Abstract
DNA is susceptible of being damaged by chemicals, UV light or gamma irradiation. Nuclear DNA damage invokes both a checkpoint and a repair response. By contrast, little is known about the cellular response to mitochondrial DNA damage. We designed an experimental system that allows organelle-specific DNA damage targeting in Saccharomyces cerevisiae. DNA damage is mediated by a toxic topoisomerase I allele which leads to the formation of persistent DNA single-strand breaks. We show that organelle-specific targeting of a toxic topoisomerase I to either the nucleus or mitochondria leads to nuclear DNA damage and cell death or to loss of mitochondrial DNA and formation of respiration-deficient ‘petite’ cells, respectively. In wild-type cells, toxic topoisomerase I–DNA intermediates are formed as a consequence of topoisomerase I interaction with camptothecin-based anticancer drugs. We reasoned that targeting of topoisomerase I to the mitochondria of top1Δ cells should lead to petite formation in the presence of camptothecin. Interestingly, camptothecin failed to generate petite; however, its derivative topotecan accumulates in mitochondria and induces petite formation. Our findings demonstrate that drug modifications can lead to organelle-specific DNA damage and thus opens new perspectives on the role of mitochondrial DNA-damage in cancer treatment.
INTRODUCTION
Topoisomerase I (Top1) is an enzyme required for the release of torsional stress built up during transcription or replication of DNA (1,2). This torsional stress is defined by an increase of negative DNA supercoils in front of the transcription or replication machinery. Top1 releases torsional stress by changing the linking number of the DNA. To change the linking number of DNA, Top1 cuts one DNA strand, passes the other through it and then re-anneals the cut strand. In Saccharomyces cerevisiae expression of the same TOP1 open reading frame (ORF) appears to account for the nuclear and mitochondrial Top1 activities. This idea was put forward because deletion of the TOP1 ORF abolishes Top1 specific activities in the nucleus as well as in mitochondria (3–5). In human cells, the mitochondrial Top1 is encoded in the nuclear genome and contains a mitochondrial localization sequence at the N-terminus of the enzyme (6,7).
Incomplete action of Top1 has been shown to be a natural source of DNA damage such as DNA single-strand breaks (SSBs). Persistent SSBs can be converted to lethal double-strand breaks (DSBs) during replication (8). Single mutations in the active center of the enzyme have been shown to strongly increase the formation of nicked DNA and to impair the viability of yeast cells (9,10). This is the case for the S. cerevisiae TOP1-103 allele in which substitution of an arginine for a lysine at amino acid 420 generates a highly toxic Top1 isoform. A strong increase in SSBs can also be mediated by drugs, which trap Top1 bound to the DNA. The ancestor of such drugs, named campthotecin [CPT; (11)], has been shown to bind to the covalent Top1–DNA complex and form a ternary complex impeding the ligation of cleaved DNA (12). CPT and its derivatives are potent anticancer drugs, which have been approved for the treatment of solid cancers (13). Severe side effects such as reversible bone marrow depression and hemorrhagic cystitis are major dose-limiting toxicities of CPT. New CPT analogs, including the prodrug CPT-11 [Irinotecan; (14)] or topotecan [TPT; (15)], have the advantage of higher antitumor activity and less toxicity. However, tumor cells tend to develop drug resistance in response to chemotherapy. Such drug resistance can be mediated by several ATP-binding cassette (ABC) transporters (16–18), as shown in yeast or human cells.
DNA damage caused by Top1 has to be repaired to avoid serious effects on genome stability and cellular viability. In S. cerevisiae the elimination of spontaneous DNA lesions, such as SSBs, is mediated by many overlapping repair pathways. Genetic and biochemical evidence suggests the involvement of homologous recombination, base excision repair (BER), nucleotide excision repair (NER) and substrate-specific enzymes such as the tyrosyl-DNA phosphodiesterase Tdp1 in the removal of the covalently bound Top1 protein from the DNA (19–21). Although the toxic effects of CPT treatment on nuclear DNA are well studied, little is known about the toxic effects of CPT treatment on mtDNA or about spontaneous mtDNA lesions mediated by Top1. However, repair enzymes and pathways that are involved in the repair of nuclear, Top1-mediated DNA lesions also participate in the repair of mitochondrial DNA lesions induced by oxidative damage (22–24). One example is the apurinic/apyrimidinic endonuclease Apn1, which bears a bipartite nuclear localization signal. Apart from its nuclear localization, Apn1 can be localized to the mitochondria upon interaction with Pir1 (25).
We reasoned that DNA nicks introduced by Top1 could serve as a tool for the development of an experimental system to target specific DNA damage to DNA-containing organelles. We show that yeast cells can be poisoned with Top1p-mediated nuclear DNA damage either by expression of truncated, toxic TOP1-103 allele or by expressing a truncated TOP1 allele and concomitant treatment of cells with the anticancer drug CPT. Moreover, we observe that targeting of the truncated, toxic mt125Top1-103 to mitochondria induces the formation of respiration-deficient petite cells and mtDNA loss. CPT treatment of cells expressing mt125Top1 protein targeted to mitochondria did not induce petite formation despite mt125Top1 purified from yeast mitochondria being sensitive to CPT treatment in vitro. In contrast to CPT, its analog TPT was capable of generating petite colonies. Consistent with this finding, TPT, but not CPT, accumulates in the mitochondria. Thus, our results suggest that mtDNA is not accessible to CPT. Our findings show that altering the chemical properties of an anticancer drug can result in organelle-specific DNA damage targeting.
MATERIALS AND METHODS
Strains and media
Experiments were performed in strain TOI-1Bu- (MATa ade2-1 trp1-1 ura3-1 his3-11,15 leu2-3,112 top1Δ::hisG can1-100) which was kindly provided by A. Aguilera (CABIMER, Sevilla). Yeast transformants were grown in liquid or solid synthetic minimal medium (SC, Difco), supplemented with amino acids and different carbon sources: 2% glucose, 3% glycerol-lactate, or 2% galactose. Where indicated, CPT (5 mg/ml stock solution in 1% DMSO, Sigma) and TPT (10 mg/ml stock solution in water, Sinova) were added to a final concentration of 20 µg/ml.
