Abstract
We have prepared triplex-forming oligonucleotides containing the nucleotide analogue 5-dimethylaminopropargyl deoxyuridine (DMAPdU) in place of thymidine and examined their ability to form intermolecular triple helices by thermal melting and DNase I footprinting studies. The results were compared with those for oligonucleotides containing 5-aminopropargyl-dU (APdU), 5-guanidinopropargyl-dU (GPdU) and 5-propynyl dU (PdU). We find that DMAPdU enhances triplex stability relative to T, though slightly less than the other analogues that bear positive charges (T << PdU < DMAPdU < APdU < GPdU). For oligonucleotides that contain multiple substitutions with DMAPdU dispersed residues are more effective than clustered combinations. DMAPdU will be especially useful as a nucleotide analogue as, unlike APdU and GPdU, the base does not require protection during oligonucleotide synthesis and it can therefore be used with other derivatives that require mild deprotection conditions.
INTRODUCTION
The formation of three-stranded DNA complexes can be exploited as a means to recognize specific DNA sequences with several applications in diagnostics and medicine (1–4). In this strategy a triplex-forming oligonucleotide (TFO) binds to a specific double-stranded DNA sequence by hydrogen bonding between its bases and exposed groups on the duplex base pairs, generating base triplets (5). Oligonucleotides composed of pyrimidine bases bind in a parallel orientation to the purine-strand of the duplex forming T.AT and C+.GC triplets (6). Although triplex formation has been demonstrated as a way of introducing mutations in cell culture and animal models (7–10) its efficiency under physiological pH and ionic conditions is limited by several intrinsic limitations. For example, the binding of a TFO is often weak, due to charge repulsion between the three polyanionic DNA strands and a requirement for protonation of third strand cytosines. Triplex formation is also restricted to oligopurine.oligopyrimidine target sites as there is no method for recognizing pyrimidine.purine base pairs using natural bases. To overcome these limitations we and others have prepared nucleotide analogues and have used them to generate oligonucleotides that form stable triplexes at mixed sequence duplex targets at physiological pH (1,11–13).
One of the simplest strategies to alleviate the charge repulsion problem has been to use nucleoside analogues that contain positively charged moieties on the sugar, the phosphate or the base (14–21). To this end the nucleoside analogues 5-aminopropargyl-dU and 2′-aminoethoxy,5-aminopropargyl-dU position protonated amines to interact with the negative phosphodiester residues within the duplex purine-strand and/or the TFO and form more stable triplets with AT base pairs than T (8,15,18,20,21). However, a drawback of these analogues is the relatively harsh conditions required to deprotect the amino groups during oligonucleotide synthesis. This limits the compatibility of such modifications with other chemical groups, such as the cross-linking agent psoralen used in gene-targeting. There is therefore a need to develop novel charged nucleosides that do not require, or require less harsh deprotection conditions.
We have previously reported the synthesis of the nucleoside analogue 5-dimethylaminopropargyl deoxyuridine (DMAPdU; Figure 1A) (22). This analogue is similar to 5-aminopropargyl dU (APdU) but contains a dimethylamine in place of the primary amine at the 5-position. In contrast to a primary amine, DMAPdU does not require protection during phosphoramidite monomer or oligonucleotide synthesis. Preliminary melting experiments have shown that although DMAPdU is not as stabilizing as APdU against an AT base pair it does lead to enhanced triplex stability (22). Here we further explore the sequence selectivity and affinity of this analogue by examining TFOs that contain single or multiple substitutions of the analogue at different positions within the TFO. We also examine its effect on the kinetics of triplex formation. The properties of DMAPdU are compared with three other uridine derivatives; 5-aminopropargyl-dU (APdU); 5-propynyl-dU (PdU), which lacks the charge at the end of the propargyl chain and 5-guanidinopropargyl-dU (GPdU), which contains a guanidine at the same position. The structures of these analogues and the proposed base triplets that they generate with an AT base pair are shown in Figure 1A.
Figure 1.
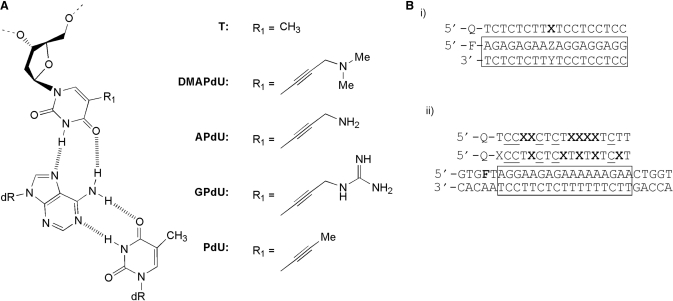
(A) Chemical structures of the X.AT triplets, where X is either T or the 5-position deoxyuridine analogues examined in this study (B) (i) Sequences of the oligonucleotides used in single substitution fluorescence melting studies. The third strand is labelled with dabcyl (Q) at the 5′-end, whilst the purine strand of the duplex (boxed) is labelled with fluorescein (F) at the 5′-end. Each analogue was examined in turn at position X against duplex targets containing each base pair at position ZY. (ii) Sequence of the oligonucleotides used in multiple substitution fluorescence melting and DNase I footprinting studies. In melting experiments the purine strand of the duplex is labelled with fluorescein attached to T adjacent to the 5′-end of the oligopurine tract (F = FAM-dT). In footprinting experiment the TFO target (F = T) is contained within the 110 base pair tyrT(43–59) DNA fragment which was labelled at the 3′-end of the EcoRI site with [α-32P]dATP. In both cases the third strand was labelled with dabcyl (Q) at the 5′-end and contained methyl C (C) in place of C. Each analogue was examined in turn as six substitutions at position X in each TFO.
