Abstract
The C-terminal domain (CTD) of RNA polymerase II regulates transcription through spatially and temporally coordinated events. Previous work had established that the CTD binds DNA but the significance of this interaction has not been determined. The present work shows that the CTD binds DNA in its unphosphorylated form, the form in which it is present in the pre-initiation complex. The CTD/DNA complex is recognized by and is phosphorylated by Cdk7 but not by Cdk9. Model-building studies indicate the structural mechanism underlying such specificity involves interaction of Cdk7 with DNA in the context of the CTD/DNA complex. The model has been tested by mutagenesis experiments. CTD dissociates from DNA following phosphorylation by Cdk7, allowing transcription initiation. The CTD then becomes accessible for further phosphorylation by Cdk9 that drives the transition to transcription elongation.
INTRODUCTION
Phosphorylation of RNA polymerase II (RNA-Pol II) carboxy-terminal domain (CTD) by cyclin-dependent kinase 7 (Cdk7) and cyclin-dependent kinase 9 (Cdk9) in association with their cyclins regulates transcription and allows the coordination of transcription with mRNA maturation. The mechanisms underlying this regulation are complex. The human CTD comprises 52 repeats of the heptapeptide Y1S2P3T4S5P6S7 (mostly consensus at the N-terminal region with divergent repeats located at the C-terminal region) and is involved in binding with several proteins. Depending on its phosphorylation state, the CTD can discriminate among its binding partners and hence is poised to organize transcription both temporally and spatially (1).
The unphosphorylated CTD binds proteins of the pre-initiation complex like the TATA-binding protein (TBP) (2) and the Mediator complex (3) and is involved in the assembly of the inactive transcription machinery on the promoter DNA. Phosphorylation of CTD Ser5 by Cdk7 disrupts those interactions resulting in an active RNA-Pol II and a scaffold complex that remains at the promoter. Phospho-Ser5 CTD is recognized by the capping enzyme (4,5). The RNA cap structure is added shortly after transcription initiation, when nascent RNA is about 25–30 nt in length.
Ser5 phosphorylated CTD RNA-Pol II is competent for initiation of transcription but is unable to enter the elongation phase and to transcribe the entire mRNA (6,7). This form of the RNA-Pol II binds the DRB (5,6-dichlorobenzimidazole 1-β-d-ribofuranoside)-sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF), the two inhibitors of the elongation phase of transcription, that lead to generation of abortive transcripts (8). Inhibition of elongation is removed following phosphorylation of DSIF and NELF by Cdk9 (9–11). Phosphorylation of CTD Ser2 by Cdk9 generates a new form of the RNA-Pol II, that is recognized by elongation factors like Elongator and allows the assembly of the elongation-competent RNA-Pol II (12,13). Moreover the Ser2-Ser5 phosphorylated CTD is recognized by splicing factors and is required for assembly of the spliceosome and for efficient splicing reactions on the elongating RNA (14). Finally, although polyadenylation, cleavage and transcription termination requires interactions of the cleavage/polyadenylation specificity factor (CPSF) and of the Cleavage-stimulation Factor (CstF) with CTD these interactions do not appear to be dependent on the phosphorylation status of the CTD (15–17).
This strict spatio-temporal organization for Cdk7 and Cdk9 action on CTD requires a mechanism discriminating between the two kinases (that are present at the same time in the same location) so that they can never act together but only sequentially.
The work reported here describes a possible mechanism starting with the observation that CTD binds DNA in its unphosphorylated form. The CTD/DNA complex is recognized by Cdk7 that can then phosphorylate Ser5. This releases the CTD from the DNA and makes it available for Cdk9 phosphorylation. A structural model for CTD/DNA recognition by Cdk7 is proposed and tested with mutagenesis studies.
MATERIALS AND METHODS
Protein production and mutagenesis
Cdk7, Cdk7/CycH and Cdk9-330/CycT1-298 were produced in baculo-viral-infected insect cells as previously described (18,19). Cdk9-330 (residues 1–330) corresponds to the Cdk9 kinase domain. CycT1-298, corresponding to residues 1–298, contains the two cyclin boxes required for Cdk9 activation.