Constructs
DNA manipulation, PCR and DNA sequencing were performed according to standard protocols. GAL1p and MET25p expression vectors: A 590-bp SpeI–SmaI fragment of pGAL1:RNH1 (A. Aguilera, CABIMER) containing the GAL1 promoter was cloned into pRS424 (26) in order to create pRS424-GAL1p. A 716bp XbaI fragment was removed from pUG34 (J.H. Hegemann, Universität Düsseldorf) and the GFP-less vector was named pUG-MET25p.
Constructs containing the full-length TOP1 ORF
The ORFs of the full-length (2307 bp) and truncated (1965 bp) wild-type TOP1 and mutant allele TOP1-103 were obtained by PCR amplification (Phusion Taq-polymerase, Finnzymes) from plasmids pWE3 and pNL30, respectively. Plasmids were kindly provided by the laboratory of Dr. G. Fink (Whitehead Institute for Biomedical Research, Cambridge, MA 02142). Primers (Invitrogen) were tagged 5′ with BglII and SmaI restriction sites and the PCR fragments were cloned into pUG23 (full-length TOP1 ORF) and pUG34 (truncated 125TOP1 ORF). The GFP-125TOP1 fusion constructs were again PCR amplified with a 5′ primer matching the first 20 bp (including a BamHI restriction site) of the GFP ORF and a 3′ primer matching the last 20 bp of the TOP1 ORF (including an SmaI restriction site). Then the corresponding PCR fragments containing the corresponding GFP-TOP1 chimera were cloned into single BglII–SmaI or BamHI–SmaI restriction sites of pRS424-GAL1p and pUG-MET25p.
Constructs containing an SV40-NLS
To tag the truncated TOP1 ORF with 33 bp of the SV40-NLS, a 3′ primer was designed matching the last 17 bp of the TOP1 ORF (omitting the stop codon) and having a 33-bp SV40 protruding tail (including a stop codon and a SmaI restriction site). Then the PCR fragments containing the corresponding GFP-TOP1 chimera were cloned into single BglII–SmaI or BamHI–SmaI restriction sites of pRS424-GAL1p and pUG-MET25p.
Constructs containing an SOD2-MLS
BamHI–SOD2–MLS–BglII or BglII–SOD2–MLS–BamHI, 60-bp PCR fragments were ligated and cloned into single BglII or BamHI restriction sites of vectors pRS424-GAL1p and pUG-MET25p, respectively. Note that upon ligation of overlapping, sticky BamHI–BglII ends in this sequence are no longer cleavable by either restriction enzyme. Then the corresponding PCR fragments containing the corresponding GFP-125TOP1 chimera were cloned into single BglII–SmaI restriction sites of pRS424-GAL1p-SOD2-MLS and pUG-MET25p-SOD2-MLS. A detailed list of all vectors is found in Table 1.
Table 1.
Plasmids
| Plasmid | Feature | Source |
|---|---|---|
| pWE3 | GAL1p::TOP1 | (9) |
| pNL30 | GAL1p::TOP1-103 | (9) |
| pUG23 | HIS3, MET25p::GFP::CYC1-T | U. Gueldener and J.H. Hegemann |
| pUG34 | HIS3, MET25p::GFP::CYC1-T | U. Gueldener and J.H. Hegemann |
| pUG-MET25p | HIS3, MET25p::CYC1-T | Present study |
| pRWE085 | HIS3, MET25p::SOD2MLS-GFP-125TOP1-103::CYC1-T | Present study |
| pRWE090 | HIS3, MET25p::GFP-125TOP1-103-SV40NLS::CYC1-T | Present study |
| pRWE091 | HIS3, MET25p::GFP-125TOP1-SV40NLS::CYC1-T | Present study |
| pRWE096 | HIS3, MET25p::SOD2MLS-GFP-125TOP1::CYC1-T | Present study |
| pRS424 | TRP1 | (48) |
| pGAL1:RNH1 | URA3, GAL1p::RNH1::CYC1-T | A. Aguilera |
| pRS424-GAL1p | TRP1, GAL1p::CYC1-T | Present study |
| pRWE108 | TRP1, GAL1p::SOD2MLS-GFP-125TOP1-103::CYC1-T | Present study |
| pRWE120 | TRP1, GAL1p::TOP1-GFP::CYC1-T | Present study |
| pRWE121 | TRP1, GAL1p::TOP1-103-GFP::CYC1-T | Present study |
| pRWE123 | TRP1, GAL1p::SOD2MLS-GFP-125TOP1::CYC1-T | Present study |
| pRWE126 | TRP1, GAL1p::GFP-125TOP1-103-SV40NLS::CYC1-T | Present study |
| pRWE129 | TRP1, GAL1p::GFP-125TOP1-SV40NLS::CYC1-T | Present study |
Northern and Southern blots
DNA fragments were derived by PCR amplification of primers 5′ATGACTATTGCTGATGCTTC3′ and 5′TTAAAACCTCCAATTTTCAT3′ (TOP1 probe), primers 5′TTGGAGAGGGCAACTTTGG3′ and 5′CAGGATCGGTCGATTGTGC3′ (25S rDNA probe), primers 5′TGTATTTTCGACTCTTTG3′ and 5′TCAGCCAAATGAAGTAAG3′ (nuclear DNA probe), primers 5′ATGTTAGATTTATTAAGA3′ and 5′TTATTGTTCATTTAATCA3′ (mtCOX2 probe). The mtCOX2 containing plasmid pJM2 was kindly provided by T. Fox (Cornell University). Radioactive probes were prepared by random primer labeling and then G50 column purified. Membranes (Hybond, Amersham) were exposed to PhosphorImager screens (Fuji) and scanned (FujiFilm FLA 5100). Quantification of the P32 signal was performed with the Image Gauge imaging software supplied (Fuji).