MATERIALS AND METHODS
Oligonucleotide synthesis
Phosphoramidites
The phosphoramidite monomers for the analogues were synthesized as previously reported (15,22). The synthesis of the phosphoramidite of 5-guanidinopropargyl-dU is described in the supplementary material. These were then dissolved in anhydrous acetonitrile to a concentration of 0.1 M. The phosphoramidites of 5-propynyl-dU, 5′-fluorescein, fluorescein-dT, 5-methyl dC and 5′-dabcyl were purchased from Glen Research or Link Technologies.
Oligonucleotides
All oligonucleotides were synthesized on an Applied Biosystems 394 automated DNA/RNA synthesizer using a standard 1.0 μmol phosphoramidite cycle of acid-catalysed detritylation, coupling, capping and iodine oxidation. Standard A, G, C and T monomers were coupled for 30 s and all other monomers were coupled for 6 min. Stepwise coupling efficiencies were determined by the automated trityl cation conductivity monitoring facility and found to be >98.0%. After oligonucleotide assembly the columns were treated with 20% diethylamine in acetonitrile for 20 min to remove cyanoethyl groups from the phosphotriester moieties and prevent addition of acrylonitrile to the N(3)-position of the modified uracil bases. The columns were then washed with acetonitrile then cleavage and deprotection of the oligonucleotides from the solid support was achieved by exposure to concentrated ammonia solution (20 h at room temperature). Analysis and purification of oligonucleotides was then carried out by reversed phase HPLC and analysed by MALDI-TOF mass spectrometry using a ThermoBioAnalysis Dynamo MALDI-TOF mass spectrometer in positive ion mode using internal dTn standards. The sequences of the oligonucleotides are shown in Figure 1B.
Fluorescence melting experiments
Melting temperatures (Tm)
Fluorescence melting experiments on the triplexes containing single and multiple substitutions of the analogues shown in Figure 1 were carried out using a Roche LightCycler as previously described (23). In these experiments each TFO was labelled at the 5′-end with a quencher (methyl red or dabcyl) while the purine strand of the duplex was either labelled at the 5′-end with fluorescein (Figure 1Bi) or internally with FAM-dT (Figure 1Bii). The triplexes were prepared in either 50 mM sodium acetate buffer (pH 5.0, 5.5 or 6.0) or 50 mM sodium phosphate (pH 7.0) both containing 200 mM sodium chloride. Melting experiments were performed in a total volume of 20 µl and contained 0.25 μM duplex and 3 μM of TFO. The complexes were first denatured by rapidly heating to 95°C and left to equilibrate for 10 min. The complexes were then cooled to 30°C at a rate of 0.2°C min−1 to eliminate hysteresis. After 5 min the complexes were heated to 95°C at the same temperature gradient. Although the slowest rate of continuous temperature change in the LightCycler is 0.1°C s−1, slower melting profiles were obtained by increasing the temperature in 1°C steps, leaving the samples to equilibrate for a set amount of time. Recordings were taken during both the heating and cooling steps to check for hysteresis. Tm values were determined from the first derivatives of the melting profiles using the software provided with the machine and usually differed by less than 0.5°C.
Non-equilibrium melting
The hysteresis between melting and annealing curves at fast rates of heating and cooling were used to estimate association and dissociation rate constants as previously described (8,24,25). For these experiments the triplexes were first heated to 95°C and then cooled slowly to 35°C at a rate of 0.2°C min−1. Melting curves were then obtained by heating the samples to 95°C, at a rate of 6°C min−1 leaving them to equilibrate for 5 min, and then cooling to 35°C at the same rate. Recordings were taken during both the heating and cooling cycles. If αc and αh are the fractions of the duplex that are occupied by the third strand in the cooling and heating curves respectively, then d(αc)/dT = d(αc)/dt × (dT/dt)−1 and d(αh)/dT = d(αh)/dt × (dT/dt)−1, where t is time and T is temperature. If k1 and k−1 are the triplex association and dissociation rate then d(αc)/dt = k1[TFO](1 − αc) −k−1αc and d(αh)/dt = k1[TFO](1 − αh) − k−1αh. By measuring d(αc)/dT, d(αh)/dT, αc and αh, the individual rate constants can be estimated at each temperature. Since the third-strand concentration is in excess in these experiments it effectively remains constant during the reaction (3 µM), yielding a pseudo first-order process with a rate constant k* given by k1[TFO].
DNase I footprinting
Footprinting reactions were performed as previously described (26).
DNA fragment
The tyrT(43–59) fragment contains a 17-base oligopurine tract between positions 43 and 59, which has the same sequence target as that used in the fluorescence melting experiments (27). A 110 base pair radiolabelled fragment containing this sequence was obtained by digesting the plasmid with EcoRI and AvaI and labelling at the 3′-end of the EcoRI site using reverse transcriptase and [α-32P]dATP. This was then separated from the remainder of the plasmid DNA on an 8% (w/v) non-denaturing polyacrylamide gel. After elution the fragment was dissolved in 10 mM Tris–HCl, pH 7.5, containing 0.1 mM EDTA to give about 10 cps/µl as determined on a hand-held Geiger counter (< nM).
