads did not affect the nuclear fluorescent intensity. As shown in Figure 4A (top panel) and corresponding histogram (Figure 4B), the fluorescent intensity in 48BR cells loaded with beads was practically the same as those in cells without beads, indicating that the beads do not interfere with the EdU-based UDS assay.
Figure 4.
EdU assay in the presence of latex beads. Normal 48BR cells were pre-labeled with 0.5-μm diameter latex beads and co-cultured with indicated XP fibroblasts (no-beads) on coverslips. Cells were then UVC-irradiated (20 J/m2), followed by 2-h incubation with 10 μM EdU. Coverslips were then processed as in Figure 1. (A) Typical photos are displayed. White and red markings, arrows and arrow heads represent 48BR and indicated XP cells, and non-S-phase and S-phase cells, respectively. (B–D) Histograms of the UDS assay with the internal standard 48BR and (B) 48BR, normal; (C) XP12BR, XP-D; and (D) XP15BR, XP-A. Bars represent frequencies as described in Figure 1, with (black, 48BR; blue, XP cell-lines) or without (white, 48BR; red, XP cell-lines) UVC irradiation. Asterisks indicate the mean values of nuclear fluorescent intensities, which correspond the UDS levels. Δ represents UDS differences with or without UVC irradiation.
Modest (XP12BR; Figure 4A middle panel and 4C) as well as severe (XP15BR; Figure 4A bottom panel and 4D) UDS-deficient fibroblasts were both easily distinguishable from co-cultured 48BR fibroblasts by their nuclear fluorescent levels. The UDS measurements with or without internal controls were nearly the same (compare Figure 2A, 2C and 2F with Figure 4B, 4D and 4C), suggesting that the prep-to-prep or coverslip-to-coverslip variation in the EdU-based assay is small; this may be an advantage when applying the EdU assay to experiments that are incompatible with the use of an internal standard (e.g. fluorescent-based high-throughput screening using GE's In-Cell-Analyzer or flow cytometry).
Further optimization of the EdU assay
The EdU-based experiments described above are sufficient for routine XP screening as well as many types of NER research; however, further precise experiments such as distinguishing between no UDS activity and very low levels may require higher sensitivity. We have attempted to increase the specific EdU uptake as well as to reduce the non-specific background. The incorporation of a thymidine analogue into the DNA is dependent on its concentration relative to endogenously synthesized thymine nucleotides in the nucleotide pool. Fluorodeoxyuridine (FdU) is an inhibitor of thymidylate synthetase, which renders the cell dependent on exogenously supplied thymidine or its analogues. We therefore used FdU and a longer incubation period (4 h) after UV irradiation. Additionally, we used stringent acid extraction with Bouin's fixative to try and reduce the background. As shown in Figure 5, although not dramatic, both background (compare white and red bars and their corresponding asterisks) and UDS-specific EdU uptake (compare black and blue bars) were improved.
DISCUSSION
Rapid and accurate assessment of UDS levels is indispensable for clinical XP diagnosis as well as NER research. In this report, we have described a novel UDS assay system that provides a very simple method for measurement of repair-replication activity in human cells.
Orthodox UDS assay methods have been based on 3H-thymidine incorporation (12,15,16). Although a variety of alternative reagents have been developed, most XP-diagnosis laboratories still measure UDS by autoradiography or direct scintillation counting of incorporated 3H-thymidine. The biggest disadvantages of using 3H-thymidine for UDS assay is its time-consuming procedures and the requirement for laboratories designated for radioactive work.
The autoradiographic process provides an accurate measurement of UDS activity as each silver grain represents an individual event of nucleotide incorporation into a repair patch. However, the experiment is complicated, requiring significant skills, and takes 1–2 weeks to be completed; furthermore, counting of silver grains under the microscope is laborious. An alternative procedure uses direct scintillation counting of 3H-thymidine incorporation (12). This method requires abrogation of S-phase DNA replication. To accomplish this, cells are brought into quiescence by serum starvation, and hydroxyurea is needed during the nucleotide incorporation period, to block replicative DNA synthesis. This technique significantly reduced the time required for the UDS assay, but quantitation of UDS is less accurate.