Protein purification
About 1 l of yeast cells were grown in glycerol-lactate containing SD minimal medium to an OD600=0.5. Protein expression was induced by shifting the cells to galactose for 1 h. After incubation in galactose medium the expression of GFP-chimeras was checked by fluorescence microscopy. Yeast crude extracts were obtained from 2 g of cells by the FASTPREP method. All following steps were carried out at 4°C. The cell pellet was resuspended in 2 ml of lysis buffer (50 mM Tris–HCl pH7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.15% NP-40) and a protease inhibitor cocktail (0.5 mM PMSF, 2 mM Benzamidine, 1 mM Leupeptin, 2 mM Chymostatin and 2.6 mM Aprotinin; Sigma) was added. Cells were mixed with glass beads (1 g glass beads per gram of cell pellet) and transferred to a FASTPREP-24 (MQ-Biogene). Cells were broken with four pulses of vortexing (4.0 m/s for 20 s), keeping the cells on ice between each pulse. Protein purification was essentially performed as described in ref. 27. In brief, a 10-ml protein sample was injected into an FPLC (AKTA explorer) and protein was eluted gradually by increasing the NaCl concentration with a linear gradient from 0.26 M to 1 M. One-milliliter fractions were collected and 50 µl of each fraction transferred to an Inmobilon membrane (Millipore) using a vacuum dot-blot apparatus (Bio-Rad BioDot SF Cell) prior to GFP detection with an anti-GFP-HRP antibody (Miltenyi Biotec). Main peaks of mt125Top1 protein elution were determined in fractions containing 0.294–0.316 M NaCl, and main peaks of n125Top1 protein elution were determined in fractions 0.362 M from 0.384 M NaCl. Two 1-ml fractions were pooled, and protein concentration and buffer exchange were achieved by a microcon YM-50 spin-column (Millipore). Samples were stored at –20°C in 50 mM Tris–HCl pH7.5 with 1 mM EDTA, 1 mM DTT, 0.5 M NaCl and 50% glycerol.
Immunoblotting
Three microliters of mt125Top1 and 15 µl of n125Top1 extracts were analyzed on an 8% Tris–glycine-SDS–polyacrylamide gel. Gels were either stained with Coomassie or GFP-125Top1 fusion proteins were transferred to a membrane (Inmobilon, Millipore) and inmuno-detected (anti-GFP-HRP antibody, Miltenyi Biotec) according to standard protocols.
DNA relaxation assays
DNA relaxation reactions were performed as described in ref. 28 with the following modifications. Partially purified enzyme preparations were incubated in 20 µl reaction volumes containing 0.3 µg of negatively supercoiled pBR322 plasmid DNA in 0.2 M Tris–HCl pH7.5, 1 mM EDTA, 0.1 M MgCl2 and 1 M KCl. The samples were incubated for 30 min at 30°C using 5 U of enzyme. Reactions were terminated by adding 1% SDS and incubated for 50 min with 0.75 μg/μl proteinase K. Products were electrophoresed in a 0.8% agarose gel in 1 × TBE buffer and plasmid DNA was visualized by staining with 0.5 μg/ml EtBr and photographed over a UV trans-illuminator. One unit was defined as the amount of enzyme needed to completely relax 0.3 µg of negatively supercoiled plasmid under standard reaction conditions.
DNA unwinding assays
Reaction conditions were as indicated as above but after a 30 min incubation at 30°C using 50 U of enzyme. CPT (0.5 mM in 1% DMSO) was added at a final concentration of 50 μM and the incubation was prolonged for another 30 min. Products were separated in 0.8% agarose in 1 × TBE buffer containing 0.5 µg/ml EtBr.
Microscopy
Cells were grown at an OD600 = 0.2, washed and incubated in fresh, prewarmed media containing 1.15 μg/μl of MitoTracker Orange (Invitrogen) for 30 min at 30°C. Then cells were washed two times with 1× PBS, and either incubated with 1 μg/μl of Hoechst33422 (Fluka) or 0.01 μg/μl DAPI (Sigma) for 30 min at 30°C. For drug-localization experiments cells were incubated with 20 µg/ml CPT or 20 µg/ml TPT for 30 min at 30°C. Finally, cells were washed again two times with 1 × PBS. Confocal images (TCS-SP5, Leica) and wide-field images (DM-6000B, Leica) were obtained at a 63× and 100× magnification, respectively.
CHEF
DNA was extracted in low melting agarose (Pronadisa) plugs as described (29). In-gel digestion of genomic and mitochondrial DNA was performed as follows: plugs were preincubated for 30 min at 4°C in 325 µl 1× restriction buffer containing 10 mM Tris–HCl, 10 mM MgCl2, 1 mM DTT and 1% BSA. Restriction buffer was replaced and upon addition of 300 U of ApaI (Takara Bio Inc., Japan) the plugs were incubated at 37°C for 24 h, another 300 U of ApaI were added and incubation was prolonged for 8 h. Plugs were imbedded into a 0.8% (1 × TAE, 0.3 µg/ml EtBr) agarose gel and fragments were separated by electrophoresis at 1.4 V/cm overnight.
RESULTS
Inducible Top1 targeting to the nucleus and mitochondria
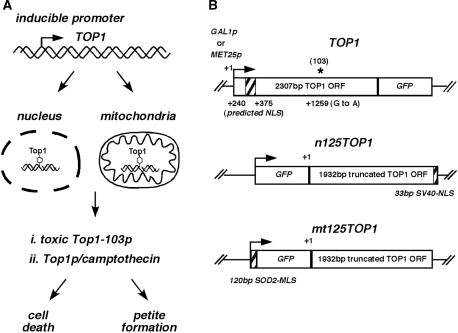
We have developed a system allowing the organelle-specific targeting of DNA damage by inducible expression of topoisomerase I (Top1), which is either directed to the nucleus or to the mitochondria. A schematic drawing is outlined in Figure 1A. Wild-type Top1 catalyzes a reversible DNA cleavage reaction by transferring a phosphodiester bond to the active site tyrosine residue (30,31). Stabilization of the Top1–DNA complexes can either be achieved by mutating the S. cerevisiae TOP1 and substituting arginine for a lysine at amino acid 420 [G to A transition at nt 1259 named TOP1-103 (9)] or interaction of the native Top1 protein with the anticancer drug CPT (12). In both cases the stabilization of a covalently attached Top1 to the DNA has been shown to result in persistent DNA single-strand breaks (SSBs). While Top1-mediated, persistent nuclear DNA damage has been shown to inhibit cell growth (9), we reasoned that persistent mitochondrial DNA damage should induce formation of respiration-deficient ‘petite’ cells due to mitochondrial dysfunction.
Figure 1.
Experimental approach. (A) Schematic presentation of different possibilities for the induction of organelle-specific Top1-mediated DNA damage. (B) Constructs used in this study. Shown are the different promoters (GAL1p, MET25p), GFP, nuclear localization sequence (NLS) and mitochondrial localization sequence (MLS). +1 refers to the start codon of the TOP1 open reading frame (ORF). A truncated TOP1 ORF was named 125TOP1, and nomenclature was changed to TOP1-103 in constructs containing a single-point mutation (103, asterisk) leading to a toxic Top1 protein.