Equilibrium reactions
A total 1.5 μl of radiolabelled DNA was mixed with 3 μl of appropriately diluted oligonucleotide and left to equilibrate at 20°C overnight. Experiments were performed in 10 mM PIPES, pH 6.0 containing 50 mM NaCl and 2.5 mM MgCl2. The final oligonucleotide concentration was varied between 3 nM and 3 µM. DNase I digestion was initiated by adding 2 µl of DNase I (typically 0.01 units/ml) dissolved in 20 mM NaCl containing 2 mM MgCl2 and 2 mM MnCl2. The reaction was stopped after 1 min by adding 4 μl DNase I stop solution [80% formamide, 10 mM EDTA, 10 mM NaOH, and 0.1% (w/v) bromophenol blue].
Association reactions
The association reactions were initiated by mixing 30 µl of appropriately diluted oligonucleotide (between 0.1 and 10 µM) with 15 µl of radiolabelled DNA. The oligonucleotides were diluted in 50 mM sodium acetate pH 5.0 containing 2.5 mM magnesium chloride. The association reaction was followed by removing 3 µl aliquots at various time points (between 30 s and 4 h) and digesting with 2 µl of DNase I (typically ∼0.05 units/ml). After 20 s the reaction was stopped by the addition of DNase stop solution.
Denaturing polyacrylamide gel electrophoresis
The products of digestion were separated on 9% polyacrylamide gels containing 8 M urea. Samples were heated to 100°C for 3 min, before loading onto the gel. Polyacrylamide gels (40 cm long, 0.3 mm thick) were run at 1500 V for about 2 h before fixing in 10% (v/v) acetic acid, transferring to Whatman 3 MM paper and drying under vacuum at 86°C for 1 h. The dried gels were subjected to phosphorimaging using a Molecular Dynamics Storm 860 phosphorimager.
Footprint quantification
The intensity of bands within each footprint was estimated using ImageQuant software. These intensities were then normalized relative to a band in the digest which is not part of the triplex target site, and which was not affected by addition of the oligonucleotides. C50 values, indicating the third strand concentration that reduced the intensity of bands in the footprint by 50%, were determined by fitting a simple binding curve to these data.
RESULTS
DNase I footprinting and fluorescence melting experiments were used to examine the triplex-forming properties of the 5-dimethylaminopropargyl dU analogue shown in Figure 1A.
Sequence specificity
The sequence specificity of TFO containing DMAPdU was assessed by determining the melting temperature (Tm) of the fluorescently labelled intermolecular triplexes shown in Figure 1Bi. In these experiments a single DMAPdU was incorporated at the centre of an 18mer TFO (indicated by X in Figure 1B) and targeted against four duplexes in which ZY was each base pair in turn. The thermal stabilities of these complexes were compared with those formed by TFOs containing T and the analogues APdU and GPdU at identical positions.
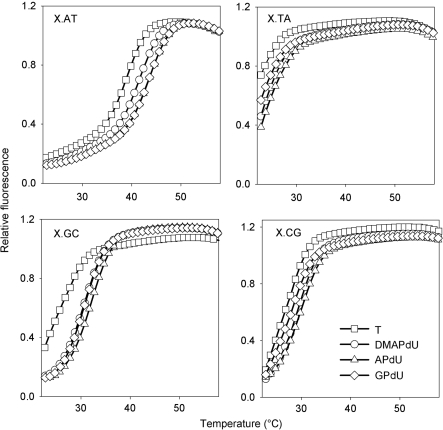
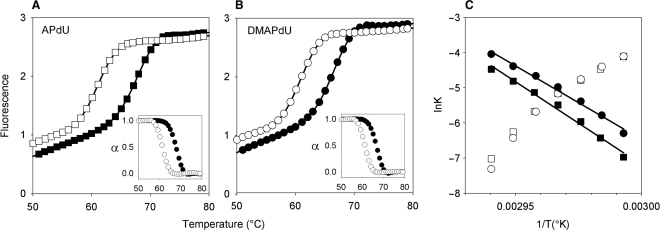
Representative melting curves for the complexes at pH 6.0 are shown in Figure 2 and Tm values derived from these and for the same complexes at pH 5.0 and 5.5 are summarized in Table 1. Examination of these data reveals that as expected all the deoxyuridine analogues increased triplex stability when positioned opposite an AT base pair in the target (top left panel). The magnitude of the stabilization was dependent on the analogue; the dimethylamino derivative DMAPdU increased the Tm by about 2.5°C, while the primary amine (APdU) and guanidine (GPdU) variants increased the Tm by about 4°C relative to T. The stabilization was also greater at higher pHs with ΔTm values nearly twice as large at pH 6.0 compared to pH 5.0. Interestingly, the pH also affected the kinetics of the modified complexes since there was a small amount of hysteresis between the melting and annealing curves at pH 5.0 but not at pH 5.5 or 6.0. In contrast, no hysteresis was observed with the T-containing TFO. This suggests that there are some slow association and/or dissociation kinetics with these modified triplexes, which are examined in greater detail below.