In contrast to the above 3H-thymidine-based methods, BrdU incorporation assay is faster and a convenient option. Indeed, BrdU has been frequently used for cell-cycle analysis applications. On the other hand, regardless of the type of experiments, denaturation of DNA is needed to detect the incorporated BrdU as the anti-BrdU antibody is too big to access the epitope in native DNA. DNA denaturation and subsequent immunofluorescence detection sometimes generate an ambiguous staining pattern (data not shown; see high SD in Figure 1E and F). This is the biggest disadvantage of the BrdU incorporation assay: nucleotide incorporation into repair patches based on DNA-repair activity is quite low and difficult to detect. For this reason, UDS based on BrdU has not been widely used.
The EdU incorporation method was originally developed for complementing known problems of S-phase population labeling by 3H-thymidine and BrdU (19). In EdU, the methyl group on the 5-position of thymidine is replaced by an alkyne group, and this analogue is incorporated into DNA during replication. Subsequent fluorescent-azide coupling, termed ‘click reaction’ is rapid, sensitive and specific (19,20); because of this property, the sensitivity and background fluorescent signal were both dramatically improved, and hence, EdU can be used for the UDS assay.
We have presented data demonstrating the usefulness of EdU for the UDS assay. Although the sensitivity of the EdU-based method is not as high as the autoradiographic technique, we easily detected the defect in an XP-D cell line with ∼40% residual UDS activity and based on our data we conservatively estimate that we can detect a level of 10–20% of normal UDS.
The EdU-based UDS assay may have an advantage in high-throughput XP diagnosis using flow cytometry. An attempt to use BrdU labeling and flow cytometry for UDS has been reported (14). Because of the improved sensitivity of EdU over BrdU, further introduction of multi-color labeling (i.e. normal internal control as well as specimen cells, with or without UV damage, all labeled with different fluorescent dyes) may provide a rapid and precise determination of the relative UDS levels.
In this report, we have demonstrated that: (i) the accuracy and resolution of the UDS assay based on EdU incorporation was comparable to the conventional autoradiographic method; we could distinguish between a normal and an XP-deficient primary fibroblast when the cells were co-cultured on a single coverslip. (ii) The assay was compatible with standard immunostaining and latex-bead-labeling, which provide proper internal controls. (iii) The time required for the assay was dramatically reduced; the entire UDS assay can be completed within half a day from the preparation of specimen coverslips. The UDS assay based on EdU could potentially become a standard technique in NER research as well as XP diagnosis.
FUNDING
The KAKENHI from Japan Society for the Promotion of Science (grant number 20810021); a Special Coordination Fund for Promoting Science and Technology from Japan Science and Technology Agency; a Butterfield Award from the Great Britain Sasakawa Foundation (to T.O.); and a grant from the Thailand Research Fund and Commission on Higher Education (to S.L.); a Global COE Program from the Ministry of Education, Culture, Sports, Sciences and Technology of Japan (to S.L., S.Y., and T.O.). Funding for open access charge: the KAKENHI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Dr. Nicolaas Jaspers for helpful comments on the manuscript. We also thank Dr. Motohiro Yamauchi for critical advice on the EdU incorporation assay.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. Washington, DC, USA: ASM Press; 2005. [Google Scholar]
- 2.Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JH. In: The Metabolic and Molecular Bases of Inherited Disease. 8th. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. New York: McGraw-Hill; 2001. pp. 677–703. [Google Scholar]