Different TOP1 constructs were designed for specific targeting to the nucleus or mitochondria (Figure 1B). The first 375 bp (125 aa) of TOP1 were deleted because sequence alignment had led to the prediction of a nuclear localization sequence (NLS) within this region (32). To assess protein localization and toxicity, the full-length TOP1 or truncated TOP1 ORFs were tagged with green fluorescent protein (GFP) at the C-terminal or N-terminal, respectively. For nuclear targeting the SV40-NLS was attached at the 3′ end of the truncated 125TOP1. For mitochondrial targeting the SOD2 mitochondrial leader sequence (MLS) was attached at the 5′ end of the GFP. To improve protein import into the mitochondria it was necessary to place two SOD2-MLS in front of the GFP [Figure S1, (33)]. Finally, expression of all constructs was placed under control of the inducible MET25 or GAL1 promoters (Table 1).
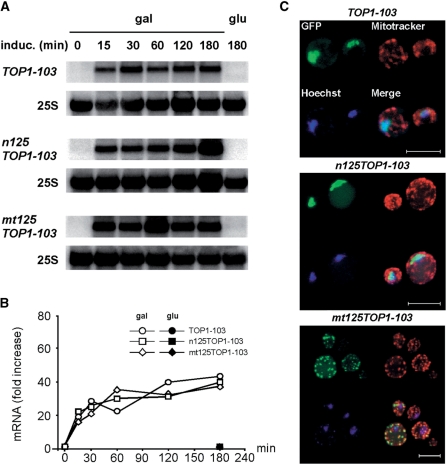
It is possible that accumulation of persistent nuclear DNA SSBs mediated by a toxic Top1-103 protein might affect mRNA transcription levels. Thus, we first analyzed mRNA expression levels of GAL1 promoter driven constructs by northern analysis (Figure 2A). TOP1-103, n125TOP1-103 and mt125TOP-103 mRNAs were not detectable in cells grown in glycerol (no induction) and glucose (repression), while shifting cells to galactose (induction) led to the rapid appearance of mRNAs. In all constructs, expression levels reached a maximum within 60 min (Figure 2B) and mRNA expression levels remained high within 180 min. No significant difference in mRNA levels was obtained comparing expression of n125TOP1-103 with mt125TOP-103, or expression of the TOP-103 constructs with a TOP1 expressing control vector (data not shown). From these results we conclude that nuclear targeting of the Top1-103 protein does not affect TOP1-103 transcription levels within 180 min of induction.
Figure 2.
Localization sequences target Top1p either to the nucleus or mitochondria. (A) Northern blot analysis of TOP1 mRNA upon galactose-induced gene expression. (B) Relative quantification of TOP1 mRNA levels. Relative TOP1 mRNA levels were normalized to 25S mRNA. The value obtained at time point 0 was set as 1. Average values of two independent experiments are shown. (C) Projection of a series of focal plane images derived from confocal fluorescence microscopy. GFP-Top1 expression was induced for 30 min in galactose (green). The nucleus and mitochondria were localized with Hoechst33342 (blue) and Mitotracker (red) dyes, respectively. White bar represents 5 µm. Control cells grown in glucose did not exhibit green fluorescence (data not shown).
To assess the cellular localization of the bio-fluorescent Top1-103 chimeras, expression was induced for 30 min in galactose and localization was monitored by confocal microscopy (Figure 2C). The Top1-103 tagged at the N-terminal with a GFP and C-terminal with a SV40-NLS (n125Top1-103) localized to nucleus (visualized by the turquoise merge of green and blue). This localization is in agreement with that of Top1 tagged at the C-terminal with a GFP (34). Importantly, GFP signal was absent from mitochondria. In contrast, expression of the truncated TOP1 bearing a SOD2-MLS (mt125Top1-103) led to an accumulation of GFP signal within the mitochondria, as seen by a punctuated pattern that overlays with the mitochondrial Mitotracker staining (visualized by the yellow merge of green and red). Notably, GFP signal in the nucleus was not detectable. Growth in glucose is sufficient to suppress mRNA expression to an extent which abolishes detection of GFP signal (data not shown). Thus, by attaching of a specific localization signal the Top1 protein can be exclusively targeted either to the nucleus or the mitochondria.
Organelle-specific targeting of a toxic 125Top1-103 protein induces cell death, or petite formation and mtDNA depletion
Growth inhibition on nonfermentable carbon sources such as glycerol is a hallmark of respiration deficient rho−, so-called ‘petite’ cells (35). In order to test the effect of nuclear or mitochondrial targeting of the Top1-103 protein on cell viability and respiration capacity we placed expression of the bio-fluorescent Top1-103 chimera under control of the MET25 promoter. This promoter has the advantage that cells can be grown in medium with different carbons sources because its expression is induced in the absence of methionine. As shown in the drop test in Figure 3, targeting of n125Top1-103 to the nucleus greatly reduced growth and viability of cells. Targeting of mt125Top1-103 to the mitochondria did not affect cell viability in fermentable medium (glucose). However, the accumulation of a red pigmentation characteristic of ade2− cells was suppressed and instead, white-colored colonies were obtained. The appearance of white colonies is an indication of respiration-deficient petite cells where mitochondria are not functional. Accordingly, the formation of petite cells was confirmed by growth inhibition in nonfermentable medium (glycerol). Thus, targeting of Top1-103-mediated SSBs to the nucleus affects cells viability, while mitochondrial targeting induces petite cells.
Figure 3.
Drop test assay to assess the effect of nuclear and mitochondrial expression of MET25p-driven constructs. Targeting of the toxic n125Top1-103 to the nucleus leads to growth inhibition in fermentable (glucose) and nonfermentable (glycerol) growth medium. Targeting of the toxic mt125Top1-103 to mitochondria leads to growth of white colonies in glucose and growth inhibition in glycerol, both of which indicate the formation of respiration deficient ‘petite’ cells.