Figure 2.
Representative fluorescence melting curves showing the interaction of the singly substituted TFOs with duplex target sites containing a variable central base pair, generating X.AT, X.TA, X.GC or X.CG base triplets where X is T (squares), DMAPdU (circles), APdU (triangles) or GPdU (diamonds). The experiments were performed in 50 mM sodium acetate pH 6.0, containing 200 mM NaCl. The complexes were heated and cooled at a rate of 0.2°C min−1.
Table 1.
Tm values determined from the melting profiles of the fluorescently labelled triplexes shown in Figure 1B
| X | pH | ZY |
|||
|---|---|---|---|---|---|
| AT | TA | GC | CG | ||
| T | 5.0 | 63.8 | 53.8 | 57.8 | 58.1 |
| 5.5 | 54.3 | 39.2 | 43.7 | 44.5 | |
| 6.0 | 39.4 | <28.0 | 27.7 | 28.3 | |
| DMAPdU | 5.0 | 65.4 (64.0) | 54.4 | 59.4 | 58.4 |
| 5.5 | 55.6 | 39.9 | 46.5 | 44.5 | |
| 6.0 | 41.8 | <28.0 | 30.9 | 29.5 | |
| APdU | 5.0 | 66.5 (64.0) | 54.8 | 60.3 | 58.7 |
| 5.5 | 56.9 | 40.3 | 47.0 | 44.9 | |
| 6.0 | 43.7 | <28.0 | 32.3 | 29.9 | |
| GPdU | 5.0 | 67.8 (64.1) | 54.6 | 60.2 | 58.5 |
| 5.5 | 57.7 | 39.6 | 47.2 | 43.9 | |
| 6.0 | 43.8 | <28.0 | 30.7 | 28.4 | |
The Tm values shown in parenthesis are those determined from the annealing profiles of the same triplexes and are included only if there is <1°C difference (hysteresis) with those obtained from melting.
Examination of the interaction of the analogues with the other three base pairs reveals that, as well as showing an increased affinity for AT, all analogues increased triplex stability when positioned against a GC base pair, with an increase of about 3.0, 4.5 and 4°C for the dimethylamine, amine and guanidine derivatives respectively. This contrasts to their interaction with CG or TA base pairs in which little or no stabilization was observed. A similar effect was previously seen with the 2′-aminoethoxy derivative of APdU suggesting that this increase in specificity is in part due to the presence of the 5-position substituent (8). No hysteresis was observed with any of the mismatched triplets.
Sequence arrangement
The effect of the arrangement of the modified bases was examined by comparing the interaction of TFOs containing six clustered substitutions or six dispersed substitutions of each analogue (indicated by X in Figure 1Bii) with the same duplex target site. DMAPdU, T, APdU, GPdU and PdU were each incorporated at identical positions in the TFOs. For these oligonucleotides 5-methyl dC was used in place of C as it has improved base stacking and a slightly higher pK, extending triplex formation to higher pH values.
Fluorescence melting
The interactions of the TFOs containing dispersed or clustered nucleotide analogues with the fluorescently labelled duplex shown in Figure 1Bii were determined. This duplex was designed to be 10 base pairs longer than the triplex in order to separate the triplex and duplex melting transitions and the fluorescein was attached to the 5-position of a T at the 5′-end of the purine strand of the TFO-binding site. The melting and annealing profiles were again obtained by heating and cooling the triplexes at a rate of 0.2°C min−1, however some hysteresis was still observed for several of the more stable triplexes at higher pH values. For those that showed hysteresis the equilibrium melting temperature was estimated from the average of the annealing and melting temperatures.
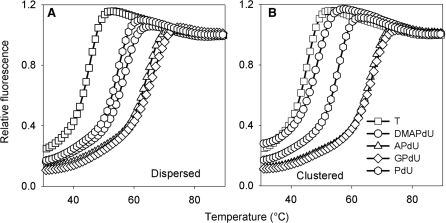
Representative melting curves for these complexes at pH 6.0 are shown in Figure 3 and the Tm values determined from the melting and annealing curves (shown in parenthesis) are presented in Table 2. It can be seen that the TFO containing dispersed substitutions of DMAPdU (left panel; circles) produces a ΔTm of 11°C relative to the triplex formed with the unmodified TFO (left panel; squares). The incorporation of more than one substitution gives greater stabilization with a ΔTm of about 2°C per substitution. Comparison with the other charged analogues reveals that the amine and guanidine modifications are more stabilizing, leading to increases in Tm of 19°C and 21°C, respectively. The oligonucleotides containing the uncharged propynyl derivative (PdU) also generate stable triplexes, though the ΔTm values are less that with DMAPdU or APdU; the difference between DMAPdU and PdU is greatest at pH 5.0. The triplex containing contiguous substitutions of DMAPdU in the TFO (right panel; circles) is stabilized less than when these residues are dispersed; this difference is greater at higher pHs. In contrast, the stabilization produced with the three other analogues is not affected by the arrangement of the modified bases and similar Tm values are produce with TFOs in which these residues are dispersed or contiguous, suggesting that steric factors may be involved in limiting the stability of triplexes with contiguous DMAPDU residues.
Figure 3.