Mutagenesis for pCdk2 were performed using the ‘Site Directed Mutagenesis Kit’ (Stratagene, La Jolla, CA, USA) and following manufacturer's instructions. Unphosphorylated Cdk2, WT-pCdk2, pCdk2 mutants, CycA, pCdk2/CycA and pCdk2/CycA mutants were purified as previously described (20).
Cdk7/CycA was obtained by immobilizing GST-Cdk7 on glutathione-sepharose and binding CycA from the CycA-expressing Escherichia coli lysate. Elution was performed with GST-3C protease that removed the GST tag from Cdk7.
pCdk2/CycH and its mutants were obtained by immobilizing GST-pCdk2 or GST-pCdk2 mutants on glutathione-sepharose and binding CycH from the CycH-expressing E. coli lysate. Elution was performed with GST-3C protease.
Human CTD (comprising all 52 repeats) was expressed as N-terminal GST-fusion protein and purified as previously described (18). pSer5-GST-CTD was obtained incubating 30 μg of GST-CTD with 0.3 μg of Cdk7/CycH in 50 mM Tris pH 8, 10 mM MgCl2 and 0.1 mM ATP for 2 h at RT. Cdk7/CycH was then removed by immobilizing pSer5-GST-CTD on glutathione-sepharose and extensively washing. pSer5-GST-CTD was eluted with 10 mM glutathione.
Kinase assays
Cdk7/CycH, Cdk9-330/CycT1-298, Cdk7/CycA, pCdk2/CycA pCdk2/CycH and pCdk2 mutants in complex with CycA or CycH activities were measured by following the incorporation of radiolabeled phosphate into substrate. GST-CTD (5 μg), alone or in complex with DNA, was incubated with 100 ng of the different Cdk/cyclin complexes in 10 μl kinase buffer [0.1 mM ATP, 10 mM MgCl2, 50 mM Tris/HCl pH 7.5, 1 μCi γ-32P-labeled ATP (MP Biomedicals)]. The reaction mixtures were incubated for 5 min at 20°C and terminated by the addition of SDS sample buffer. Samples were analyzed on SDS–PAGE gels and visualized by autoradiography. All reactions were performed in duplicate.
For the GST–CTD/DNA complex, GST-CTD samples (5 μg = 60 pmol) were previously mixed with 200 pmol of double-stranded DNA for single-point experiments or with 12, 60, 300 or 1500 pmol for titration experiments. Oligonucleotides (45 nt each: GTTACTTCTATGCCTGATTACGTCAGTTTCCCCAAGTGGGCCCGG and CCGGGCCCACTTGGGGAAACTGACGTAATCAGGCATAGAAGTAAC) were annealed in 10 mM Tris pH 8, 50 mM NaCl prior to incubation with GST–CTD in 20 mM Tris pH 8, 20 mM NaCl, 1 mM MgCl2 in ice for 30 min. Same conditions were used to test the phosphorylating activity of Cdk7/CycH on Cdk2.
Pixels were quantified using the ImageJ software (NIH) and converted into MBq using the equation: pixels = A ln(Mbq) + B. A and B are parameters derived from experimental calibration. Data were normalized with respect to the enzyme concentration.
Electrophoresis mobility shift assay
Two complementary oligonucleotides (45 nt each) were mixed in 10 mM Tris pH 8, 50 mM NaCl and 1 mM EDTA and annealed by heating to 95°C and slow cooling to RT. GST–CTD was phosphorylated by incubation with 100 ng of Cdk7/CycH or with 100 ng of Cdk9/CycT1-298 or with 100 ng of Cdk7/CycH and 100 ng of Cdk9/CycT1-298 in 50 mM Tris pH 8, 0.1 mM ATP and 10 mM MgCl2 for 2 h. Binding to DNA was tested by mixing 40 ng of DNA (2 pmol) with 0.4, 2, 10 or 50 pmol of the differently phosphorylated GST–CTD in 20 mM Tris pH 8, 20 mM NaCl, 40 mM KCl, 2 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 4% glycerol. Samples were incubated on ice for 30 min and then loaded on a 6% native acrylamide gel. Gels were stained using SYBR Green (Invitrogen).
Computational modelling
The CTD peptide 1YSPTSPSY8 was manually built using restraints from the NMR model of its complex with DNA (21). It was subsequently docked into a 6 bp dsDNA by superimposing Tyr1 and Tyr8 with the two quinoxaline rings of triostin A (PDB 185D and 1VS2). The complex was optimized using restraints from the NMR model of the CTD/DNA complex and energy minimized using Haddock (22,23).