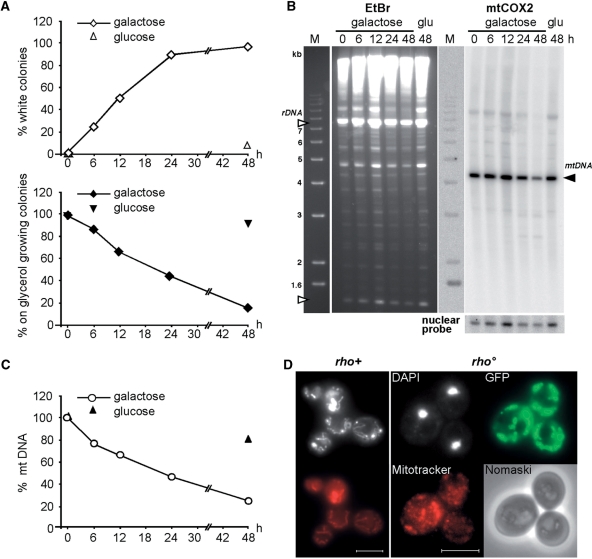
Respiration-deficient petite cells result from the loss of nuclear encoded functions, which are essential for the mitochondrial respiration capacity, or from mtDNA rearrangement, mutation or loss. Targeting of the toxic mt125Top1-103 to mitochondria results in mtDNA damage, which is predicted to impede mtDNA replication and to promote mtDNA loss. In order to quantify the extent of respiration-deficient cells, the mt125TOP1-103 construct placed under the control of the GAL1 promoter was expressed up to 48 h, and petite formation was assayed by visual inspection of white-versus red-pigmented colonies as well as growth inhibition in nonfermentable medium at the indicated time-point (Figure 4A). Within 48 h of mt125Top1-103 expression, conversion of wild-type into petite cells was nearly complete. About 97% of colonies expressing mt125Top1-103 turned white (7% in the control) and only 15% of the colonies were proficient to form colonies in nonfermentable medium (99% in the control). We wondered if the observed time-dependent increase in petite cells was due to the formation of respiration-deficient but mtDNA-containing rho– or due to the appearance of mtDNA-less rho° cells, respectively. Therefore, DNA was extracted in agarose plugs, in-gel digested with ApaI and analyzed by Southern blot (Figure 4B). After hybridization with a probe specific for the mitochondrial COX2 gene, a clear drop in mtDNA was seen after 48 h expression of the mt125Top1-103 construct (compare signals obtained for nuclear and mtDNA after 48 h of cellular growth in galactose or glucose). Quantification of the mtDNA content in respect to nuclear DNA (Figure 4C) showed that the overall mtDNA content was reduced to about 25% within 48 h of growth in galactose. To confirm that prolonged mt125Top1-103 expression induces the formation of rho° cells, cells expressing mt125Top1-103 for 48 h were stained with the DNA-intercalating agent DAPI and analyzed by fluorescence microscopy. In vivo staining of mtDNA by DAPI was practically absent in a high percentage of cells (see example in Figure 4D and Figure S2) confirming that mt125Top1-103 expression leads to mtDNA less rho° cells.
Figure 4.
Mitochondrially targeted mt125Top1-103 leads to mtDNA loss and the formation of rho° petite. (A) Time course of mt125Top1-103 dependent petite formation. Cells were grown in galactose-containing liquid medium (expression). At the indicated times cells were retrieved, grown on glucose containing plates (no expression) and replica-plated onto glycerol. Petite cells were determined by color (white; open diamonds) and their inability to grow on glycerol (black diamonds). Average values of two independent experiments are shown. (B) CHEF analysis of the mtDNA copy number. In-plug isolated, total cellular DNA was ApaI digested, gel electrophoresed and analyzed by Southern blot. Shown are the agarose gel before Southern blotting (rDNA fragments, open triangle) and the signals obtained using a specific probe against mitochondrial (right top, black triangle) and nuclear DNA (right bottom) after Southern blotting. (C) Determination of the relative mtDNA copy number. The mtCOX2 hybridization signal obtained at time point 0 was normalized to the nuclear probe and set as 100%. Average values of two independent experiments are shown. (D) Wide-field fluorescence microscopy analysis of petite cells after 48 h of mt125Top1-103 expression. DAPI (white) and Mitotracker (red) staining of rho+ (w/o induction) and rho° (48 h of induction) is shown. White bar represents 5 µm.
CPT is not capable of interacting with mitochondrial Top1p
As outlined before, another approach to induce persistent SSBs takes advantage of the anticancer drug CPT. To test the effect of CPT on organelle-specific Top1 targeting, MET25-promoter-driven constructs were used. Drop test assays revealed that upon targeting of n125Top1 to the nucleus, cellular growth was completely inhibited in the presence of CPT (Figure 5A). Interestingly, targeting of mt125Top1 to the mitochondria neither led to the appearance of white colonies nor impeded cellular growth in nonfermentable medium. This result can be interpreted in two ways: the mitochondrial mt125Top1 is not functional, or that CPT cannot interact with the mt125Top1 protein. In order to test for these possibilities, the mt125Top1 and n125Top1 proteins were purified from top1Δ yeast cells. The proteins were enriched in a two-step protocol by selective ammonium-sulfate precipitation and by FPLC using a Mono S column as described previously [(27), for detailed description see ‘Materials and Methods’ section)]. Western blot analysis using an anti-GFP antibody led to a strong fluorescent signal accompanied by a coloration of the membrane close to 114.4 kDa (Figure 5B top, arrows), which approximately corresponds to the expected molecular weight of the protein chimeras (expected molecular weight: n125Top1 = 104.98 kDa, mt125Top1 =103.66 kDa). Coomassie-staining of a protein gel run in parallel revealed enrichment of a specific band (bottom, arrow) migrating at about the same size, as well as the presence of a few contaminant proteins. From these results we conclude that the two-step purification protocol led to highly enriched Top1 proteins. In order to test for CPT sensitivity of the nuclear and mitochondrial Top1 proteins, a plasmid relaxation assay was carried out and the products were analyzed in the presence and absence of EtBr (36) (Figure 5C). Incubation of supercoiled plasmid DNA with nuclear and mitochondrial Top1 led to the formation of relaxed topoisomers as visualized in an agarose gel without EtBr (lanes 2 and 3). Importantly, electrophoresis of DNA treated with nuclear and mitochondrial Top1 proteins in the presence of EtBr shows an increase of relaxed plasmid (RL, lanes 6 and 8). In the presence of CPT in the reaction, a fraction of the relaxed plasmid was converted into nicked, open circular plasmid DNA (OC, lanes 7 and 9). Thus, the nuclear n125Top1 protein is sensitive to CPT in vivo and in vitro, but the mitochondrial mt125Top1 protein is only sensitive to CPT in vitro.
Figure 5.