Representative fluorescence melting curves showing the interaction of the multiply substituted dispersed (A) or clustered (B) TFOs with their intended duplex target site. The TFOs contained T (squares), DMAPdU (circles), APdU (triangles), GPdU (diamonds) or PdU (hexagons). The experiments were performed in 50 mM sodium acetate pH 6.0, containing 200 mM NaCl. The complexes were heated and cooled at a rate of 0.2°C min−1.
Table 2.
Tm values determined from the melting profiles of the fluorescently labelled triplexes shown in Figure 1C
| X | pH | Dispersed | Clustered |
|---|---|---|---|
| T | 5.0 | 65.2 | |
| 6.0 | 45.5 (44.3) | ||
| 7.0 | <30 | ||
| PdU | 5.0 | 77.6 | 78.4 |
| 6.0 | 54.9 | 55.7 (53.8) | |
| 7.0 | 40.5 (<30) | 41.1 (<30) | |
| DMAPdU | 5.0 | 80.9 | 76.3 |
| 6.0 | 56.8 (54.4) | 47.9 (46.5) | |
| 7.0 | 39.5 (<30) | <30 | |
| APdU | 5.0 | 83.1 | 84.1 |
| 6.0 | 64.8 (62.7) | 65.7 (64.5) | |
| 7.0 | 46.2 (39.1) | 46.7 (43.7) | |
| PGdU | 5.0 | 84.1 | 84.3 |
| 6.0 | 66.8 (64.0) | 67.0 (65.6) | |
| 7.0 | 48.8 (37.3) | 48.0 (43.0) |
The Tm values shown in parenthesis are those determined from the annealing profiles of the same triplexes and are included only if there is >1°C difference (hysteresis) with those obtained from melting.
Footprinting
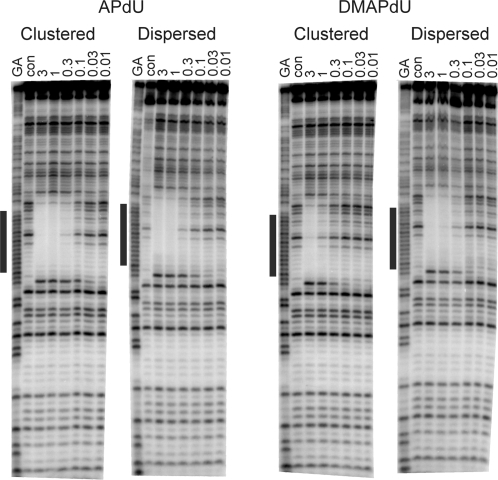
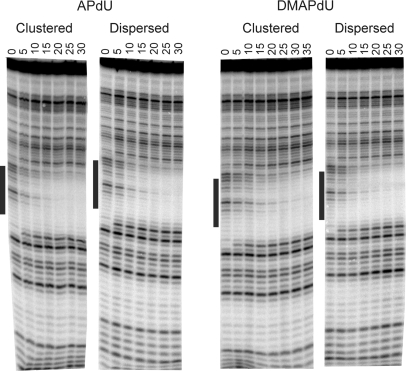
Triplex formation was further examined by DNase I footprinting using the same quencher-labelled TFOs. Representative cleavage patterns for the interaction of the clustered and dispersed DMAPdU- and APdU-containing TFOs with this fragment are shown in Figure 4. Clear footprints are evident for each TFO at the intended target site. It is can be seen that the footprints produced by the clustered and dispersed APdU-containing oligonucleotides persist to similar concentrations as the dispersed DMAPdU-containing oligonucleotide. In contrast, the footprint for the clustered DMAPdU-containing oligonucleotide requires higher oligonucleotide concentrations, confirming that it binds less strongly to the target site. C50 values, representing the TFO concentration that reduces the intensity of bands within the footprint by 50%, calculated from quantitative analysis of these footprints, together with those for by the other modified oligonucleotides are presented in Table 3, in which it can be seen that there is a 2-fold difference between the dispersed and clustered DMAPdU containing oligonucleotides.
Figure 4.
DNase I cleavage patterns showing the interaction of the clustered or dispersed APdU or DMAPdU-containing oligonucleotides with a DNA fragment containing their intended target duplex target site. The TFO concentration (µM) is shown at the top of each lane. The experiments were performed in 10 mM PIPES buffer pH 6.0 containing 50 mM sodium chloride. The complexes were equilibrated overnight at 20°C before digestion with DNase I. The lanes labelled ‘GA’ and ‘con’ represent Maxam–Gilbert markers specific for purines and DNase I cleavage of duplex DNA in the absence of TFO, respectively. The black bar represents the intended TFO target site.
Table 3.
Kd values and kinetic parameters obtained for the oligonucleotides shown in Figure 1Bii
| X | Clustered |
Dispersed |
||
|---|---|---|---|---|
| Kd (µM) | k1 (µM min−1) | Kd (µM) | k1 (µM min−1) | |
| PdU | 0.5 ± 0.2 | – | 0.4 ± 0.2 | – |
| DMAPdU | 1.2 ± 0.2 | 0.020 ± 0.001 | 0.6 ± 0.3 | 0.026 ± 0.002 |
| APdU | 0.1 ± 0.02 | 0.042 ± 0.007 | 0.2 ± 0.02 | 0.034 ± 0.001 |
| GPdU | 0.2 ± 0.1 | – | 0.2 ± 0.1 | – |
Kd values were determined at pH 6.0, while k1 was determined at pH 5.0.