Cdk7/CycH complex was modeled on pCdk2/CycA structure (PDB 1QMZ) using Cdk7 and CycH structures (PDB 1UA2 and 1KXU). CTD/DNA complex was docked into Cdk7/CycH by superimposing 5SP6 to the corresponding amino acids of the substrate peptide co-crystallized with pCdk2/CycA (PDB 1QMZ). The quaternary complex was energy minimized using Haddock.
RESULTS
Unphosphorylated CTD binds DNA but CTD phosphorylated by Cdks does not bind DNA
Previous work has shown that the CTD repeats bind DNA by intercalating their tyrosines between bases [Ka = 100 M−1 in the interaction of the YSPTSPSYSPTSPSY peptide with salmon testis DNA (21,24,25)]. How this binding might be modified by CTD phosphorylation is not known. CTD binding to DNA was analysed by band shift using unphosphorylated GST–CTD or GST-CTD phosphorylated by Cdk7/CycH, Cdk9/CycT1-298 or by both Cdk7/CycH and Cdk9/CycT1-298. Cdk7/CycH specifically phosphorylates CTD-Ser5 while Cdk9/CycT acts on CTD-Ser2 (26–28).
Unphosphorylated CTD was found to bind DNA (Figure 1A, lanes 2–5). Binding of CTD to DNA was abolished following Ser5 phosphorylation by Cdk7/CycH (Figure 1A, lanes 6–9) or following Ser2/Ser5 phosphorylation by Cdk7/CycH and Cdk9/CycT1 (Figure 1A, lanes 14–17). Binding to DNA was reduced when CTD was phosphorylated on Ser2 by Cdk9/CycT1 (Figure 1A, lanes 10–13).
Figure 1.
Effect of phosphorylation on CTD binding to DNA and effect of DNA binding on CTD phosphorylation by Cdk7 and Cdk9. (A) DNA binding by different CTD phospho-isoforms. CTD phospho-isoforms were generated by incubating CTD with the respective Cdk in kinase buffer for 2 h at RT. Increasing amounts of CTD and its phospho-isoforms were then incubated with DNA in binding buffers for 30 min at 4°C and then loaded on a 6% native acrylamide gel. Unphosphorylated CTD binds DNA (lanes 2–5). No binding could be detected using CTD phosphorylated by Cdk7 (lanes 6–9) or by both Cdk7 and Cdk9 (lanes 14–17). Binding affinity is strongly decreased when CTD is phosphorylated by Cdk9 (lanes 10–13). (B) CTD and CTD/DNA phosphorylation by Cdk7 and Cdk9. CTD/DNA complex was generated by incubating CTD with a 44 bp double-stranded DNA in binding buffer for 30 min. Kinase assay was performed by incubating 100 ng of each kinase and 5 μg of CTD, either alone or in complex with increasing amount of DNA, in kinase buffer for 5 min at 20°C. CTD phosphorylation by Cdk7 (lane 1) is increased when CTD is in complex with DNA (lanes 2–5). Cdk9 prefers CTD (lane 6) over CTD/DNA (lanes 7–10) as substrate. Cdk2 phosphorylation by Cdk7 is not affected by DNA (lanes 11–15). Once phosphorylated, Cdk2 is able to phosphorylate Cdk7 (faint band just above Cdk2 band). Cdk7 is produced in baculo-viral-infected insect cells and reproducibly purified as 70% monophosphorylated and 30% bisphosphorylated (18).
In its non-phosphorylated form the CTD is able to bind DNA and this would be the form present in the context of the pre-initiation complex. During transcription initiation the CTD is phosphorylated primarily on Ser5 by Cdk7. This event could promote initiation of transcription not only disassembling the pre-initiation complex but also disengaging CTD from DNA.
DNA-bound CTD is the preferred substrate of Cdk7 but not of Cdk9
RNA-Pol II in the pre-initiation complex is phosphorylated primarily by Cdk7 although both Cdk7 and Cdk9 are present in every stage of transcription. Does binding to DNA lock CTD in a conformation recognised by Cdk7 but not by Cdk9?