CPT has no effect on mitochondrially targeted mt125Top1 protein. (A) Drop test analysis of cell viability in cells expressing 125TOP1 constructs in the presence of CPT. (B) Western analysis of n125Top1 and mt125Top1 purified from yeast. Protein size was determined using an anti-GFP antibody and HRP substrate (top left) and inferred from a coloration of the membrane upon incubation with the antibody (top right, arrows). Coomassie blue staining shows a significant enrichment for 125Top1 proteins (bottom, arrow). (C) Biochemical characterization of 125Top1p activity by in vitro DNA relaxation assays. Arrows indicate supercoiled (SC), relaxed (RL), linear (L) and open circular (OC) forms of plasmid analyzed in the presence (lanes 1 to 3) or absence (lanes 4 to 10) of EtBr. Both n125Top1 and mt125Top1 led to the formation of topoisomers (lanes 2 and 3) and to open circular DNA in the presence of CPT (lanes 7 and 9).
Topotecan leads to petite formation and accumulates in yeast mitochondria
While yeast cells are sensitive to CPT treatment, it has been shown that CPT analogs which are soluble in aqueous solutions are much less toxic to yeast cells (37). One such analog is the anticancer drug TPT that induces apoptosis of mammalian tumor cells (38). From microscopic studies it has been inferred that TPT is enriched in mitochondria of mammalian cells (39). We reasoned that the water solubility and positive charge of the TPT molecule (15) could also allow the drug to penetrate mitochondria of S. cerevisiae. Indeed, expression of mt125Top1 in the presence of TPT led to the formation of petite cells (Figure 6A) indicating that TPT reaches the mitochondria in an active form. We determined petite formation by replica-plating of single colonies grown on galactose onto glycerol-containing medium. By counting the colonies growing on glycerol, we found that expression of mt125Top1 in the presence of TPT led to induction of more than 95% petite thus being as potent in generating petite as expression of mt125Top1-103.
Figure 6.
Topotecan is capable of inducing petite and accumulates in mitochondria. (A) Drop test assay to test the effect of TPT treatment on yeast growth. The indicated constructs were expressed under control of the GAL1 promoter and cells were grown in plates without or with 20 µg/ml TPT. Note that petite induced in galactose by expression of mt125Top1-103 under control of the GAL1p grew slower than petite induced in galactose by expression of mt125Top1-103 under control of the MET25p (Figure 3). (B) Intracellular localization of CPT and TPT. Using the settings for the detection of DAPI, the intracellular TPT and CPT localization was determined by their auto-fluorescence. Construct pRWE129 was expressed for GFP-localization of the nucleus. CPT is mainly enriched in vacuoles while TPT fluorescence and Mitotracker staining co-localized (white arrow). Cells without Mitotracker straining were used as control to discard the possibility that we detect TPT auto-fluorescence applying the Mitotracker settings. White bar represents 5 µm.
To enforce the idea that impaired penetration of CPT through mitochondrial membranes impedes the formation of ternary Top1-mtDNA-CPT complex in yeast cells, we determined the subcellular localization of CPT and TPT in yeast cells (Figure 6B). Therefore, we took advantage of the autofluorescence of CPT and TPT to visualize their subcellular localization (40). Without CPT or TPT background signals were barely visible. However, in CPT-treated cells we could see an accumulation of fluorescence in the vacuoles while a clear accumulation of fluorescence in mitochondria was observed in TPT-treated cells. The mitochondrial TPT fluorescence was not dependent on the Mitotracker staining because it could be detected in the absence of Mitotracker. Thus, our results show that TPT accumulates in yeast mitochondria and that the formation of a toxic, ternary complex between Top1-TPT and mtDNA leads to petite formation.
DISCUSSION
The goal of this study has been the development of an experimental system allowing the induction of organelle-specific DNA damage. We show that this can be achieved by inducible expression of a toxic TOP1-103 allele tagged either with a nuclear or mitochondrial localization sequence. Consequently, nuclear DNA damage led to cell death while mtDNA damage led to loss of mtDNA and rho° petite formation. In addition (by inducible expression of a TOP1), we determined that CPT and its derivative TPT are mainly targeting DNA damage to nuclear or mitochondrial DNA, respectively. This finding shows that modifying a DNA-damaging drug might have an important impact on its intracellular distribution.
Addition of multiple SOD2-MLS and partial truncation of a NLS within the N-terminal part of Top1 appeared to be crucial for the targeting of Top1 to mitochondria, while targeting of the N-terminal truncated S. cerevisiae Top1 protein to the nucleus could be achieved by adding an SN40 nuclear localization sequence (Figure S1). The N-terminal truncated Top1 targeted to the nucleus remained as sensitive as wild-type Top1 to CPT both in vivo and in vitro. Interestingly, deletion of an even larger portion of the human Top1 N-terminal domain [up to residue 190, (41)] has been shown to result in a truncated enzyme retaining full catalytic activity. From this result we infer that the basic structural properties of the truncated 125Top1 remain intact. Worthy of note, in addition to DNA nicking and ligation, the partially enriched nuclear 125Top1 had an unexpected in vitro activity. This activity was seen by the formation of linear plasmid DNA in a CPT-independent manner (Figure 5C). In the FPLC salt gradient, the n125Top1 protein always eluted several fractions ahead of the mt125Top1 protein (data not shown). The additional activity of the n125Top1 protein fraction might be due to a contaminant, or n125Top1 may be subjected to post-translational protein modifications in the nucleus, which do not occur in the mitochondria. Previously described cis- or trans-acting activities present in the N-terminal region of Top1 include binding sites for interacting proteins (42), and putative target sequences for protein phosphorylation (43).
Although targeting of the 125Top1 protein to the nucleus is cytotoxic in the presence of CPT, mitochondrial targeting of 125Top1 failed to generate petite cells by CPT treatment. One possibility is that CPT-mediated mtDNA damage might be very efficiently repaired. Since cells lacking the BER enzyme ApnI did not generate petite upon CPT treatment (data not shown), it is less likely that CPT-mediated DNA damage is efficiently repaired by BER. Other possibilities are that CPT is excluded from mitochondria by drug transporters or that CPT is unable to pass the mitochondrial membranes. Upon CPT treatment, Top1-induced DNA nicks have been shown to account for the formation of replication-mediated DNA DSBs (44). Monitoring the inhibition of yeast growth it has been shown that the biological activity of CPT is at least 10-fold higher than that of TPT [(37) and data not shown]. Thus, it is unlikely that CPT-mediated mtDNA damage would be more effectively repaired than TPT-mediated mtDNA damage. Our finding that CPT accumulates in the vacuoles and TPT in the mitochondria favor the possibility that the water and organic solvents insoluble CPT (11) is unable to penetrate the mitochondrial membrane. To easily pass mitochondrial membranes molecules have to be positively charged and water-soluble such as ethidium bromide (EtBr), lipophilic carcinogens (22,45,46) or the water-soluble, positively charged CPT analog TPT (15). Consequently, only nuclear DNA appears to be susceptible to the toxic effects mediated by CPT treatment. If the mitochondrial membrane exhibits a barrier to the passage of CPT, depending on their chemical structure, this might also be true for other CPT analogs.