Kinetics of triplex formation
We have previously shown that the increased affinity of the 2′-aminoethoxy derivative of APdU for AT base pairs is due to the slow rate of dissociation of this triplet, with little effect on the association kinetics (8). We have therefore determined the effect of DMAPdU and APdU on the kinetics of triplex formation.
Fluorescence melting experiments
Hysteresis between melting and annealing profiles occurs when the reaction is not at thermodynamic equilibrium and indicates the presence of slow steps in the association or dissociation reactions. This hysteresis can be used to estimate individual association and dissociation rate constants as previously described (8,24,25). Initially we examined the singly modified fluorescently labelled triplexes shown in Figure 1Bi for which heating and cooling these complexes at a rate of 6°C min−1 generated a suitable degree of hysteresis for kinetic analysis. Annealing and melting profiles for each of the triplexes are shown in Figure 5A. Each analogue generates a similar degree of hysteresis, suggesting similar slow steps in the reaction of each of these analogues with an AT base pair. Arrhenius plots derived from these curves are shown in Figure 5B and are typical of those seen with other triplexes; a negative slope is seen for the dissociation reaction along with a positive slope for the association reaction. The apparent negative activation energy of the association reaction indicates that we are not observing the primary kinetic event and is consistent with the suggestion that triplex formation occurs via a nucleation-zipper mechanism (28). Examining these Arrhenius plots reveals that the association rate constants for each analogue are similar but the dissociation rate constants are in different regions of the plot. By extrapolating these lines to 37°C we estimate half-lives of 3.6 × 106 and 3.7 × 107 s for the DMAPdU and APdU triplexes, respectively. This suggests that the difference in stability between the two triplexes arises from difference in their dissociation rates.
Figure 5.
Representative annealing (filled symbols) and heating profiles (open symbols) for the interaction of the singly substituted APdU (A) and DMAPdU (B) containing-oligonucleotides with their intended AT-containing duplex target. The profiles were obtained at a rate of 6°Cmin−1. Fraction folded plots for each triplex are included as insets. The experiments were performed in 50 mM sodium acetate pH 5.0, containing 200 mM NaCl. (C) Representative Arrhenius plots for the association (k1; open symbols) and dissociation (k−1; filled symbols) constants for the APdU (squares) and DMAPdU (circles) oligonucleotides.
Similar experiments were performed with the oligonucleotides containing multiple substitutions of each analogue, however the annealing curves obtained at the faster rate of cooling, which is necessary to generate hysteresis, did not show the usual sigmoidal shape, exhibiting a long transition at low temperatures. These TFOs were therefore not examined any further by fluorescence melting.
Footprinting experiments
The association rates of the DMAPdU and APdU-modified oligonucleotides were determined by performing DNase I footprinting reactions at different times after mixing the TFOs with the radiolabelled DNA fragment containing the target shown in Figure 1Bii. Representative cleavage patterns for the association of the clustered and dispersed DMAPdU and APdU-containing TFOs are presented in Figure 6. The time-dependent appearance of a footprint for each oligonucleotide is very similar, suggesting that there are no large differences in their association rates. These observed rate constants are dependent on the total nucleic acid concentration (kobs = k−1 + k1[NA]) and k1 values were determined from the concentration dependence of the apparent rate (kobs) constants. The association rate constants obtained for the four oligonucleotides are presented in Table 3. For each analogue there is no significant difference between the association rates of the oligonucleotides containing dispersed or clustered substitutions.
Figure 6.
DNase I cleavage patterns of the tyrT(43–59) DNA fragment in the presence of 3 µM of either the clustered or dispersed APdU or DMAPdU-containing oligonucleotides. The experiments were performed in 50 mM sodium acetate pH 5.0 containing 200 mM sodium chloride and 2.5 mM MgCl2 and the complexes were incubated at 20°C before digesting with DNase I. The time (min) after adding the TFO is indicated at the top of each gel lane. The black bar represents the intended TFO target site.
DISCUSSION
Nucleoside analogues that are synthetically compatible with other useful TFO modifications, such as intercalators and cross-linkers, are highly desirable for generating heavily modified triplexes for a range of applications. In this article we have examined the specificity, affinity and kinetics of the charged thymine analogue DMAPdU, which contains a dimethylamine group at the 5-position of the base. Unlike other charged thymine analogues such as APdU, this analogue does not require protection during oligonucleotide synthesis (15,21).
Fluorescence melting and DNase I footprinting experiments reveal that DMAPdU increases triplex stability relative to T. The increased triplex stability is not as great as seen with other charged analogues such as APdU or GPdU but is superior to that produced by PdU, which contains only an uncharged propyne substituent in the 5-position. We suggest two explanations for its slightly lower affinity: firstly addition of the methyl groups could sterically hinder interaction with the target site. Secondly it is possible that hydrogen bond donor interactions from the appended amino groups may contribute to the stabilization afforded by APdU and GPdU, consistent with the suggestion that 5-(3-hydroxyprop-1-ynyl)-2′-deoxyuridine also produces good triplex stabilization (29). However, the extent of triplex stabilization is dependent on the arrangement and position of DMAPdU substitutions; oligonucleotides in which the substitutions are dispersed bind more tightly than when they are clustered. This effect could arise either from steric clash between the methyl groups of closely positioned substitutions or competition for protonation between adjacent residues. In contrast, the stabilization afforded by APdU and GPdU does not depend on the arrangement of the modified nucleotides at this target sequence. This is surprising since GPdU also contains a bulky, albeit planar, group at the same position. The superiority of dispersed over clustered substitutions is similar to our previous observations with other positively charged analogues (20), but contrasts with the results for nucleotides bearing 2′-aminoethoxy substituents (9,14,19,30).