- 3.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 4.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleijer WJ, Laugel V, Berneburg M, Nardo T, Fawcett H, Gratchev A, Jaspers NG, Sarasin A, Stefanini M, Lehmann AR. Incidence of DNA repair deficiency disorders in western Europe: Xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. DNA Repair (Amst) 2008;7:744–750. doi: 10.1016/j.dnarep.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Stefanini M, Lagomarsini P, Giliani S, Nardo T, Botta E, Peserico A, Kleijer WJ, Lehmann AR, Sarasin A. Genetic heterogeneity of the excision repair defect associated with trichothiodystrophy. Carcinogenesis. 1993;14:1101–1105. doi: 10.1093/carcin/14.6.1101. [DOI] [PubMed] [Google Scholar]
- 9.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Sarasin A, Stary A. New insights for understanding the transcription-coupled repair pathway. DNA Repair (Amst) 2007;6:265–269. doi: 10.1016/j.dnarep.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann AR, Stevens S. A rapid procedure for measurement of DNA repair in human fibroblasts and for complementation analysis of xeroderma pigmentosum cells. Mutat. Res. 1980;69:177–190. doi: 10.1016/0027-5107(80)90187-6. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann AR, Thompson AF, Harcourt SA, Stefanini M, Norris PG. Cockayne's syndrome: correlation of clinical features with cellular sensitivity of RNA synthesis to UV irradiation. J. Med. Genet. 1993;30:679–682. doi: 10.1136/jmg.30.8.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent F, Ceraline J, Goldblum S, Klein-Soyer C, Bergerat JP. A new flow cytometric method to follow DNA gap filling during nucleotide excision repair of UVc-induced damage. Cytometry. 2001;45:96–101. doi: 10.1002/1097-0320(20011001)45:2<96::aid-cyto1151>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Kleijer WJ, van der Sterre ML, Garritsen VH, Raams A, Jaspers NG. Prenatal diagnosis of xeroderma pigmentosum and trichothiodystrophy in 76 pregnancies at risk. Prenat. Diagn. 2007;27:1133–1137. doi: 10.1002/pd.1849. [DOI] [PubMed] [Google Scholar]
- 16.Stefanini M, Giliani S, Nardo T, Marinoni S, Nazzaro V, Rizzo R, Trevisan G. DNA repair investigations in nine Italian patients affected by trichothiodystrophy. Mutat. Res. 1992;273:119–125. doi: 10.1016/0921-8777(92)90073-c. [DOI] [PubMed] [Google Scholar]
- 17.Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat. Res. 2002;509:165–174. doi: 10.1016/s0027-5107(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 18.Droy BF, Miller MR, Freeland TM, Hinton DE. Immunohistochemical detection of CCl4-induced, mitosis-related DNA synthesis in livers of trout and rat. Aquat. Toxicol. 1988;13:155–166. [Google Scholar]
- 19.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breinbauer R, Kohn M. Azide-alkyne coupling: a powerful reaction for bioconjugate chemistry. Chembiochem. 2003;4:1147–1149. doi: 10.1002/cbic.200300705. [DOI] [PubMed] [Google Scholar]
- 21.Arlett CF, Green MH, Priestley A, Harcourt SA, Mayne LV. Comparative human cellular radiosensitivity: I. The effect of SV40 transformation and immortalisation on the gamma-irradiation survival of skin derived fibroblasts from normal individuals and from ataxia-telangiectasia patients and heterozygotes. Int. J. Radiat. Biol. 1988;54:911–928. doi: 10.1080/09553008814552321. [DOI] [PubMed] [Google Scholar]
- 22.Arlett CF, Plowman PN, Rogers PB, Parris CN, Abbaszadeh F, Green MH, McMillan TJ, Bush C, Foray N, Lehmann AR. Clinical and cellular ionizing radiation sensitivity in a patient with xeroderma pigmentosum. Br. J. Radiol. 2006;79:510–517. doi: 10.1259/bjr/83726649. [DOI] [PubMed] [Google Scholar]
- 23.Moriwaki S, Stefanini M, Lehmann AR, Hoeijmakers JH, Robbins JH, Rapin I, Botta E, Tanganelli B, Vermeulen W, Broughton BC, et al. DNA repair and ultraviolet mutagenesis in cells from a new patient with xeroderma pigmentosum group G and cockayne syndrome resemble xeroderma pigmentosum cells. J. Invest. Dermatol. 1996;107:647–653. doi: 10.1111/1523-1747.ep12584287. [DOI] [PubMed] [Google Scholar]
- 24.Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AR. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer KH, Coon HG, Petinga RA, Barrett SF, Rahe AE, Robbins JH. Genetic heterogeneity in xeroderma pigmentosum: complementation groups and their relationship to DNA repair rates. Proc. Natl Acad. Sci. USA. 1975;72:59–63. doi: 10.1073/pnas.72.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]