TPT accumulates in mitochondria and induces respiration-deficient petite cells when S. cerervisiae mitochondria are provided with Top1. We found that TPT treatment of wild-type cells fails to induce petite (data not shown). This result implies that Top1 is not present in S. cerevisiae mitochondria and opens the possibility that other enzymatic activities, such as Top3, substitute for the lack of Top1 in this organelle. In mammalian cells the mitochondrial accumulation of TPT could account in part for its anticancer activity. In line with this finding, the chemical properties of the CPT derivate SN-38 would predict that it is capable of passing the mitochondrial membrane barrier. Indeed, treatment of carcinoma-derived cell lines with SN-38 has been shown to induce the loss of mitochondrial membrane potential as well as mitochondrial membrane permeability transition (47). If we anticipate that mtDNA is a main target for Top1-mediated DNA damage, it would be evident that a functional mtTop1 is required for the full anticancer activity of water-soluble CPT derivatives. Human cells encode for a mitochondria-specific Top1 (6) and it would be very interesting to check if cells lacking this enzymatic activity would be more resistant to TPT treatment. One might speculate that mutations in the human mtTop1 could impair its interaction with TPT leading to increased resistance of cancer cells against therapeutic treatment. Tissue-specific mtTop1 expression levels might also alter the impact of TPT treatment.
We present a novel method for the organelle-targeted induction of Top1p-mediated DNA damage. This system can be easily applied to studies in human cells and provides the means to further advance our knowledge about mitochondria-specific DNA damage repair and checkpoint responses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Drs A. Aguilera, H. Gaillard, D. Fitzgerald, R. McFarlane, F. Monje-Casas and R. Stuckey for helpful discussion and critical reading of the manuscript; Drs G. Fink, J. Fox and J. Hegemann for providing us with strains and plasmids; and M. Hernandez and Dr P. Dominguez for assistance with FPLC-protein purification and microscopy.
FUNDING
Research grant from the Spanish Ministry of Education and Science (MEC; BIO2006-08051 to R.W. and SAF2003-00204 to M.C.L.) and Junta de Andalucía (CVI102). M.C.L. was a recipient of an FPI PhD training grant from the Spanish Ministry of Science and Education. Funding for open access charge: Spanish Ministry of Education and Science (grant BIO2006-8051).
Conflict of interest statement. None declared.
REFERENCES
- 1.Goto T, Laipis P, Wang JC. The purification and characterization of DNA topoisomerases I and II of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1984;259:10422–10429. [PubMed] [Google Scholar]
- 2.Roca J. The mechanisms of DNA topoisomerases. Trends Biochem. Sci. 1995;20:156–160. doi: 10.1016/s0968-0004(00)88993-8. [DOI] [PubMed] [Google Scholar]
- 3.Ezekiel UR, Towler EM, Wallis JW, Zassenhaus HP. Evidence for a nucleotide-dependent topoisomerase activity from yeast mitochondria. Curr. Genet. 1994;27:31–37. doi: 10.1007/BF00326576. [DOI] [PubMed] [Google Scholar]
- 4.Tua A, Wang J, Kulpa V, Wernette CM. Mitochondrial DNA topoisomerase I of Saccharomyces cerevisiae. Biochimie. 1997;79:341–350. doi: 10.1016/s0300-9084(97)80028-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Kearney K, Derby M, Wernette CM. On the relationship of the ATP-independent, mitochondrial associated DNA topoisomerase of Saccharomyces cerevisiae to the nuclear topoisomerase I. Biochem. Biophys. Res. Commun. 1995;214:723–729. doi: 10.1006/bbrc.1995.2345. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Barcelo JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, Pommier Y. Human mitochondrial topoisomerase I. Proc. Natl Acad. Sci. USA. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Meng LH, Pommier Y. Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie. 2007;89:474–481. doi: 10.1016/j.biochi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- 9.Levin NA, Bjornsti MA, Fink GR. A novel mutation in DNA topoisomerase I of yeast causes DNA damage and RAD9-dependent cell cycle arrest. Genetics. 1993;133:799–814. doi: 10.1093/genetics/133.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Megonigal MD, Fertala J, Bjornsti MA. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J. Biol. Chem. 1997;272:12801–12808. doi: 10.1074/jbc.272.19.12801. [DOI] [PubMed] [Google Scholar]
- 11.Wall ME, Wani MC. Camptothecin and taxol: from discovery to clinic. J. Ethnopharmacol. 1996;51:239–253. doi: 10.1016/0378-8741(95)01367-9. discussion 253–234. [DOI] [PubMed] [Google Scholar]
- 12.Kjeldsen E, Svejstrup JQ, Gromova II, Alsner J, Westergaard O. Camptothecin inhibits both the cleavage and religation reactions of eukaryotic DNA topoisomerase I. J. Mol. Biol. 1992;228:1025–1030. doi: 10.1016/0022-2836(92)90310-g. [DOI] [PubMed] [Google Scholar]
- 13.Adams DJ. The impact of tumor physiology on camptothecin-based drug development. Curr. Med. Chem. Anticancer Agents. 2005;5:1–13. doi: 10.2174/1568011053352596. [DOI] [PubMed] [Google Scholar]
- 14.Sawada S, Okajima S, Aiyama R, Nokata K, Furuta T, Yokokura T, Sugino E, Yamaguchi K, Miyasaka T. Synthesis and antitumor activity of 20(S)-camptothecin derivatives: carbamate-linked, water-soluble derivatives of 7-ethyl-10-hydroxycamptothecin. Chem. Pharm. Bull. 1991;39:1446–1450. doi: 10.1248/cpb.39.1446. [DOI] [PubMed] [Google Scholar]
- 15.Underberg WJ, Goossen RM, Smith BR, Beijnen JH. Equilibrium kinetics of the new experimental anti-tumour compound SK&F 104864-A in aqueous solution. J. Pharm. Biomed. Anal. 1990;8:681–683. doi: 10.1016/0731-7085(90)80102-u. [DOI] [PubMed] [Google Scholar]