Examining the sequence selectivity of the DMAPdU-containing TFOs reveals that, as previously observed with the 2′-aminoethoxy derivative of APdU, this nucleotide produces enhanced selectivity for YR over RY base pairs relative to T (18). A similar effect is also observed with APdU and GPdU. Analysis of the kinetics of the APdU.AT and DMAPdU triplets highlights that the difference in affinity arises from the faster dissociation of DMAPdU relative to APdU, with little effect on the rates of association. This analogue would therefore be useful in any application that requires a TFO to exhibit similar association kinetics to APdU.
DMAPdU produces more stable triple helices than equivalent oligonucleotides containing T, though these are less stable than substitutions with APdU and GPdU or the doubly substituted bis-amino-U (18,21). However the main useful advantage of this nucleotide analogue is that the base does not require protection during oligonucleotide synthesis. It can therefore be used along with other triplex-stabilizing derivatives that require mild deprotection conditions. Most studies for enhancing triplex stability have examined the effect of a single modified nucleotide or conjugate, while the major advances in the technology will probably require a combination of various stabilizing agents. This will only be possible if the different modified nucleotides have compatible deprotection reactions during oligonucleotide synthesis. A number of oligonucleotide conjugates, such as the addition of psoralen (31), camptothecin (32) or other intercalators (33), require mild deprotection conditions and therefore cannot be used along with many of the triplex-stabilizing nucleotides derivatives that have been devised. The addition of the dimethylaminopropargyl group to the 5-position of dU will also be fully compatible with 2′-modifications such as aminoethoxy (30) or methoxyethyl which stabilize triplexes or give enhanced biological stability. DMAPdU will therefore be useful for producing TFOs that contain combinations of modified nucleotides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants from the BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 2.Helene C, Toulme JJ. Specific regulation of Gene-expression by antisense, sense and antigene nucleic-acids. Biochim. Biophys. Acta. 1990;1049:99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- 3.Kalish JM, Glazer PM. Targeted genome modification via triple helix formation. Ann. NY Acad. Sci. 2005;1058:151–161. doi: 10.1196/annals.1359.023. [DOI] [PubMed] [Google Scholar]
- 4.Potaman VN. Applications of triple-stranded nucleic acid structures to DNA purification, detection and analysis. Expert Rev. Mol. Diagn. 2003;3:481–496. doi: 10.1586/14737159.3.4.481. [DOI] [PubMed] [Google Scholar]
- 5.Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 6.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 7.Re RN, Cook JL, Giardina JF. The inhibition of tumor growth by triplex-forming oligonucleotides. Cancer Lett. 2004;209:51–53. doi: 10.1016/j.canlet.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Rusling DA, Broughton-Head VJ, Tuck A, Khairallah H, Osborne SD, Brown T, Fox KR. Kinetic studies on the formation of DNA triplexes containing the nucleoside analogue 2'-O-(2-aminoethyl)-5-(3-amino-1-propynyl)uridine. Org. Biomol. Chem. 2008;6:122–129. doi: 10.1039/b713088k. [DOI] [PubMed] [Google Scholar]
- 9.Seidman MM, Puri N, Majumdar A, Cuenoud B, Miller PS, Alam R. The development of bioactive triple helix-forming oligonucleotides. Ann. NY Acad. Sci. 2005;1058:119–127. doi: 10.1196/annals.1359.020. [DOI] [PubMed] [Google Scholar]
- 10.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 11.Buchini S, Leumann CJ. Stable and selective recognition of three base pairs in the parallel triple-helical DNA binding motif. Angew. Chem. Int. Ed. Engl. 2004;43:3925–3928. doi: 10.1002/anie.200460159. [DOI] [PubMed] [Google Scholar]
- 12.Li JS, Shikiya R, Marky LA, Gold B. Triple helix forming TRIPside molecules that target mixed purine/pyrimidine DNA sequences. Biochemistry. 2004;43:1440–1448. doi: 10.1021/bi035648d. [DOI] [PubMed] [Google Scholar]
- 13.Rusling DA, Powers VEC, Ranasinghe RT, Wang Y, Osborne SD, Brown T, Fox KR. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alam MR, Majumdar A, Thazhathveetil AK, Liu ST, Liu JL, Puri N, Cuenoud B, Sasaki S, Miller PS, Seidman MM. Extensive sugar modification improves triple helix forming oligonucleotide activity in vitro but reduces activity in vivo. Biochemistry. 2007;46:10222–10233. doi: 10.1021/bi7003153. [DOI] [PubMed] [Google Scholar]