Cdk7 and Cdk9 were tested for their ability to phosphorylate the CTD by itself and in complex with DNA. The CTD/DNA complex is a better substrate for Cdk7 than the CTD alone (Figure 1B, lanes 1–5) but the CTD alone is a better substrate for Cdk9 than the CTD/DNA complex (Figures 1B, lanes 6–10). This suggests that CTD bound to DNA confers substrate discrimination for the two Cdks allowing preferential phosphorylation by Cdk7 and reducing phosphorylation by Cdk9.
Cdk2 phosphorylation by Cdk7/CycH was not affected by DNA (Figure 1B, lanes 11–15), confirming that Cdk7 is not a DNA-activated kinase and that the effect observed on CTD phosphorylation has to be ascribed to the CTD–DNA complex formation and to the substrate presentation to Cdk7.
A model for Cdk/CTD/DNA complex
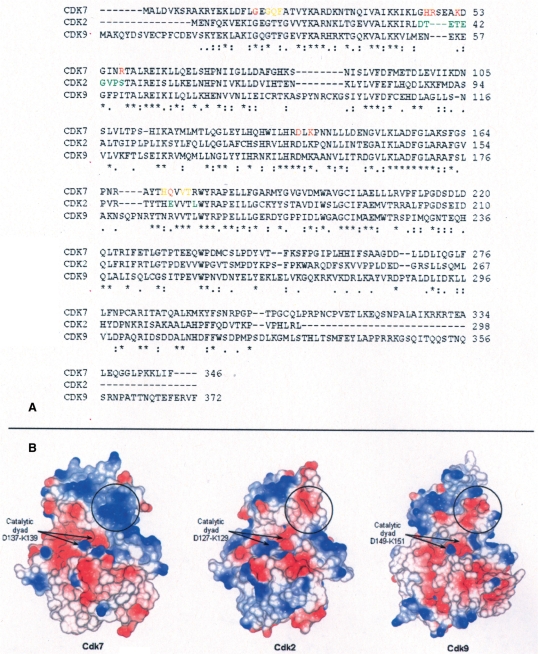
To understand these different recognition properties, a model of the CTD/DNA complex was built based on the NMR model of a dsDNA hexamer in complex with the YSPTSPSY CTD peptide (21) and on the crystal structures of DNA–triostin A complex (PDB 185D and 1VS2) (29–31). Triostin A is a bis-intercalating antibiotic mimicking the binding mode to DNA of single CTD repeats (24). In the model of the DNA/CTD complex, Ser5 is exposed but Ser2 is buried because of its proximity to the buried tyrosine of the CTD (Figure 2A). The CTD is in a beta-turn conformation generated by the DNA-intercalating tyrosines (21).
Figure 2.
CTD/DNA complex and model of interaction with Cdk7/CycH. (A) Two different views of the CTD/DNA complex (left) and their surface representation (right). DNA is in yellow, CTD in magenta, Ser5 in blue and Ser2 in grey. Ser5 is exposed while Ser2 is poorly accessible. (B) CTD/DNA docked inside Cdk7/CycH active site. DNA is in yellow, CTD in magenta, Cdk7 in green and CycH in cyan. Polar interactions are represented as dotted lines. CTD Ser5 is in contact with Cdk7 catalytic Asp137 and Lys139. A basic stretch of amino acids in Cdk7 contacts DNA backbone.
The DNA/CTD complex was subsequently docked in the Cdk7/CycH complex using as a guide information from the crystal structure of Cdk2 in complex with a Ser-Pro containing substrate peptide. The quaternary complex was energy minimized. Ser5 could be easily placed inside Cdk7 active site and in contact with catalytic amino acids Asp137 and Lys139. Pro6 is in the proline pocket generated by Val173, Val174 and Thr175 typical of Ser-Pro directed kinases like Cdks (Figure 2B). The quaternary complex was submitted to Protein Interfaces, Surfaces and Assemblies service PISA at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (32). Output values for solvent-accessible surface areas buried and energies of complex formation are given in Table 1. Buried surface area for CycH contacting Cdk7 (2470 Å2) is typical of a stable protein–protein interaction (3250 Å2 for the Cdk2/CycA complex and 1740 Å2 for the Cdk9/CycT1 complex). Buried surface area for CTD contacting Cdk7 (503 Å2) is typical of enzyme substrate interactions (930 Å2 for the Cdk2/substrate peptide complex) and is reinforced by the contacts with DNA. Polar contacts with distance <3.5 Å between Cdk7 and the CTD/DNA complex are given in Table 2. No intermolecular bad contacts were present in the quaternary complex.