- 16.Reid RJ, Kauh EA, Bjornsti MA. Camptothecin sensitivity is mediated by the pleiotropic drug resistance network in yeast. J. Biol. Chem. 1997;272:12091–12099. doi: 10.1074/jbc.272.18.12091. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa T. In: Nature Encyclopedia of the Humane Genome, Cooper DN, editor. Vol. 4. London: Nature Publishing Group; 2003. pp. 154–160. [Google Scholar]
- 18.Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, Robey R, Pommier Y, Fojo T, Bates SE. Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999;59:5938–5946. [PubMed] [Google Scholar]
- 19.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl Acad. Sci. USA. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl Acad. Sci. USA. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandavilli BS, Santos JH, Van Houten B. Mitochondrial DNA repair and aging. Mutat. Res. 2002;509:127–151. doi: 10.1016/s0027-5107(02)00220-8. [DOI] [PubMed] [Google Scholar]
- 23.Bogenhagen DF, Pinz KG, Perez-Jannotti RM. Enzymology of mitochondrial base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:257–271. doi: 10.1016/s0079-6603(01)68105-4. [DOI] [PubMed] [Google Scholar]
- 24.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Vongsamphanh R, Fortier PK, Ramotar D. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell. Biol. 2001;21:1647–1655. doi: 10.1128/MCB.21.5.1647-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki M, Takagi E, Kojima K, Izuta S, Yoshida S. Rapid purification and structural study of DNA topoisomerase I from human Burkitt lymphoma Raji cells. J. Biochem. 1989;106:742–744. doi: 10.1093/oxfordjournals.jbchem.a122926. [DOI] [PubMed] [Google Scholar]
- 28.Knab AM, Fertala J, Bjornsti MA. A camptothecin-resistant DNA topoisomerase I mutant exhibits altered sensitivities to other DNA topoisomerase poisons. J. Biol. Chem. 1995;270:6141–6148. doi: 10.1074/jbc.270.11.6141. [DOI] [PubMed] [Google Scholar]
- 29.Naumov GI, Naumova ES, Lantto RA, Louis EJ, Korhola M. Genetic homology between Saccharomyces cerevisiae and its sibling species S. paradoxus and S. bayanus: electrophoretic karyotypes. Yeast. 1992;8:599–612. doi: 10.1002/yea.320080804. [DOI] [PubMed] [Google Scholar]
- 30.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 31.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 32.Alsner J, Svejstrup JQ, Kjeldsen E, Sorensen BS, Westergaard O. Identification of an N-terminal domain of eukaryotic DNA topoisomerase I dispensable for catalytic activity but essential for in vivo function. J. Biol. Chem. 1992;267:12408–12411. [PubMed] [Google Scholar]
- 33.Galanis M, Devenish RJ, Nagley P. Duplication of leader sequence for protein targeting to mitochondria leads to increased import efficiency. FEBS Lett. 1991;282:425–430. doi: 10.1016/0014-5793(91)80529-c. [DOI] [PubMed] [Google Scholar]
- 34.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 35.Williamson D. The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 2002;3:475–481. doi: 10.1038/nrg814. [DOI] [PubMed] [Google Scholar]
- 36.Bailly C. DNA relaxation and cleavage assays to study topoisomerase I inhibitors. Methods Enzymol. 2001;340:610–623. doi: 10.1016/s0076-6879(01)40445-9. [DOI] [PubMed] [Google Scholar]
- 37.Del Poeta M, Chen SF, Von Hoff D, Dykstra CC, Wani MC, Manikumar G, Heitman J, Wall ME, Perfect JR. Comparison of in vitro activities of camptothecin and nitidine derivatives against fungal and cancer cells. Antimicrob. Agents Chemother. 1999;43:2862–2868. doi: 10.1128/aac.43.12.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legarza K, Yang LX. Novel camptothecin derivatives. In Vivo. 2005;19:283–292. [PubMed] [Google Scholar]
- 39.Croce AC, Bottiroli G, Supino R, Favini E, Zuco V, Zunino F. Subcellular localization of the camptothecin analogues, topotecan and gimatecan. Biochem. Pharmacol. 2004;67:1035–1045. doi: 10.1016/j.bcp.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 40.Ziomkowska B, Kruszewski S, Cyrankiewicz M. Deactivation rate of camptothecin determined by factor analysis of steady-state fluorescence and absorption spectra. Optica. Applicata. 2006;XXXVI:10. [Google Scholar]
- 41.Christensen MO, Barthelmes HU, Boege F, Mielke C. Residues 190-210 of human topoisomerase I are required for enzyme activity in vivo but not in vitro. Nucleic Acids Res. 2003;31:7255–7263. doi: 10.1093/nar/gkg927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo YY, Wang C, Beck WT. A novel nuclear localization signal in human DNA topoisomerase I. J. Biol. Chem. 2000;275:41107–41113. doi: 10.1074/jbc.M003135200. [DOI] [PubMed] [Google Scholar]
- 43.Hackbarth JS, Galvez-Peralta M, Dai NT, Loegering DA, Peterson KL, Meng XW, Karnitz LM, Kaufmann SH. Mitotic phosphorylation stimulates DNA relaxation activity of human topoisomerase I. J. Biol. Chem. 2008;283:16711–16722. doi: 10.1074/jbc.M802246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Moustacchi E. Cytoplasmic ‘petite’ induction in recombination-deficient mutants of Saccharomyces cerevisiae. J. Bacteriol. 1973;115:805–809. doi: 10.1128/jb.115.3.805-809.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luedtke NW, Liu Q, Tor Y. On the electronic structure of ethidium. Chemistry. 2005;11:495–508. doi: 10.1002/chem.200400559. [DOI] [PubMed] [Google Scholar]
- 47.Ikegami T, Matsuzaki Y, Al Rashid M, Ceryak S, Zhang Y, Bouscarel B. Enhancement of DNA topoisomerase I inhibitor-induced apoptosis by ursodeoxycholic acid. Mol. Cancer Ther. 2006;5:68–79. doi: 10.1158/1535-7163.MCT-05-0107. [DOI] [PubMed] [Google Scholar]
- 48.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.