- 15.Bijapur J, Keppler MD, Bergqvist S, Brown T, Fox KR. 5-(1-propargylamino)-2′-deoxyuridine (U-P): a novel thymidine analogue for generating DNA triplexes with increased stability. Nucleic Acids Res. 1999;27:1802–1809. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuenoud B, Casset F, Husken D, Natt F, Wolf RM, Altmann KH, Martin P, Moser HE. Dual recognition of double-stranded DNA by 2′-aminoethoxy-modified oligonucleotides. Angew. Chem. Intl. Ed. Engl. 1998;37:1288–1291. doi: 10.1002/(SICI)1521-3773(19980518)37:9<1288::AID-ANIE1288>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Gowers DM, Bijapur J, Brown T, Fox KR. DNA triple helix formation at target sites containing several pyrimidine interruptions: stabilization by protonated cytosine or 5-(1-propargylamino)dU. Biochemistry. 1999;38:13747–13758. doi: 10.1021/bi9911637. [DOI] [PubMed] [Google Scholar]
- 18.Osborne SD, Powers VEC, Rusling DA, Lack O, Fox KR, Brown T. Selectivity and affinity of triplex-forming oligonucleotides containing 2′-aminoethoxy-5-(3-aminoprop-1-ynyl)uridine for recognizing AT base pairs in duplex DNA. Nucleic Acids Res. 2004;32:4439–4447. doi: 10.1093/nar/gkh776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puri N, Majumdar A, Cuenoud B, Miller PS, Seidman MM. Importance of clustered 2′-O-(2-aminoethyl) residues for the gene targeting activity of triple helix-forming oligonucleotides. Biochemistry. 2004;43:1343–1351. doi: 10.1021/bi035808l. [DOI] [PubMed] [Google Scholar]
- 20.Rusling DA, Le Strat L, Powers VEC, Broughton-Head VJ, Booth J, Lack O, Brown T, Fox KR. Combining nucleoside analogues to achieve recognition of oligopurine tracts by triplex-forming oligonucleotides at physiological pH. FEBS Lett. 2005;579:6616–6620. doi: 10.1016/j.febslet.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 21.Sollogoub M, Darby RAJ, Cuenoud B, Brown T, Fox KR. Stable DNA triple helix formation using oligonucleotides containing 2′-aminoethoxy,5-propargylamino-U. Biochemistry. 2002;41:7224–7231. doi: 10.1021/bi020164n. [DOI] [PubMed] [Google Scholar]
- 22.Brennan L, Peng GM, Srinivasan N, Fox KR, Brown T. 2'-O-dimethylaminoethoxyuridine and 5-dimethylaminopropargyl deoxyuridine for AT base pair recognition in triple helices. Nucleos. Nucleot. Nucleic Acids. 2007;26:1283–1286. doi: 10.1080/15257770701530525. [DOI] [PubMed] [Google Scholar]
- 23.Darby RAJ, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernal-Mendez E, Leumann CJ. Stability and kinetics of nucleic acid triplexes with chimaeric DNA/RNA third strands. Biochemistry. 2002;41:12343–12349. doi: 10.1021/bi025937m. [DOI] [PubMed] [Google Scholar]
- 25.Rougée M, Faucon B, Mergny JL, Barcelo F, Giovannangeli C, Garestier T, Hélène C. Kinetics and thermodynamics of triple-helix formation – effects of ionic-strength and mismatches. Biochemistry. 1992;31:9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]
- 26.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: A method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Brown PM, Madden CA, Fox KR. Triple-helix formation at different positions on nucleosomal DNA. Biochemistry. 1998;37:16139–16151. doi: 10.1021/bi981768n. [DOI] [PubMed] [Google Scholar]
- 28.Alberti P, Arimondo PB, Mergny JL, Garestier T, Helene C, Sun JS. A directional nucleation-zipping mechanism for triple helix formation. Nucleic Acids Res. 2002;30:5407–5415. doi: 10.1093/nar/gkf675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazier JA, Shibata T, Townsley J, Taylor BF, Frary E, Williams NH, Williams DM. Amino-functionalized DNA: the properties of C5-amino-alkyl substituted 2′-deoxyuridines and their application in DNA triplex formation. Nucleic Acids Res. 2005;33:1362–1371. doi: 10.1093/nar/gki254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puri N, Majumdar A, Cuenoud B, Natt F, Martin P, Boyd A, Miller PS, Seidman MM. Minimum number of 2′-O-(2-aminoethyl) residues required for gene knockout activity by triple helix forming oligonucleotides. Biochemistry. 2002;41:7716–7724. doi: 10.1021/bi025734y. [DOI] [PubMed] [Google Scholar]
- 31.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C. Sequence-specific photoinduced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc. Natl Acad. Sci. USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arimondo PB, Thomas CJ, Oussedik K, Baldeyrou B, Mahieu C, Halby L, Guianvarc'h D, Lansiaux A, Hecht SM, Bailly C, Giovannangeli C. Exploring the cellular activity of camptothecin-triple-helix-forming oligonucleotide conjugates. Mol. Cell Biol. 2006;26:324–33. doi: 10.1128/MCB.26.1.324-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garbesi A, Bonazzi S, Zanella S, Capobianco ML, Giannini G, Arcamone F. Synthesis and binding properties of conjugates between oligodeoxynucleotides and daunorubicin derivatives. Nucleic Acids Res. 1997;25:2121–2128. doi: 10.1093/nar/25.11.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.