Table 1.
Solvent-accessible surface areas buried upon complex formation and energies of interactions in the Cdk7/CycH/CTD/DNA model
| Molecule A | Molecule B | Buried surface area (Å2) | ΔG (Kcal/mol) |
|---|---|---|---|
| Cdk7 | CycH | 2470 | −12.4 |
| Cdk7 | CTD (YSPTSPSY) | 503 | −1.9 |
| Cdk7 | DNA (6 bp) | 798 | −7.1 |
| CTD (YSPTSPSY) | DNA (6 bp) | 1011 | −1.3 |
Table 2.
Polar contacts between Cdk7 and the CTD/DNA complex
| Cdk7 | CTD/DNA | Distance |
|---|---|---|
| Gly21 O | Thr4 OG | 3.1 |
| His47 NE2 | PO-backbone | 3.4 |
| Arg48 NH1 | PO-backbone | 3.2 |
| Arg48 NH2 | PO-backbone | 3.2 |
| Lys52 NZ | PO-backbone | 2.8 |
| Arg57 NH1 | PO-backbone | 3.3 |
| Arg57 NH2 | PO-backbone | 2.9 |
| Asp137 OD1 | Ser5 OG | 3.4 |
| Asp137 OD2 | Ser5 OG | 3.0 |
| Lys 139 NZ | Ser5 OG | 3.4 |
| Gln 172 NE2 | DNA base | 2.9 |
| Gln 172 OE1 | Ser7 OG | 3.3 |
The CTD/DNA complex inserts between the two kinase lobes, both of which contribute to the binding via van der Waals and polar interactions (Figure 2B). The catalytic dyad Asp137-Lys139, responsible for the phospho-transfer from ATP to the substrate, is conserved in all Cdks as well as Gly21 that interacts with CTD via its peptide backbone. Other interactions involve Gln172 and four residues of the β3-αC loop that is longer and more basic in Cdk7 than in the other Cdks (residues 45–57 in Cdk7; Figures 2B and 3). The interaction between the β3-αC loop and DNA spans four bases of the same DNA strand (Table 2). It is largely responsible for the calculated energy of interaction between Cdk7 and the CTD/DNA complex and gives the correct orientation to the CTD/DNA complex to accommodate Ser5 inside Cdk7 active site. These interactions could increase Cdk7 affinity for CTD/DNA selecting it as a better substrate rather than the CTD alone.
Figure 3.
Primary and tertiary structure comparison for Cdk7, Cdk2 and Cdk9. (A) Sequence alignment. Cdk7 amino acids contacting the CTD/DNA complex (distance <3.5 Å) are shown in red (polar contacts) and orange (van der Waals contacts). The model of CTD/DNA recognition by Cdk7 was tested using Cdk2 mutants. Mutated residues are shown in green in the Cdk2 sequence. (B) Electrostatic surface representation. Black circles delimitate β3αC loops of the three kinases. Differently from Cdk2 and Cdk9, the Cdk7 β3αC loop is positively charged.
Docking of Ser2 into the catalytic site of Cdk7 generates extensive clashes between the CTD/DNA complex and Cdk7 activation segment and αD, αF and αG helices (labeled in Figure 2B). Ser2 is next to the intercalating Tyr1 and is poorly accessible.
The model identifies Ser5 phosphorylation by Cdk7 as the most likely site of modification of the CTD/DNA complex. Cdk9 has a different β3-αC loop (residues 52–61) and a different surface potential that would disfavor its interaction with CTD/DNA (Figures 2 and 3). When promoting transition from the pre-initiation to the initiation stage of transcription, Cdk7 is part of the basal transcription factor TFIIH. TFIIH has a significantly higher CTD phosphorylating activity than the Cdk7/CycH complex (33). TFIIH contains DNA-binding factors, like the N-terminal zinc-finger domain of MAT1 or the helicases XPB and XPD, which could participate in increasing affinity and activity of TFIIH for its substrate CTD/DNA. It is possible that, in comparison to Cdk7/CycH, the TFIIH complex would show an even greater preference for CTD/DNA over CTD.
The CTD/DNA complex can be a substrate for Cdk7/CycH for phosphorylation on Ser5 but it will be a poor substrate for Cdk7/CycH for phosphorylation on Ser2 and for Cdk9/CycT for phosphorylation on both Ser5 and Ser2.
Cdk7 β 3-αC loop is sufficient to favor phosphorylation of CTD/DNA over CTD
The β3-αC loop is flexible in many Cdk/cyclin complexes and appears to be a region that allows response to different cyclins and inhibitors. In order to verify the role of Cdk7 β3-αC loop in favoring phosphorylation activity on CTD/DNA over CTD, a Cdk2 mutant (named Cdk2-β3αC) was constructed substituting part of its β3-αC loop 38DTET—EGVPS46 with the corresponding sequence in Cdk7 GHRTEAKDGVNR. Cdk2 was used to compare with Cdk7 because it can be activated in vitro by both its physiological partner CycA and by CycH, the physiological Cdk7 cyclin, and because the E. coli expression system made the preparation of mutants easier than with the baculovirus system for Cdk7. Likewise, Cdk7 is activated in vitro by both CycH and CycA. Comparison of those two Cdks, in complex with one or the other cyclin, could uncover any involvement of cyclins in the preferential recruitment of CTD/DNA over CTD. The Cdk2- β3αC mutant was further mutagenized to substitute part of Cdk2 activation segment 162EVVTL166 with the corresponding sequence in Cdk7 QVVTR. This mutant, named Cdk2-β3αCact, contains two additional mutations: Glu to Gln (Gln172 in Cdk7) predicted to interact with the CTD/DNA complex and Leu to Arg (Arg176 in Cdk7) that is unique in Cdk7 and found to be involved in recognition of the other Cdks when Cdk7 acts as Cdk-activating kinase (CAK) (34).
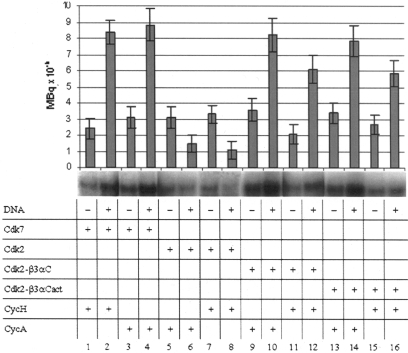
Both Cdk7/CycH and Cdk7/CycA show a greater activity in phosphorylating CTD/DNA than the CTD alone (Figure 4, lanes 1–4). However, Cdk2/CycA and Cdk2/CycH show a small preference for CTD as substrate (Figure 4, lanes 5–8). The effect is less pronounced than that observed for Cdk9/CycT1 (Figure 1B). The observation that similar effects are observed with Cdks in complex with different cyclins leads to the conclusion that cyclins are not involved in substrate selection.
Figure 4.
CTD and CTD/DNA phosphorylation by Cdk7, Cdk2 and Cdk2 mutants in complex with either CycH or CycA. All reactions were performed in duplicate. Root mean square deviations are shown as error bars. Samples were loaded on 4–12% gradient SDS acrylamide gels and visualized by autoradiography.
Cdk2-β3αC/CycH and Cdk2-β3αC/CycA behave like Cdk7/CycH and Cdk7/CycA showing that Cdk7-β3αC loop is sufficient to favor CTD/DNA over CTD phosphorylation (Figure 4, lanes 9–12). No substantial differences could be detected between Cdk2-β3αC and Cdk2-β3αCact in complex with both CycH and CycA (Figure 4, lanes 9–12 compared with lanes 13–16). Cdk7 Gln172 and Arg176 appear not to be essential for preferential CTD/DNA phosphorylation. In the model, Arg176 was not involved in any interaction with CTD/DNA. Gln172, predicted to interact with CTD/DNA, is substituted by Glu in Cdk2 and by Arg in Cdk9 both capable of partially reproducing Gln172 interactions shown in Table 2.
The basic Cdk7 β3-αC loop, complementing the nucleic acid backbone, directs the kinase action towards the CTD–DNA complex.
Previous CTD phosphorylation by Cdk7 does not alter CTD-phosphorylating activity of Cdk9
Cdk9 phosphorylates CTD Ser2 following CTD Ser5 phosphorylation by Cdk7. It is tempting to speculate that a docking site for CTD pSer5 on the Cdk9 surface would help Cdk9 in selecting such substrate. I therefore tested if prior phosphorylation of the CTD by Cdk7 made it a better substrate for Cdk9. Comparison of Cdk9 activity on unphosphorylated CTD and pSer5-CTD (obtained by previous incubation with Cdk7) did not show significant differences (Figure 5). The effect of phosphorylation by Cdk7 is not to make the CTD a better substrate for Cdk9. CTD binding and release from DNA and initiation transcription factors could be the event governing the right timing of Cdk7 and Cdk9 action on CTD.
Figure 5.
Cdk9 activity on CTD and pSer5-CTD. (A) CTD and pSer5-CTD were loaded on SDS–PAGE. Phosphorylation by Cdk7/CycH shifts the CTD band upwards. (B) Kinase assay. Cdk9/CycT1 phosphorylates equally well CTD and pSer5-CTD (generated by previous incubation with Cdk7/CycH). No further incorporation of 32P could be detected incubating pSer5-CTD with Cdk7/CycH indicating that the previous reaction went to completeness.
DISCUSSION
CTD binds dsDNA, but not ssDNA, in an unspecific non-DNA sequence-dependent manner (24,35). The work reported here highlights this binding property as specific for unphosphorylated CTD. Phosphorylation by Cdk7 abolishes binding and phosphorylation by Cdk9 strongly reduces binding. In the CTD/DNA complex, Ser2 is very close to the DNA backbone and addition of a phosphate group to it is not compatible with DNA binding. The beta-turn conformation of the CTD bound to DNA is centered on Ser5 and its phosphorylation would impede maintenance of such a compact structure. The present work shows that the preference for prior phosphorylation by Cdk7 resides in the availability of Ser5 exposed to solvent in the CTD/DNA complex while Ser2 is buried and is not accessible. In the absence of DNA, phosphorylation of Ser2 by Cdk9 proceeds with equal efficiency regardless of whether Ser5 is phosphorylated or not. However, phosphorylation by Cdk7 on Ser5 results in a CTD that can no longer bind DNA and hence making Ser2 available for phosphorylation by Cdk9.
CTD-Ser7 has recently been found to become phosphorylated during the transcription cycle. Ser7 is buried in the CTD–DNA model and access to it is not compatible with CTD binding to DNA. Consequently, CTD phosphorylated on Ser7 should not be able to bind DNA. This is in agreement with the observation that Ser7 phosphorylation is enriched on RNA-Pol II moving toward the end of genes (36) and is involved in 3' processing of transcripts (37). CTD-Tyr1 is phosphorylated by the c-Abl kinase (38), although a physiological role for this event has not been elucidated. Phosphorylation on CTD-Thr4 has not been reported so far. In the proposed CTD–DNA model, Tyr1 is intercalating between DNA base pairs and is not accessible, while Thr4 is partially exposed to the solvent.
Cdk7 and Cdk9 have different phosphorylating propensities for different CTD repeats. Cdk7 phosphorylates equally well N-terminal repeats (mostly consensus) and C-terminal repeats (divergent from consensus sequence), while Cdk9 has a strong preference for consensus repeats and weak activity on C-terminal repeats (39,40). pSer2-CTD, generated by Cdk9 action, has then some unphosphorylated repeats still able to bind DNA, while phosphorylation on Ser5 throughout the entire CTD by Cdk7 completely abolishes DNA binding. This can be ascribed to the compact beta-turn conformation, required for DNA binding and largely determined by Pro6 and tyrosines intercalation, that cannot accommodate a phosphate group on Ser5, which in contrast would force CTD to change conformation.
CTD/DNA complex is a better substrate for Cdk7 than free CTD. The model of the Cdk7/cyclin H/CTD/DNA complex indicates that DNA recognition is achieved by the Cdk7 β3-αC loop, and this observation is supported by mutagenesis experiments. When bound to DNA, CTD phosphorylation by Cdk9 is strongly reduced. In the CTD/DNA complex, Ser2 is poorly accessible so that, following DNA binding, Cdk9 should be unable to phosphorylate it. The observation that some phosphorylation did occur in the presence of Cdk9 indicates that, in these experimental conditions, some CTD repeats are not involved in DNA binding.
At the pre-initiation stage CTD is bound to DNA and to several transcription factors (41). Those interactions hide CTD to Cdk9 via steric hindrance and/or electrostatic repulsion. Only after such interactions have been removed by Cdk7 phosphorylation of CTD, Cdk9 can have access to it. Cdk7 does not make CTD a better substrate for Cdk9 but only makes it available for further modifications. After initiation, transcription pauses before entering elongation phase. This pause, required for RNA capping before elongation, could be ascribed to a delay in Cdk9 phosphorylation of CTD. Initial CTD phosphorylation by Cdk9 is a slow event while subsequent phosphate additions are more rapid (40,42).
FUNDING
This work was supported by the Federation of European Biochemical Societies [Long-Term Fellowship ref. FEY]. Funding for open access charge: Istituto di Ricerche di Biologia Molecolare “P. Angeletti”.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
I am grateful to Prof. Louise Johnson for useful discussions throughout this work and for critical reading of the manuscript and to Dr Alessandro Grottesi and Dr Pietro Roversi for their help with computational analysis.
REFERENCES
- 1.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 2.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 3.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu. Rev. Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 4.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl Acad. Sci. USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobor MS, Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- 7.Oelgeschlager T. Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J. Cell. Physiol. 2002;190:160–169. doi: 10.1002/jcp.10058. [DOI] [PubMed] [Google Scholar]
- 8.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 2001;276:12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- 11.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 13.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 14.Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem. Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 15.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 16.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 18.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure. 2004;12:2067–2079. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb, its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell. Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 21.Khiat A, Lamoureux M, Boulanger Y. Structural differences between the free and bound states of the DNA-bisintercalating peptide YSPTSPSY. J. Med. Chem. 1996;39:2492–2498. doi: 10.1021/jm9503254. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 1993;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 23.de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki M. The heptad repeat in the largest subunit of RNA polymerase II binds by intercalating into DNA. Nature. 1990;344:562–565. doi: 10.1038/344562a0. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Shullenberger DF, Long EC. Aromatic stacking and bending of the DNA helix by the individual repeat units of the carboxy-terminal domain of RNA polymerase II. Biochem. Biophys. Res. Commun. 1994;198:712–719. doi: 10.1006/bbrc.1994.1103. [DOI] [PubMed] [Google Scholar]
- 26.Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 27.Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Addess KJ, Feigon J. Sequence specificity of quinoxaline antibiotics. 1. Solution structure of a 1:1 complex between triostin A and [d(GACGTC)]2 and comparison with the solution structure of the [N-MeCys3,N-MeCys7]TANDEM-[d(GATATC)]2 complex. Biochemistry. 1994;33:12386–12396. doi: 10.1021/bi00207a005. [DOI] [PubMed] [Google Scholar]
- 30.Wang AH, Ughetto G, Quigley GJ, Hakoshima T, van der Marel GA, van Boom JH, Rich A. The molecular structure of a DNA-triostin A complex. Science. 1984;225:1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]
- 31.Wang AH, Ughetto G, Quigley GJ, Rich A. Interactions of quinoxaline antibiotic and DNA: the molecular structure of a triostin A-d(GCGTACGC) complex. J. Biomol. Struct. Dyn. 1986;4:319–342. doi: 10.1080/07391102.1986.10506353. [DOI] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Yankulov KY, Bentley DL. Regulation of the CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lolli G, Johnson LN. Recognition of Cdk2 by Cdk7. Proteins. 2007;67:1048–1059. doi: 10.1002/prot.21370. [DOI] [PubMed] [Google Scholar]
- 35.Peterson CL, Kruger W, Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 36.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 37.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baskaran R, Escobar SR, Wang JY. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth Differ. 1999;10:387–396. [PubMed] [Google Scholar]
- 39.Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe'ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- 40.Pinhero R, Liaw P, Bertens K, Yankulov K. Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur. J. Biochem. 2004;271:1004–1014. doi: 10.1111/j.1432-1033.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- 41.Douziech M, Forget D, Greenblatt J, Coulombe B. Topological localization of the carboxyl-terminal domain of RNA polymerase II in the initiation complex. J. Biol. Chem. 1999;274:19868–19873. doi: 10.1074/jbc.274.28.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]